Abstract
Staphylococcus aureus is an important pathogen, causing a wide range of infections including sepsis, wound infections, pneumonia, and catheter-related infections. In several pathogens ClpP proteases were identified by in vivo expression technologies to be important for virulence. Clp proteolytic complexes are responsible for adaptation to multiple stresses by degrading accumulated and misfolded proteins. In this report clpP, encoding the proteolytic subunit of the ATP-dependent Clp protease, was deleted, and gene expression of ΔclpP was determined by global transcriptional analysis using DNA-microarray technology. The transcriptional profile reveals a strong regulatory impact of ClpP on the expression of genes encoding proteins that are involved in the pathogenicity of S. aureus and adaptation of the pathogen to several stresses. Expression of the agr system and agr-dependent extracellular virulence factors was diminished. Moreover, the loss of clpP leads to a complete transcriptional derepression of genes of the CtsR- and HrcA-controlled heat shock regulon and a partial derepression of genes involved in oxidative stress response, metal homeostasis, and SOS DNA repair controlled by PerR, Fur, MntR, and LexA. The levels of transcription of genes encoding proteins involved in adaptation to anaerobic conditions potentially regulated by an Fnr-like regulator were decreased. Furthermore, the expression of genes whose products are involved in autolysis was deregulated, leading to enhanced autolysis in the mutant. Our results indicate a strong impact of ClpP proteolytic activity on virulence, stress response, and physiology in S. aureus.
The Clp proteases were first identified in Escherichia coli and consist of an ATPase specificity factor (ClpA or ClpX in E. coli; ClpX, ClpC, or ClpE in Bacillus subtilis) and a proteolytic domain (ClpP) that contains a consensus serine protease active site (33). In E. coli, ClpP-mediated proteolysis is regulated by heat shock and removes abnormal proteins that accumulate during stress conditions, recycles amino acids from nonessential proteins during starvation, and contributes to the clearance of truncated peptides from stalled ribosomes by the SsrA-tagging system (34, 65). Moreover, it has been shown that Clp proteases play a significant role in certain processes regulating cellular functions via proteolysis (33, 36, 45). For example, in E. coli ClpXP is involved in degradation of regulator proteins including the alternative sigma factor SigS, the UmuD SOS protein, and different phage proteins (23, 31, 34). Regulatory proteolysis is presumably determined by certain amino acid sequences which serve as a degradation signal. Flynn et al. have identified more than 50 proteins in E. coli as potential substrates for proteolysis by ClpXP (19). Further substrate proteins with regulatory functions were identified in other bacteria such as CtrA in Caulobacter crescentus, sigma s and FlhC/FlhD in Salmonella enterica serovar Typhimurium, PopR in Streptomyces lividans, and HdiR in Lactococcus lactis (44, 69, 73, 74). In B. subtilis Clp-specific target proteins were recognized which are involved in peptidoglycan synthesis, competence, sporulation, and heat shock regulation (30, 48, 50).
In addition, several studies indicate that ClpP proteolytic activity is critical for virulence of pathogenic bacteria, including S. enterica serovar Typhimurium, Streptococcus pneumoniae, Listeria monocytogenes, and Staphylococcus aureus (26, 29, 37, 58, 63, 73, 75). Interestingly, ClpP seems to be essential for survival of L. monocytogenes in murine macrophages (28, 29). Furthermore, clpX and clpP mutants of S. aureus, respectively, were attenuated in a murine abscess model (26), and ClpC plays an important role for long-term survival and for intracellular replication of this pathogen (12, 25). More recently, Frees et al. found that clpP deletion (ΔclpP) in S. aureus 8325-4 resulted in impaired virulence properties (26). In this study, the global regulatory agr locus was repressed in the ΔclpP strain, giving rise to a reduced α-toxin and extracellular protease activity. Moreover, the ΔclpP strain was more sensitive against hydrogen peroxide and not able to replicate intracellularly. The authors suggested that the reduced virulence of the ΔclpP strain is most likely due to reduced agr-regulated virulence gene expression rather than to decreased stress resistance (26). In addition, there are indications that Rot (repressor of toxin) in complex with RNAIII is a substrate of Clp-dependent degradation regulating serine protease sspA expression (27). All these reports suggest that ClpP proteolytic activity is important not only for cell physiology but also for regulation of virulence properties of pathogenic bacteria.
In order to get a more comprehensive picture of the role of ClpP protease on global transcription in S. aureus and how it relates to physiology and virulence, a ΔclpP strain was constructed in strain S. aureus 8325, and global gene expression was studied by comparative DNA microarray hybridization. We report here that clpP deletion affects the expression of genes belonging to specific regulons which are involved in adaptation to changes in the physiological state of the cell as well as in virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strain DH5α and S. aureus strains were grown in Luria-Bertani broth (LB) unless otherwise indicated. The recombinant E. coli and S. aureus clones were cultivated under selective pressure with either ampicillin (100 μg ml−1), chloramphenicol (10 μg ml−1), or erythromycin (10 μg ml−1), respectively. For growth curves and RNA isolation, overnight cultures of S. aureus were diluted to an optical density at 600 nm (OD600) of 0.01 in LB and were incubated at different temperatures (20°C, 30°C, 37°C, 42°C, and 45°C) under aerobic conditions with orbital shaking (180 rpm). Samples were collected in intervals during the first 10 h. Samples for RNA isolation were collected in the exponential growth phase (OD600 of 1.0).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | λ− φ80dlacΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK−) supE44 thi-1 gyrA relA1 | 68 |
| S. aureus | ||
| RN4220 | NCTC 8325-4-r (restriction mutant, with 11-bp deletion in rsbU) | 49 |
| 8325 | NCTC 8325 (wild-type, with 11-bp deletion in rsbU) | Laboratory stock |
| 8325ΔclpP | clpP deletion strain of 8325 | This study |
| 8325ΔclpP+ | 8325ΔclpP, containing pHPS9clpP | This study |
| 8325ΔagrA | agrA deletion strain of 8325 | This study |
| 8325ΔagrC | agrC deletion strain of 8325 | This study |
| Plasmids | ||
| pBT2 | Shuttle vector; Apr in E. coli; Cmr in S. aureus | 9 |
| pEC1 | Apr EmrermB fragment in pUC18 | 9 |
| pBT2ΔclpP | Deletion vector for clpP; ermB fragment flanked by fragments upstream and downstream of clpP in pBT2; Apr in E. coli; Emr and Cmr in S. aureus | This study |
| pHPS9 | Shuttle vector; Cmr in E. coli; Emr in S. aureus | 35 |
| pHPS9KclpP | pHPS9, containing clpP fragment for ΔclpP complementation | This study |
Construction of the S. aureus ΔclpP mutant.
For construction of a ΔclpP mutant in S. aureus, two PCR fragments of 1,046 bp and 943 bp, encompassing the up- and downstream regions of the clpP gene in strain 8325, corresponding to SA0724 of strain N315, were amplified using primer with added HindIII and BamHI restriction sites for the upstream fragment and PstI and EcoRI for the downstream fragment (see Table S2 in the supplemental material). The plasmid pEC1 containing the ermB resistance cassette was digested with BamHI and PstI. The up- and downstream fragments and the ermB cassette were ligated in one step into the temperature-sensitive shuttle vector pBT2, which was digested with EcoRI and HindIII (9). Construction of this deletion vector was carried out in E. coli DH5α and subsequently introduced into S. aureus strain 8325 by transduction with phage φ85 (72). In this strain gene inactivation was carried out as described by Brückner (9). Successful homologous recombination and loss of the plasmid were proven by Southern blot hybridization and PCR.
Complementation of the ΔclpP strain.
For complementation of the ΔclpP strain, an 824-bp PCR fragment containing the entire SA0723 locus and the ribosome-binding site was amplified by PCR using primer with added BamHI and EcoRI restriction sites and ligated into the shuttle vector pHPS9 (35). The plasmid was transformed into RN4220 by electroporation and transduced into the ΔclpP strain by using phage φ85. Clones were selected using erythromycin and chloramphenicol.
RNA preparation and Northern blot analysis.
Total RNA was isolated from S. aureus cultures in the exponential growth phase (OD600 of 1.0). Bacteria were harvested with the addition of RNA Protect (QIAGEN, Hilden, Germany) according to the manufacturer's instruction. Prior to RNA isolation bacteria were lysed using glass beads in a Fast Prep shaker (Qbiogene, Heidelberg, Germany) for 45 s at a speed of 6.5 units. RNA was isolated using a QIAGEN RNeasy kit according to the standard QIAGEN RNeasy protocol. Ten to twenty micrograms of total RNA was used to perform a denaturing agarose gel electrophoresis and Northern blot hybridization as described previously (2). The probes were generated by PCR by using the primer sets listed in Table S2 in the supplemental material and were labeled by use of an ECL kit (Amersham Biosciences, Freiburg, Germany). Hybridization was performed as described in the manufacturer's instructions. The signals were quantified by densitometric scanning.
Semiquantitative reverse transcription-PCR (RT-PCR).
Reverse transcription was performed using 2 μg of DNase I-treated RNA samples, a random hexamer primer mix and Superscript III TM reverse transcriptase (Invitrogen, Karlsruhe, Germany) at 50°C for 1 h. The cDNA was adjusted to 40 μl with double-distilled water and amplified in different PCRs (including negative controls) with primers specific for the corresponding genes (for primers, see Table S2 in the supplemental material).
Microarray analysis.
S. aureus N315 full genome microarrays containing PCR products of 2,334 genes were used for microarray analysis (Scienion, Berlin, Germany). Each slide contained 6,336 features corresponding to duplicate copies of each open reading frame (ORF) and several controls. Total RNA for DNA microarray analysis was isolated from cultures in the exponential growth phase at an OD600 of 1.0 at 37°C. Reverse transcription and fluorescent labeling reactions were performed using 10 μg of total RNA using random primers and Superscript III reverse transcriptase (Invitrogen), and cDNA was concomitantly labeled using the dyes Cy3 and Cy5 according to the manufacturer's instructions (Scienion). RNA obtained from four different biological experiments was utilized, and a reverse labeling (dye switch) experiment was performed to minimize bias due to differential dye bleaching or incorporation of the Cy3 and Cy5 dyes during the RT reaction. Microarray hybridization (16 h at 50°C) and washing of the slides were performed according to the manufacturer's instructions. Hybridized slides were scanned using a Genepix 4000B laser scanner (Axon Instruments Inc., Union City, CA). Bioinformatic analyses on the slide hybridization results of each single experiment were performed using Genepix Pro3.0 (Axon Instruments Inc.). Data of each image were normalized to the mean ratio of means of all features. Different experiments were normalized to each other using Expressionist software, version 3.1 (Genedata, Martinsried, Germany). Mean values and standard deviations of gene expression ratios based on two spot replicates on each microarray and four different hybridization experiments were calculated in Microsoft Excel XP.
Triton X-100-induced autolysis assays.
Autolysis assays were performed as described by Mani et al. (56). Bacteria were grown in tryptic soy broth (TSB) containing 1 M NaCl to an OD600 of ∼0.7 at 37°C with shaking at 250 rpm. After one wash with phosphate-buffered saline (PBS), cells were resuspended in the same volume of 0.05 M Tris-HCl buffer (pH 7.5) containing 0.1% Triton X-100 and were incubated at 30°C with shaking. The optical density was measured in intervals. Results were normalized to an OD600 at time zero, and percent lysis was calculated.
Physiological analysis.
For analysis of physiological changes in the mutant, an API-Staph test system was used according to the manufacturer's instructions (bioMérieux, Nürtingen, Germany).
Fibronectin binding assays.
The capability of the S. aureus strains to bind fibronectin was measured by using a radiometric assay described by Hussain et al. (42).
Infection experiments and gentamicin-lysostaphin protection assays.
Gentamicin-lysostaphin protection assays were performed as described by Agerer et al. (1). Briefly, overnight cultures of S. aureus were diluted 1:100 in TSB and were cultured to an OD600 of 1.0. Bacteria were harvested and washed twice with PBS. For gentamicin-lysostaphin protection assays, 293T cells (2 × 105 cells/well) were infected with bacteria at a multiplicity of infection of 20. After 2 h of coincubation at 37°C, the culture medium was replaced by Dulbecco's modified Eagle's medium-10% calf serum containing 50 μg/ml gentamicin and 20 μg/ml lysostaphin, and cells were further incubated for 45 min. Cells were washed with PBS, and intracellular bacteria was released by incubation in 1% saponin in PBS for 20 min at 37°C. Samples were diluted in PBS and plated on TSB agar plates for determination of the recovered CFU.
Scanning electron microscopy.
For scanning electron microscopy, staphylococci were grown overnight in TSB medium on polystyrene chamber slides at 37°C. After the medium was decanted, the slides were washed three times with 1× PBS, mounted on aluminum stubs, and shadowed with gold. For visualization, a scanning electron microscope (Zeiss DSM962) was used at 15 kV.
RESULTS AND DISCUSSION
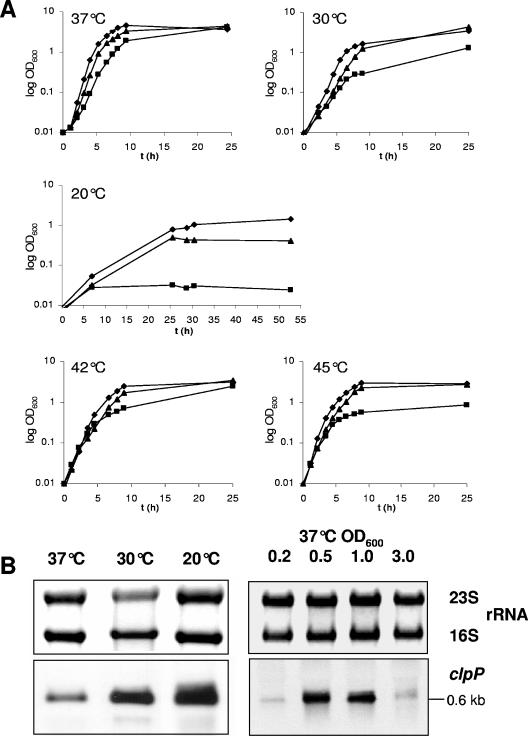
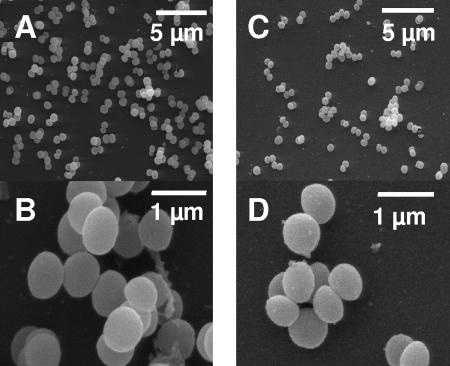
Clp proteases are responsible for degradation of misfolded proteins under certain stress conditions (31). Recently, it has been reported that S. aureus ClpP is required for growth at reduced and elevated temperatures on solid medium (26). To obtain a more detailed view of the ability of a ΔclpP strain to cope with nonpermissive temperatures, growth experiments in liquid culture were performed by comparing growth at different temperatures of the ΔclpP strain to the parent strain 8325 and the complemented mutant ΔclpP+ strain. The ΔclpP strain showed a growth defect at all temperatures tested (37°C, 30°C, 20°C, 42°C, and 45°C) (Fig. 1A). The temperature sensitivity of the mutant was especially observed at reduced temperatures. At 20°C the mutant grew for 6 h with a similar growth rate as the wild type; however, subsequently the cells ceased growth (Fig. 1A). To investigate transcriptional activation of clpP at reduced temperatures, Northern blot analysis was performed after growth of the wild-type strain at 37°, 30°, and 20°C, respectively. A threefold induction of clpP could be observed at 30°C, and transcription of the clpP gene was increased fourfold at 20°C (Fig. 1B). These results suggest an increase of ClpP protease activity at lower temperatures which might be an essential response of S. aureus to survive under these conditions. Low temperatures, similar to heat shock, cause extensive protein denaturation and subsequent aggregation (24). Likewise, in the cyanobacterium Synechococcus sp., ClpP1 is essential for adaptation and growth at 25°C (64). The growth kinetics suggests that ClpP is even more important for growth at low temperatures than at higher temperatures. Importantly, during the first hours of growth, there was no significant difference in the doubling time of mutant and wild-type cells, but growth stops in the logarithmic growth phase (Fig. 1A). This is probably due to an accumulation of misfolded and aggregated proteins that prevents further expression of functional proteins. Notably, the morphology of colonies of the ΔclpP strain showed a reduction in size (∼0.8-fold) compared to the wild-type strain 8325 (Fig. 2). After prolonged incubation at 37°C, the wild type became slightly yellowish while the mutant remained white (data not shown). A slightly different cell surface of the ΔclpP strain was observed by scanning electron microscopy, and it appeared to be more rough and irregular (Fig. 2). All effects in the deletion mutant could be restored by complementation (data not shown).
FIG. 1.
(A) Growth kinetics of S. aureus 8325 wild-type (♦), 8325ΔclpP (▪), and 8325ΔclpP+ (▴) strains grown at 37°C, 30°C, 20°C, 42°C, and 45°C. The results are representative of three independent experiments. (B) Northern blot analysis of clpP transcription in S. aureus 8325 at various temperatures (left) and at various time points during the growth phase (at indicated OD600 values) at 37°C.
FIG. 2.
Scanning electron microscopy of strain 8325 wild-type (A and B) and the isogenic ΔclpP mutant (C and D). Cells of the ΔclpP strain show a rougher and more irregular surface and decreased cell size than the wild-type strain. Preparation of samples was performed as described in Materials and Methods.
Global transcriptional profile of ΔclpP mutant of strain 8325.
There is increasing evidence that the Clp protease complex is involved in not only the degradation of misfolded proteins under stress conditions but also the regulation of protein expression and secretion (32, 36). In several bacterial pathogens, including S. aureus, virulence is strongly influenced by the activity of ClpP (26). To learn more about the regulatory role of ClpP in S. aureus, transcriptome analysis was performed by comparing exponentially growing (OD600 of 1.0) ΔclpP mutant and parental strain 8325 using an S. aureus full genome chip. We decided to analyze gene expression at this time point because clpP transcription was maximal in the logarithmic growth phase (Fig. 1B). Moreover, it has been demonstrated by DNA microarray analysis that ∼97% of all genes are expressed at the end of the exponential growth phase (70). The experiments presented here revealed a reduced transcription of 227 ORFs in the ΔclpP strain, whereas transcription of 197 ORFs was increased. The expression of genes belonging to several regulons which play a role in virulence, oxidative stress, redox state, SOS response, metal homeostasis, and anaerobic growth were affected by the deletion of clpP. The expression data of the different categories are described and discussed in the following sections.
Virulence factor expression.
Expression of 46 virulence-associated genes was differentially regulated in the ΔclpP strain (Table 2). Genes that encode adhesins, including those encoding fibrinogen-binding proteins (clfA and clfB), the fibronectin binding proteins (fnbA and fnbB), and the elastin-binding protein (epbS), were induced in the mutant strain. Many exoenzymes were down-regulated, including alpha-toxin encoded by hla, V8 serine protease encoded by sppA, the serine proteases encoded by the spl operon, the metalloproteinase encoded by aur, a lipase precursor (encoded by lip), the cysteine proteinase staphopain (encoded by SA1725), a staphylococcal nuclease (encoded by nuc), and glycerol ester hydrolase (encoded by geh). Other virulence factors, such as those encoded by the cap operon (including 16 genes, capA-P) and an immunoglobulin G-binding protein (sbi), were down-regulated, whereas fmtB, isaB, and SA2447 (encoding a hypothetical protein, similar to streptococcal hemagglutinin protein) were up-regulated (Table 2). In addition, transcription of the ica operon (icaADBC), encoding products responsible for synthesis of the polysaccharide intercellular antigen, which is involved in biofilm formation of staphylococci, was strongly down-regulated in the mutant (Table 2).
TABLE 2.
Virulence-associated factors of S. aureus differentially expressed in the ΔclpP strain
| N315 ORF | Gene name | Description or predicted function | Expression ratio of WT/ΔclpPa |
|---|---|---|---|
| Up-regulated factors | |||
| Adhesins | |||
| SA0742 | clfA | Fibrinogen-binding protein A, clumping factor (LPXTG) | 0.3 |
| SA1268 | ebhB | Hypothetical protein, similar to streptococcal adhesin emb | 0.5 |
| SA1312 | ebpS | Elastin binding protein | 0.4 |
| SA2161 | Hypothetical protein, attachment to host cells and virulence | 0.4 | |
| SA2423 | clfB | Clumping factor B (LPXTG) | 0.4 |
| SA2290 | fnbB | Fibronectin-binding protein homolog (LPXTG) | Up |
| SA2291 | fnbA | Fibronectin-binding protein homolog (LPXTG) | Up |
| Toxin, SA1811 | hlb | Truncated beta-hemolysin | 0.4 |
| Other | |||
| SA0891 | Hypothetical protein; similar to ferrichrome ABC transporter | 0.5 | |
| SA1964 | fmtB | FmtB protein (LPXTG) | 0.4 |
| SA1979 | Hypothetical protein, similar to ferrichrome ABC transporter | 0.5 | |
| SA2337 | feoB | Ferrous iron transport protein B homolog | 0.3 |
| SA2356 | isaA | Immunodominant antigen A | 0.5 |
| SA2431 | isaB | Immunodominant antigen B | 0.4 |
| SA2447 | hsa | Hypothetical protein, similar to streptococcal hemagglutinin protein (LPXTG) | 0.3 |
| Down-regulated factors | |||
| Adhesins | |||
| SA0587 | mntC | Lipoprotein; streptococcal adhesin PsaA homologue | 2.5 |
| SA2459 | icaA | Intercellular adhesion protein A | 4.3 |
| SA2460 | icaD | Intercellular adhesion protein D | 9.0 |
| SA2461 | icaB | Intercellular adhesion protein B | 2.0 |
| SA2462 | icaC | Intercellular adhesion protein C | 2.9 |
| Toxins | |||
| SA1007 | hla | Alpha-hemolysin precursor | 3.8 |
| SA1813 | Hypothetical protein; similar to leukocidin chain lukM precursor | 4.3 | |
| Exoenzymes | |||
| SA0022 | Hypothetical protein; similar to 5′ nucleotidase (LPXTG) | 3.2 | |
| SA0309 | geh | Glycerol ester hydrolase | 3.9 |
| SA0746 | nuc | Staphylococcal nuclease | 5.0 |
| SA0901 | sspA | Serine protease; V8 protease; glutamyl endopeptidase | 4.1 |
| SA1628 | splD | Serine protease SplD | 3.6 |
| SA1629 | splC | Serine protease SplC | 7.7 |
| SA1630 | splB | Serine protease SplB | 3.6 |
| SA1725 | Staphopain, cysteine proteinase | 3.4 | |
| SA2430 | aur | Zinc metalloproteinase aureolysin | 18.8 |
| SA2463 | lip | Triacylglycerol lipase precursor | 3.7 |
| Other | |||
| SA0144 | capA | Capsular polysaccharide synthesis enzyme Cap5A | 5.6 |
| SA0145 | capB | Capsular polysaccharide synthesis enzyme Cap5B | 4.6 |
| SA0146 | capC | Capsular polysaccharide synthesis enzyme Cap8C | 4.1 |
| SA0147 | capD | Capsular polysaccharide synthesis enzyme Cap5D | 4.4 |
| SA0148 | capE | Capsular polysaccharide synthesis enzyme Cap8E | 3.3 |
| SA0149 | capF | Capsular polysaccharide synthesis enzyme Cap5F | 3.2 |
| SA0150 | capG | Capsular polysaccharide synthesis enzyme Cap5G | 2.6 |
| SA0151 | capH | Capsular polysaccharide synthesis enzyme Cap5H | 2.6 |
| SA0152 | capI | Capsular polysaccharide synthesis enzyme Cap5I | 2.5 |
| SA0153 | capJ | Capsular polysaccharide synthesis enzyme Cap5J | 2.1 |
| SA0154 | capK | Capsular polysaccharide synthesis enzyme Cap5K | 2.6 |
| SA0155 | capL | Capsular polysaccharide synthesis enzyme Cap5L | 2.5 |
| SA0156 | capM | Capsular polysaccharide synthesis enzyme Cap5M | 2.7 |
| SA0157 | capN | Capsular polysaccharide synthesis enzyme Cap5N | 2.1 |
| SA0158 | capO | Capsular polysaccharide synthesis enzyme Cap8O | 3.0 |
| SA0159 | capP | Capsular polysaccharide synthesis enzyme Cap5P | 2.1 |
| SA0252 | lrgA | Holin-like protein LrgA | 11.2 |
| SA0253 | lrgB | Holin-like protein LrgB | 12.7 |
| SA0566 | Hypothetical protein; similar to iron-binding protein | 2.5 | |
| SA0841 | Hypothetical protein; similar to cell surface protein Map-w | 2.3 | |
| SA1709 | Hypothetical protein; similar to ferritin | 3.1 | |
| SA2206 | sbi | Immunoglobulin G-binding protein SBI | 4.3 |
Ratio of gene expression of wild-type (WT) versus the ΔclpP mutant strain. Values of ≥2 indicate decreased expression, and values of ≤0.5 indicate increased expression in the ΔclpP strain compared to the wild type. Up, increased transcription in the ΔclpP strain confirmed by RT-PCR.
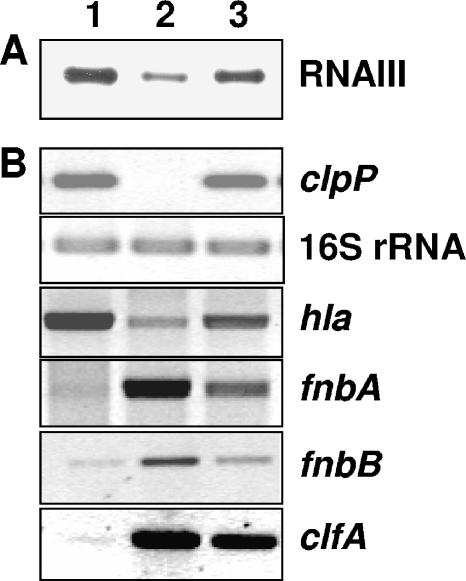
Since many of the deregulated virulence factors are regulated by the global regulatory agr system (61), we investigated the expression of RNAIII, the effector molecule of the agr system, by Northern blot analysis. As shown in Fig. 3A, the RNAIII transcript levels were about threefold decreased in the ΔclpP strain, confirming the results by Frees et al. (26, 27). Thus, the observed changes in the expression of agr-regulated genes could be the direct result of down-regulation of RNAIII effector molecule levels. In addition, transcription of sarA, a global repressor of protease expression (13, 47), was up-regulated 2.5-fold in the ΔclpP strain. The strong repression of the metalloprotease aureolysin gene aur (18.8-fold) in particular might be due to the overexpression of sarA, as it has been shown that aur is most sensitive to repression by SarA (47). The mechanism of how ClpP regulates expression of agr and sarA remains unknown, especially if AgrA, AgrC, or SarA is a substrate of proteolytical cleavage by ClpP. Recently, Frees et al. (27) suggested that agr and ClpXP act epistatically on extracellular gene expression and that possibly Rot, a repressor of toxin expression, links the agr regulatory system with the ClpXP machinery, where Rot is targeted by ClpXP in the presence of accumulating RNAIII (27). However, further work has to be done to unravel the role of ClpP in the network of virulence factor regulation in S. aureus. Possibly, different regulators are substrates of ClpP-dependent proteolysis, as has been suggested for Rot (27). The microarray expression data of extracellular proteases could be corroborated by reduced proteolysis on milk-agar plates (data not shown). Moreover, we confirmed the down-regulation of hla expression which has been shown previously (26) by RT-PCR (Fig. 3B). Down-regulation of hla is most probably the result of low RNAIII expression. In addition, sarT, a repressor of hla, was up-regulated in the mutant, which might contribute to the decreased hla expression levels.
FIG. 3.
Transcriptional analysis of selected genes in 8325 wild-type (lane 1), ΔclpP (lane 2), and ΔclpP+ (lane 3) strains. RNA was isolated from exponentially growing cells (OD600 of 1.0). (A) Northern blot analysis of RNAIII expression by hybridization with an RNAIII-specific probe. (B) Semiquantitative RT-PCR for transcriptional analysis of hla, fnbA, fnbB, and clfA. As a control, expression of 16S rRNA and clpP was determined.
Regulation of virulence factor expression in S. aureus is extremely complex, involving at least four two-component systems (agrAC, arlRS, saeRS, and srrAB), several transcription factors (encoded by sarA, sarS, sarT, sarR, and rot), and an alternative sigma factor (σB) (reviewed in reference 61). Here, we show that deletion of clpP exerted a strong impact on transcription of virulence-associated genes, many of which are under the control of global regulatory systems. The transcription of the agr system, arlRS, and sigB was down-regulated, whereas sarA and sarT were up-regulated in the ΔclpP strain. However, not all data of our study fit into the current concept of regulatory events leading to expression of a distinct virulence gene. For example, the arlRS system acts divergently to agr in the regulation of virulence determinants including hla, hlb, lip, and sspA, whose transcription is increased in an arlS mutant, as well as RNAIII transcription (22). In our study, arlRS expression was decreased by a factor of 3, and agrAC and RNAIII expression was also decreased by a factor of 2 to 4. This suggests that the impact of ArlRS on agr-regulated gene expression was superseded by other regulatory processes or that the level of ArlRS expression was still sufficient to depress RNAIII production. Alternatively, the reported regulatory impact of ArlRS on agr may reflect the fact that it was mainly investigated in strain 8325-4; however, Fournier and Klier (21) stated that in strain 8325, the strain used in this study, regulation might be different than in strain 8325-4 (21). Recently, Liang et al. investigated the Arl regulon by DNA microarray analysis (53). It was shown that ArlRS up-regulates the transcription of agrBDCA as well as hld located within the regulatory RNAIII in strain WCUH29. These results are in contrast to previous reports showing a repressive effect of ArlRS on agr RNAII and RNAIII expression (22). Further work has to be done to clarify the exact role of ArlRS on gene regulation in different genetic backgrounds. The strong impact of the clpP deletion on certain regulatory pathways of virulence factor expression clearly indicates a link between ClpP protease activity and regulation of virulence traits.
Internalization of the ΔclpP strain in 293T cells.
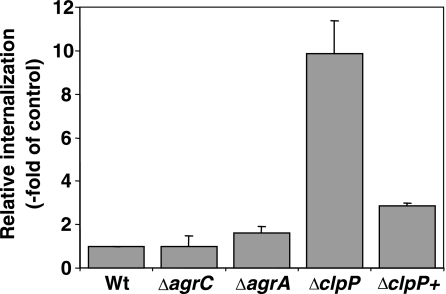
As several adhesins including fibronectin binding proteins A and B (encoded by fnbA and fnbB) were up-regulated in the ΔclpP strain (Fig. 3), we tested the ability of the ΔclpP strain to invade human epithelial cells. Interestingly, the rate of internalization by 293T cells increased about ∼10-fold compared to the parent strain (Fig. 4). The isogenic strains 8325ΔagrA and 8325ΔagrC, which were taken as controls, showed no significant differences in internalization rate, indicating an agr-independent mechanism responsible for increased internalization of the ΔclpP strain. The fibronectin-binding proteins FnbA and FnbB serve as the main surface proteins of S. aureus that mediate adherence to host cells by binding of fibronectin, which interacts with β1α5-integrins on the surface of host cells. In turn, β1α5-integrin clustering triggers the uptake of S. aureus by a zipper-like mechanism (1, 71). Gene expression data of both fnbA and fnbB were excluded from microarray analysis due to differences in homology between N315 and 8325 DNA sequences. Thus, the expression of these genes was analyzed by RT-PCR, revealing an induction of expression of fnbA and fnbB by threefold compared to the wild type (Fig. 3). Furthermore, fibronectin binding capacity was analyzed. The ΔclpP mutant showed a 2.3-fold increased capability to bind fibronectin in comparison to the wild type (data not shown). These results suggest that at least one reason for the increased internalization rate could be the overexpression of FnbA and FnbB. Recently, Frees et al. (24) investigated the intracellular replication of a clpP mutant of S. aureus strain 8325-4 in MAC-T cells, a bovine mammary epithelial cell line. ΔclpP cells were not able to replicate intracellularly, as indicated by bioluminescence (25). In contrast to that study, where the internalization rate was not affected by the clpP deletion, we clearly observed a significant increase in the internalization rate of ΔclpP cells compared to the wild type. Since we used the human kidney cell line 293T and our strain background was 8325, it has to be clarified whether the observed differences are due to the S. aureus strain background or the host cell line.
FIG. 4.
Internalization of ΔclpP mutant cells was increased in 293T cells. Relative internalization of different isogenic mutants of strain 8325 (ΔagrA, ΔagrC, ΔclpP, and complemented ΔclpP+ strains) is compared to internalization of 8325 wild type (Wt). Means ± standard deviations of four experiments are given.
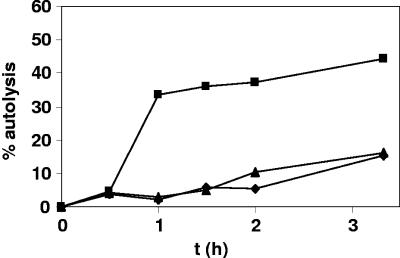
Autolysis.
Expression of regulators of murein hydrolases (encoded by lytSR, lrgAB, arlSR, and rat) was mostly decreased in the ΔclpP strain, while transcription of lytM was increased (Tables 2 to 4). To determine the effect of clpP deletion on autolysis, an assay was performed treating cells with Triton X-100. The ΔclpP strain showed a strong induction of autolysis starting after 30 min of growth compared to wild-type and ΔclpP+ strains, confirming the microarray data (Fig. 5). The two-component system lytSR is involved in regulation of peptidoglycan hydrolases. In S. aureus a lytS mutant showed increased autolysis, altered levels of hydrolase activity, and a rough cell surface (10). lrgA and lrgB are positively regulated by lytSR, and their products show similarities to a bacteriophage murein hydrolase transporter family of proteins known as holins, which negatively affect peptidoglycan hydrolases (10). Interestingly, as mentioned above, expression of arlSR, encoding a two-component system (TCS) involved in autolysis, was also reduced in the clpP mutant (20, 53). Recently, DNA microarray analysis revealed a down-regulation of lytSR and lrgAB by ArlRS (53). Thus, it is tempting to speculate that decreased arlRS expression in the mutant contributes to the enhanced autolysis in the ΔclpP strain. In addition, the transcriptional regulator rat is described to be a repressor of autolysis and belongs to the MarR and SarA protein families (43). This type of repressor was also down-regulated in the clpP mutant. Altogether, the transcriptional profile of genes involved in autolysis may reflect the strong influence of ClpP protease activity on the regulation of autolysis in S. aureus.
TABLE 4.
Putative YycFG-controlled genes of S. aureus differentially expressed in the ΔclpP strain
| N315 ORF | Gene name | Description or predicted functiona | Expression ratio of WT/ΔclpPb | Position (orientation)c | Putative YycF-binding sequenced |
|---|---|---|---|---|---|
| Up-regulated | |||||
| SA0265e | lytM | Peptidoglycan hydrolase | 0.2 | −142 (+) | TGTAATGACAATGTAAT |
| SA0674e | Putative anion-binding protein | 0.5 | −16 (+) | TGTAATCAAATTGTAAT | |
| SA1221e | Thioredoxin reductase | 0.3 | −113 (−) | TGTTAAGAAAATGTAAA | |
| SA1305e | hu | DNA-binding protein II | 0.4 | −58 (+) | TGTAATGCTTGTGTTAA |
| SA1312e | ebpS | Elastin binding protein | 0.4 | −22 (+) | TGTAAAATCATTGTAAT |
| SA1898e | HP; similar to SceD precursor | 0.5 | −113 (+) | TGTAATCACTGTGTAAA | |
| SA2093e | ssaA | Secretory antigen precursor SsaA homolog | 0.2 | −266 (−) | TGTTACAAATTTGTAAT |
| −138 (−) | TGTTAACGTTTTGTAAT | ||||
| SA2097e | HP; similar to secretory antigen precursor SsaA | 0.4 | −123 (−) | TGTTATTGATTTGTAAA | |
| SA2285 | aap | HP; similar to accumulation-associated protein | 0.3 | −34 (+) | TGTAAATTCACTGTAAG |
| SA2290 | fnbB | Fibronectin-binding protein homolog | Up | −121 (−) | TGTTAACTTTATGTATA |
| SA2353e | HP; similar to secretory antigen precursor SsaA | 0.4 | −166 (−) | TGTTATCATAATGTAAT | |
| SA2356e | isaA | Immunodominant antigen A | 0.5 | −140 (+) | TGTAAAGAAAGTGTAAT |
| SA2447-SA2440f | hsa | HP; similar to streptococcal hemagglutinin protein | 0.3-0.5 | −388 (−) | TGTAATATATGTGTAAT |
| SA2481e | Predicted sulfur transferase | 0.3 | −24 (+) | TGTTATAAGCATGTTAA | |
| Down-regulated | |||||
| SA0129e | sasD | HP | 4.5 | −16 (+) | TGTAATCAAATTGTAAT |
| SA0616-SA0617f | vraF | ABC transporter ATP-binding protein | 2.1-2.4 | −65 (+) | TGTTAGTCATATGTTAA |
| SA0682e-SA0681f | Putative di-tripeptide ABC transporter | 2.4-3.1 | −246 (−) | TGTTATTTTAATGTAAC | |
| SA0913e-SA0910f | qoxA | Putative quinol oxidase polypeptide II QoxA | 2.2-2.6 | −53 (+) | TGTAAATATTGTGTAAT |
| SA1945e-SA1944f | Mannose-6 phosphate isomerase Pmi homolog | 3.0-3.3 | −179 (+) | TGTTAAAGTACTGTAAA | |
| YycF consensus sequencee | TGTWAHNNNNNTGTWAH |
HP, hypothetical protein.
Ratio of gene expression of wild-type (WT) versus the ΔclpP strain. Values of ≥2 indicate decreased expression, and values of ≤0.5 indicate increased expression in the ΔclpP strain compared to the wild type. Up, increased transcription in the ΔclpP mutant confirmed by RT-PCR.
Position of the putative YycF-binding sequence relative to the translational start site in base pairs. Orientation (+/−) is given relative to the transcription of the respective gene.
Boldface, 100% conserved residues; italics, nucleotides conserved in more than half of all sequences.
Putative YycF-binding sequence (16). lytM, ssaA, and isaA were experimentally confirmed.
First and last ORF of putative transcription unit.
FIG. 5.
Autolysis of whole cells of S. aureus 8325 wild-type (♦), 8325ΔclpP (▪), and complemented mutant ΔclpP+ (▴) strains by Triton X-100. The results are expressed as lysis percentages as described in Materials and Methods. The average of two independent experiments is shown.
Heat shock regulation.
The loss of ClpP leads to accumulation of misfolded proteins similar to stress conditions, resulting in an increased demand for chaperones and proteases which are typically induced under heat shock conditions. The transcription of CtsR and HrcA, the main regulators of the heat shock response, is completely derepressed, as are the genes of the corresponding heat shock regulon (Table 3) as described for clpP mutants of S. aureus and S. pneumoniae (25, 66). In S. aureus the HrcA regulon (hrcA-grpE-dnaK-dnaJ and groESL) is embedded within the CtsR regulon (ctsR-mcsA-mcsB-clpC, clpB, and the HrcA regulon) (11). Hence, its derepression could be the result of inactivation of the CtsR repressor. In B. subtilis HrcA requires GroE to adopt its active conformation. Decreased levels of free GroE by association with misfolded proteins under heat shock conditions lead to inactivation of HrcA and a derepression of transcription of the HrcA regulon (59). The activity of the repressor CtsR is modulated by McsA and McsB and results in targeted degradation of CtsR by ClpCP in response to several stresses (14, 50). McsA contains a CXXC motif which might serve as a sensor of oxidative conditions. In B. subtilis elevated temperatures and oxidative stress conditions (H2O2, paraquat, NO, and diamide) give rise to an inactivation of CtsR and a derepression of transcription of corresponding genes (3, 52, 60). In S. aureus CtsR accumulates in cells lacking ClpP due to limited degradation by the Clp proteolytic machinery (25). As transcription of heat shock genes controlled by CtsR was induced in the clpP mutant, this would imply that CtsR accumulates in an inactive conformation in the ΔclpP strain and is not able to bind to the promoter region of those genes.
TABLE 3.
Genes encoding putative regulators of S. aureus differentially expressed in the ΔclpP strain
| N315 ORF | Gene name | Description or predicted functiona | Putative transcription unitb (5′→3′) | Expression ratio of WT/ΔclpPc |
|---|---|---|---|---|
| Up-regulated | ||||
| SA0017 | yycF (vicR) | Two-component response regulator | yycF (0.5)-yycG (0.4) | 0.5 |
| SA0298 | HP; similar to regulatory protein PfoR | 0.5 | ||
| SA0480 | ctsR | Repressor of class III stress genes | ctsR (0.3)-SA0481 (0.1)-SA0482 (0.1)-clpC (0.1) | 0.3 |
| SA0573 | sarA | Staphylococcal accessory regulator A | 0.4 | |
| SA1041 | pyrR | Pyrimidine operon repressor chain A | pyrR (0.5)-pyrP (0.4)-pyrB (0.4)-pyrC (0.4)-pyrAA (0.4)-pyrAB (0.5)-pyrF (0.5)-pyrE (0.4) | 0.5 |
| SA1098 | codY | Transcription pleiotropic repressor CodY | 0.5 | |
| SA1139 | glpP | Glycerol uptake operon antiterminator | 0.5 | |
| SA1174 | lexA | SOS regulatory LexA protein | 0.4 | |
| SA1411 | hrcA | Heat-inducible transcriptional repressor | hrcA (0.4)-grpE (0.4)-dnaK (0.3)-dnaJ (0.3) | 0.4 |
| SA1897 | HP; similar to transcriptional activator TenA | SA1897 (0.3)-thiD (0.4)-thiM (0.5)-thiE (0.4) | 0.3 | |
| SA2286 | sarT | SarA homologue | 0.5 | |
| SA2320 | HP; similar to regulatory protein PfoR | SA2320 (0.3)-SA2319 (0.4)-SA2318 (0.2) | 0.3 | |
| SA2418 | HP; similar to two-component RR | SA2418 (0.5)-SA2417 (0.4) | 0.4 | |
| Down-regulated | ||||
| SA0250 | lytS | Two-component sensor HK | lytS (2.1)-lytR (2.1) | 2.1 |
| SA0322 | HP; similar to transcription regulator, MarA family | SA0322 (3.5)-svrA (2.8) | 3.5 | |
| SA0454 | purR | pur operon repressor homologue | 2.1 | |
| SA0641 | rat | HP; similar to transcriptional regulator | 3.8 | |
| SA1248 | arlR | Two-component RR | arlR (3.6)-arlS (2.1) | 3.6 |
| SA1509 | COG1327: predicted transcriptional regulator | 2.4 | ||
| SA1748 | HP; similar to transcription regulator, GntR family | SA1748 (2.6)-SA1747 (ND*)-SA1746 (2.1)-SA1745 (2.3)-SA1744 (2.2) | 2.6 | |
| SA1801 | Antirepressor | 4.3 | ||
| SA1843 | agrC | Accessory gene regulator C | agrB (ND*)-agrD (ND*)-agrC (2.4)-agrA (1.9) | 2.4 |
| SA1869 | sigB | Sigma factor B | rsbU (4.2)-rsbV (ND*)-rsbW (4.5)-sigB (4.1) | 4.1 |
| SA2089 | sarR | SarA homologue | 2.3 | |
| SA2108 | HP; similar to transcription regulator, RpiR family | 2.0 | ||
| SA2180 | nreB | HP; similar to two-component HK | nreA (3.1)-nreB (3.2)-nreC (2.4) | 3.2 |
HP, hypothetical protein.
Values in parentheses indicate relative expression levels of genes organized in one putative operon. ND*, ORF not represented on microarray used.
Ratio of gene expression of wild-type (WT) versus the ΔclpP strain. Values of ≥2 indicate decreased expression, and values of ≤0.5 indicate increased expression in the ΔclpP strain compared to the wild type.
Transcription of regulatory proteins was strongly affected by clpP deletion.
The genes of five TCSs were differentially expressed in the clpP mutant compared to the wild type: four were down-regulated, including lytSR, arlRS, agrAC, and a TCS with homology in sequence and orientation with nreBC of S. carnosus (Table 3). In contrast, the essential YycG/YycF TCS was up-regulated. Furthermore, the expression of 10 putative regulators was reduced, including those encoded by rat/mgr and sarR; an antirepressor encoded by SA1801; and two putative transcriptional regulators, encoded by SA0322 (MarA family) and SA1748 (GntR family). In addition, transcription of 10 transcriptional regulators was increased, including those encoded by ctsR, hrcA, sarA, sarT, codY, and lexA; a putative transcriptional regulator similar to TenA, encoded by SA1897; and a hypothetical protein similar to the regulator protein PfoR, encoded by SA2320. The genes of the sigB operon and the sigB-dependent asp23 were down-regulated. Notably, although strain 8325 is regarded as a functional sigB mutant due to an 11-bp deletion in rsbU, sigB transcription could be detected, suggesting a residual SigB activity in strain 8325. Likewise, Palma and Cheung (62) detected a reduced (by up to 50%) but still present expression of SigB-dependent genes in an rsbU mutant of the wild-type strain FDA486 (62). The observed strong influence of clpP deletion on transcription of regulators suggests that ClpP proteolytic activity may serve as an important mechanism to control gene expression in S. aureus. Therefore, a genome-wide in silico sequence analysis was performed using known consensus sequences of regulatory proteins including Fur (ferric uptake regulator), PerR, MntR, LexA, Fnr/ArcR, and YycFG to assess the impact of ClpP on expression of genes belonging to several regulons. This analysis revealed a strong impact of clpP deletion on the expression of genes that may be part of these regulons. However, it has to be stressed that the in silico recognition sequence search was solely based on known or putative consensus sequences and that for most members of specific regulons no experimentally confirmed data are available. For those genes for which regulator binding has been experimentally confirmed, this information was included in the analysis. The conclusion that ClpP might be involved in the regulation of the transcription of members of the Fur, PerR, MntR, LexA, Fnr/ArcR, and YycFG regulons was based on the observation that a significantly higher portion of ORFs with a putative recognition sequence of one of these regulators upstream of the translational start was deregulated (between 33 and 63%) than the overall percentage of deregulated genes (approximately 19% of all ORFs).
Impact of ClpP on expression of genes of the essential YycFG regulon.
The highly conserved YycF/YycG (VicR/VicK) TCS has been demonstrated to be essential in several gram-positive bacteria by regulation of cell wall biosynthesis and cell division (16, 41, 46, 57). In S. aureus a mutation in yycF results in a lethal phenotype at nonpermissive temperatures, and its essentiality has been proven by regulated expression of yycF using a conditional mutant system (16, 51, 57). In our experiments, deletion of clpP increased transcription of the yycFG locus. Regulation of yycFG transcription is presently unknown. Autoregulation can be ruled out as no YycF-specific recognition sequence can be found in the upstream region. Interestingly, we could identify a putative Crp/Fnr-like consensus sequence 62 bp upstream of the translation start of yycF (see below). The YycF-specific DNA-binding sequence consists of two repetitive hexamers: [TGT(A/T)A(A/T/C)-5N-TGT(A/T)A(A/T/C)] identified in B. subtilis and S. aureus (16, 41). In S. aureus N315 the consensus sequence could be found upstream of 31 ORFs (16). For three genes (lytM, ssa, and isaA) binding of the response regulator to the consensus was demonstrated recently (16). In the ΔclpP strain 16 putative members of the described yycFG regulon were deregulated, including the three experimentally confirmed genes lytM, ssa, and isaA (Table 4). In addition, we identified four additional putative yycFG-regulated genes: aap, hsa, fnbB, and vraF. Fourteen out of the 20 genes were up-regulated in the mutant including genes involved in cell wall synthesis (lytM, ssa, and two ssa homologous genes, SA2097 and SA2353) and virulence (isaA, ebpS, SA1898, fnbB, and hsa). Overall, 57% of all putative YycFG-regulated genes were deregulated in the ΔclpP strain.
ClpP controls metal ion homeostasis and oxidative stress proteins.
In B. subtilis and S. aureus, genes involved in iron and manganese homeostasis are regulated by three Fur homologous repressors, Fur, PerR, and Zur, and in addition by the DtxR homolog MntR. Fur is a transcriptional repressor controlling genes involved in iron uptake. Fur-regulated genes possess a so-called Fur box located upstream of the start codon. To find putative Fur-regulated genes, we used the postulated Fur box GATAATGATWATCATTATC for a consensus sequence search (40). We found that the expression of 6 out of 12 genes with a putative Fur box in the N315 genome was differentially regulated in the ΔclpP strain (Table 5). Interestingly, all of these genes including the Fur-dependent iron transporters feoB and feoB2 and a gene coding for a thioredoxin-homologous protein (SA2162) were up-regulated in the ΔclpP strain, indicating a lower Fur repressor activity in the clpP mutant.
TABLE 5.
Putative Fur-controlled genes of S. aureus differentially expressed in the ΔclpP strain
| N315 ORF | Gene name | Description or predicted function | Expression ratio of WT/ΔclpPa | Position (orientation)b | Putative Fur boxc |
|---|---|---|---|---|---|
| SA0162 | aldA | Aldehyde dehydrogenase homolog | 0.3 | −234 (−) | CTTGAGAATAATTCTCATTAAA |
| SA1982d-SA1980e | feoB2 | Putative transporter | 0.3-0.4 | −24 (+) | AATGATAATGATTCTTATTATC |
| SA1979 | Putative ferrichrome ABC transporter | 0.4 | −41 (−) | ATTGATAACAATTATCATTGTC | |
| SA2001 | Putative oxidoreductase, aldo/keto reductase family | 0.5 | −135 (+) | ATTGATAATTATGATAATCATA | |
| SA2162d | Putative thioredoxin reductase | 0.4 | −92 (+) | ATTGATAATTATTATCATTTAA | |
| SA2337d | feoB | Ferrous iron transport protein | 0.3 | −20 (+) | AGTGATAATGATTATTATTTCT |
| Fur consensus sequence (40) | NNNGATAATGATTATCATTATC |
Ratio of gene expression of wild-type versus the ΔclpP strain. Values of ≥2 indicate decreased expression, and values of ≤0.5 indicate increased expression in the ΔclpP strain compared to the wild type.
Position of the putative Fur box relative to the translational start site in base pairs. Orientation (+/−) is given relative to the transcription of the respective gene.
Boldface, 100% conserved residues; italics, nucleotides conserved in more than half of all sequences.
Predicted Fur box (40).
First and last ORF of putative transcription unit.
Furthermore, we analyzed the transcription of putative PerR-regulated genes. PerR controls as a Mn-dependent repressor a peroxide defense regulon. Members of this regulon, like catalase and peroxidases, detoxify reactive oxygen species (ROS); others, like ferritin or MrgA, store iron. Using an adapted consensus sequence postulated by Horsburgh et al. (39), we found 36 putative PerR-regulated genes in the genome of strain N315; 12 out of these 36 genes were deregulated in the ΔclpP strain (Table 6). Ten out of the 12 genes were up-regulated in the mutant, including the known PerR-controlled genes ahpCF, nfrA, and trxB. However, transcription of ftnA (ferritin) was decreased, suggesting incomplete derepression of the PerR regulon in the ΔclpP strain or other yet unknown regulatory mechanisms. In S. aureus and other bacteria, peroxide defense mechanisms and iron homeostasis are linked with manganese (Mn) transport that is controlled by MntR. MntR regulates as a Mn-dependent repressor the expression of two transport systems, mntABC and mntH (38). It has been proposed that mntABC represents the major Mn transport system in S. aureus that is regulated by several metal-depending repressors including PerR. Expression of mntABC has been shown to be induced at high Mn concentrations, while expression of mntH is repressed (38). Manganese acts in a dual way as an antioxidant and as a cofactor of enzymes like catalases, superoxide dismutases, and peroxidases. It is assumed that manganese protects S. aureus against ROS as a scavenger of either superoxide (O2−) or hydrogen peroxide (H2O2) (38). Therefore, these bacteria possess a basal protection against ROS and are not required to activate the energy-dependent PerR defense regulon. Consistently, it was shown that Mn(II) acts as a repressor of PerR (38). However, under high oxidative stress conditions, the Mn-based defense mechanism becomes inadequate, giving rise to induction of the H2O2-sensitive PerR regulon. The deletion of clpP has a drastic effect on the expression of genes of the PerR, Fur, and MntR regulons, which strongly suggests that ClpP proteolytic activity is a key element in the defense against ROS under aerobic growth conditions. As mntABC (SA0587 to SA0589) expression was decreased in the clpP mutant, this would suggest that manganese transport is affected by the clpP deletion. Consequently, a decreased intracellular Mn level could contribute to oxidative stress conditions and derepression of the PerR regulon. The exact role of ClpP in coping with oxidative stress remains to be defined; however, the observed deregulation of oxidative stress-related regulons underlines the importance of functional ClpP activity for oxidative stress response. Importantly, Frees et al. (25) reported that the ClpP mutant in strain 8325-4 was more sensitive to hydrogen peroxide than the wild type (25).
TABLE 6.
Putative PerR-controlled genes of S. aureus differentially expressed in the ΔclpP strain
| N315 ORF | Gene name | Description or predicted function | Putative transcription unit (5′→3′) | Expression ratio of WT/ΔclpPa | Position (orientation)b | Putative PerR boxc,e |
|---|---|---|---|---|---|---|
| SA0229 | Conserved hypothetical protein | SA0230d-SA0229 | 0.4 | −238 (−) | AATTAAATTATTATTTT | |
| SA0298 | Putative regulatory protein PfoR | 0.5 | −118 (−) | ATAATAATTATTATTAA | ||
| SA0366e | ahpC | Alkyl hydroperoxide reductase subunit F | ahpC-ahpF | 0.3-0.4 | −59 (+) | ATTAGAATTATTATAAT |
| SA0367e | nfrA | Putative nitro/flavin reductase | 0.3 | −93 (+) | AGTTCAATTATTAACTT | |
| SA0719e | trxB | Thioredoxine reductase | 0.4 | −634 (+) | CATATAATTATTATTAT | |
| SA0891 | Putative ferrichrome ABC transporter | 0.5 | −390 (+) | AGATTAATTATTAAATA | ||
| SA0914 | chiB | Putative chitinase B | 2.8 | −137 (−) | GAAATAATTATTATTTT | |
| SA1268 | ebhB | Similar to streptococcal adhesin | ebhB-ebhA | 0.5 | −252 (+) | TTTATAATTATTATAAA |
| SA1407 | Conserved hypothetical protein | 0.3 | 15 (+) | CTTTCAATTATTATTAA | ||
| SA1617 | Similar to latent nuclear antigen | SA1617-SA1621 (SA1620d) | 0.1-0.4 | −191 (+) | TTTACAATTATTAAATT | |
| SA1709 | ftnA | Putative ferritin | 3.1 | −77 (+) | ATTATAATTATTATTAT | |
| SA1897 | Putative transcriptional activator TenA | SA1897-thiD-thiM-thiE | 0.3-0.5 | −261 (+) | TATAGAATTATTATTTA | |
| PerR consensus sequence (39) | ATTATAATTATTATAAT |
Ratio of gene expression of wild-type (WT) versus the ΔclpP strain. Values of ≥2 indicate decreased expression, and values of ≤0.5 indicate increased expression in the ΔclpP strain compared to the wild type.
Position of the putative PerR recognition sequence relative to the translational start site in base pairs. Orientation (+/−) is given relative to the transcription of the respective gene.
Boldface, 100% conserved residues; italics, nucleotides conserved in more than half of all sequences.
ORF not represented on the microarray.
Identification of putative LexA-regulated genes.
LexA regulates genes involved in repair of DNA damage. In E. coli LexA and the LexA homologous repressor HdiR have been recognized as substrates of ClpP-derived proteolysis (19, 69). Recently, a LexA-dependent regulation of fibronectin-binding protein B has been reported in S. aureus (6). In order to assess the impact of clpP deletion on the expression of putative LexA-regulated genes, a consensus sequence search using the B. subtilis recognition sequence (CGAACRNRYGTTCG) was performed (76). Without variation of the recognition motif, no putative LexA-regulated gene within the N315 genome could be identified. However, if we used a sequence adapted to GAAC-N4-GTTC, we recognized 12 out of 20 putative LexA-dependent genes which were differentially regulated in the mutant (Table 7). Importantly, all of these 12 genes were up-regulated in the mutant, including both known LexA-regulated genes, recA and fnbB. Moreover, putative LexA-regulated genes like umuC, uvrA, and lexA itself were up-regulated in the ΔclpP strain. In addition, we found the LexA recognition motif upstream of two genes belonging to the PerR regulon (ahpC and the ferric ABC transporter SA0891 gene). The expression of the fibronectin-binding protein fnbB was determined by RT-PCR as the DNA microarray experiments did not allow a clear prediction (Fig. 3B). The expression data indicate a derepression of the LexA-regulated SOS-DNA repair regulon, which might be the consequence of increased DNA damage due to the reduced capability of the ΔclpP strain to cope with oxidative stress and to remove unfolded proteins.
TABLE 7.
Putative LexA-controlled genes of S. aureus differentially expressed in the ΔclpP strain
| N315 ORF | Gene name | Description or predicted function | Expression ratio of WT/ΔclpPa | Position (orientation)b | Putative LexA-binding sequencec |
|---|---|---|---|---|---|
| SA0366 | ahpC | Alkyl hydroperoxide reductase subunit F | 0.3 | −308 (+) | CGAACAAATATTCT |
| SA0714d | uvrA | Exinuclease ABC subunit A | 0.4 | −65 (+) | CGAAAGATTTAGAT |
| SA0891 | Putative to ferrichrome ABC transporter | 0.5 | −354 (+) | TGAACAATTGTTGT | |
| SA0993 | uvrC | Excinuclease ABC subunit C | 0.5 | −79 (+) | CGAAGATGTTGATT |
| SA1128d | recA | RecA | 0.4 | −86 (+) | CGAACAAATATTCG |
| −129 (−) | CGAACAAACGTGCT | ||||
| SA1174d | lexA | SOS regulatory LexA protein | 0.4 | −58 (+) | CGAACAAATGTTTG |
| SA1180 | Similar to exonuclease SbcD | 0.5 | −15 (+) | CGAACAAATGTTCT | |
| SA1196d | umuC | Similar to DNA-damage repair protein | 0.5 | −35 (−) | CGAACACGTGTTCT |
| SA2090d | fnbB | Fibronectin-binding protein homolog | Up | −58 (+) | CGAACAATATAGAA |
| −86 (−) | TGAAAAAAAGCGAG | ||||
| SA2091d | fnb | Fibronectin-binding protein homolog | Up | −59 (+) | CGAACAATATAGAC |
| SA2375 | Similar to dihydroorotate dehydrogenase | 0.4 | −223 (−) | TGAACAATGGTTAG | |
| SA2473 | Hypothetical protein | 0.4 | −205 (−) | TGAACGTTGGTTAC | |
| LexA consensus sequenced | GAAC-N4-GTTC |
Ratio of gene expression of wild-type versus the ΔclpP strain. Values of ≥2 indicate decreased expression, and values of ≤0.5 indicate increased expression in the ΔclpP strain compared to the wild type. Up, increased transcription in the ΔclpP mutant confirmed by RT-PCR.
Position of the putative LexA recognition sequence relative to the translational start site in base pairs. Orientation (+/−) is given relative to the transcription of the respective gene.
Boldface, 100% conserved residues; italics, nucleotides conserved in more than half of all sequences.
Anaerobic growth.
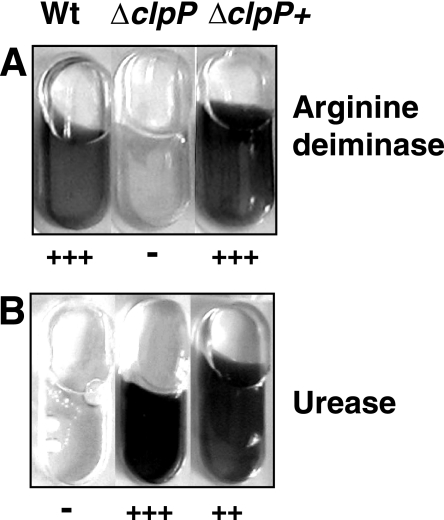
The physiological examination of the ΔclpP strain by using the API Staph test system showed that arginine deiminase activity was reduced (Fig. 6A). Arginine deiminase is encoded by arcA, which is located in an operon (arcABCDR) whose transcription is induced under anaerobic conditions and which is controlled by catabolite repression (15, 78). The arginine deiminase system is used by many prokaryotes to produce ATP under anaerobic conditions by catalyzing the conversion of arginine to ornithine, ammonia, and CO2. Expression of the arc operon is controlled by regulatory proteins of the Crp/Fnr family (54). S. aureus and other gram-positive bacteria carry a gene coding for a Crp/Fnr homologous protein (arcR), located downstream of arcA in the N315 genome. A consensus sequence search using the ArcR recognition sequence of Bacillus licheniformis within all deregulated genes of the ΔclpP strain resulted in the identification of 19 genes that carry an arcR consensus sequence upstream of the transcriptional start site resembling the B. licheniformis ArcR binding site TGTGA-N6-TCACG (55) (Table 8). Among these, 15 genes were down-regulated, and 10 of them are preferentially expressed under anaerobic conditions, including those encoding arginine deiminase (arcA), formate acetyltransferase (pflB), lactate dehydrogenase (lctE), nitrite extrusion protein (narK), and alcohol-acetaldehyde dehydrogenase (adhE). Overall, expression of almost two-thirds (19 out of 30) of all genes with a putative ArcR consensus sequence in front of the translational start were influenced by clpP deletion, suggesting a significant impact of ClpP on regulation of ArcR-dependent gene expression. Moreover, the transcription of other genes that are involved in anaerobic growth was affected in the ΔclpP strain. The TCS NreBC regulates anaerobic respiration in Staphylococcus carnosus by controlling transcription of the nitrate reductase operon (narGHIJ) and nitrite reductase (nir) (18). In S. aureus N315 we could identify a putative TCS with high homology to nreABC of S. carnosus (SA2181 to SA2179). In the ΔclpP strain nreABC as well as the nar and nir operons were down-regulated (see Table S1 in the supplemental material). Consequently, the ΔclpP strain showed a growth defect under anaerobic conditions on solid medium (data not shown). All these data indicate that ClpP is essential for growth and survival of S. aureus under anaerobic conditions, probably due to regulating the activity of the arginine deiminase pathway and, furthermore, nitrate and nitrite respiration.
FIG. 6.
Arginine deiminase (A) and urease (B) activity of 8325 wild-type (Wt), ΔclpP, and complemented mutant ΔclpP+ strains after 4 h of incubation (urease) or after 16 h of incubation (arginine deiminase). API Staph test was performed according to the manufacturer's instructions (BioMérieux). +++, ++, and − indicate very high, high, and no enzymatic activity, respectively.
TABLE 8.
Putative ArcR-controlled genes of S. aureus differentially expressed in the ΔclpP strain
| N315 ORF | Gene name | Description or predicted functiona | Expression ratio in WT/ΔclpPb | Position (orientation)c | Putative ArcR-binding sequenced |
|---|---|---|---|---|---|
| Down-regulated | |||||
| SA0143 | adhE | Alcohol-acetaldehyde dehydrogenase | 6.0 | −22 (+) | TTGTGAAATAATTCACAA |
| SA0218-SA0219e | pflB | Formate acetyltransferase | 7.8-15.7 | −79 (+) | ATGTGAAAAAAATCACAA |
| SA0232 | lctE | l-Lactate dehydrogenase | 12.1 | −210 (+) | ATGTGAAATAAATCACAA |
| SA0293 | nirC | HP; similar to formate transporter NirC | 4.4 | −55 (−) | TTGTGAATAATTTCACAA |
| SA0295 | HP; outer membrane protein precursor | 5.7 | −141 (+) | ATGTGATAGGTCTCCCAT | |
| SA0562 | adh1 | Alcohol-dehydrogenase I | 3.5 | −289 (+) | TTGTGAATTAATTCACAT |
| SA0641 | rat | Transcriptional regulator | 3.8 | −69 (+) | TTGTGAATTAATAAACAA |
| SA1272 | ald | Alanine dehydrogenase | 3.8 | −21 (−) | TTGTGAATAATTTCACAA |
| SA1813 | lukM | HP; similar to leukocidin chain lukM precursor | 4.3 | −49 (−) | ATGTGAATAATATCACAA |
| SA2156 | lctP | l-Lactate permease lctP homolog | 5.5 | −107 (+) | TTGTGAAAAAAATCACAT |
| SA2176 | narK | Nitrite extrusion protein | 3.6 | −151 (−) | TTGTGAAAAAGTGAACAT |
| SA2189-SA2188e | nirR | HP; similar to NirR | 7.7-10.1 | −47 (−) | TTGTGAAAAGAATCACAT |
| SA2268 | HP | 17.8 | −112 (+) | TTGTGAAATACATCACAA | |
| SA2428 | arcA | Arginine deiminase | 3.4 | −62 (+) | ATGTGAATATAATCACAT |
| SA2430 | aur | Zinc metalloproteinase aureolysin | 18.8 | −215 (−) | TTGTGAAAATATTAACAA |
| Up-regulated | |||||
| SA0017-SA0018e | yycFG | Response regulator/histidine kinase | 0.4-0.5 | −62 (−) | TTGTGTAAAAAATCACAG |
| SA0175 | Conserved HP | 0.4 | −42 (−) | TTGTGAAAATAATCACAA | |
| SA2311 | HP; similar to NAD(P)H-flavin oxidoreductase | 0.5 | −168 (+) | TTGTGAAAAATATCACAA | |
| SA2373-SA2371e | HP | 0.5 | −95 (+) | TTTTGAATATAATCACAA | |
| ArcR consensus sequencef | TGTGAA-N5-TCACA |
HP, hypothetical protein.
Ratio of gene expression of wild-type (WT) versus the ΔclpP strain. Values of ≥2 indicate decreased expression, and values of ≤0.5 indicate increased expression in the ΔclpP strain compared to the wild type.
Position of the putative ArcR-binding sequence relative to the translational start site in base pairs. Orientation (+/−) is given relative to the transcription of the respective gene.
Boldface, 100% conserved residues; italics, nucleotides conserved in more than half of all sequences.
First and last ORF of putative transcription unit.
In B. licheniformis (TGTGA-N6-TCACG) (55).
Urease activity.
It was striking that the urease activity test revealed a strong induction in the ΔclpP strain after 4 h of incubation, whereas the parental strain and the ΔclpP+ strain did not show any activity at this time point (Fig. 6B). This observation is clearly consistent with the microarray data showing an induction of the complete ure operon (SA2081; ureABCEFGD) in the ΔclpP strain by 5- to 10-fold at an OD600 of 1.0 (see Table S1 in the supplemental material). These results were also confirmed by RT-PCR (data not shown). Recently, induction of the ure operon was described in S. aureus biofilms and in a rot mutant (5, 67). In the ΔclpP strain no alteration of transcription of rot could be detected. As urease catalyzes the hydrolysis of urea to form ammonia and CO2, it has been suggested that a high urease activity may indicate attempts of bacteria to neutralize acidic environments. For example, urease activity of Helicobacter pylori is essential to colonize the acidic environment present in the stomach (17). Alternatively, the ure operon is induced in response to nitrogen starvation, e.g., in B. subtilis and Corynebacterium glutamicum (4, 7). Interestingly, carbamoyl phosphate synthetase transcription (pyrAA [SA1045] and pyrAB [SA1046]) was induced twofold in the mutant, but other enzymes of the urea cycle were not. Thus, increased levels of carbamoyl phosphate may be sufficient to generate a higher concentration of urea that is toxic for the cell and, consequently, has to be inactivated by urease. Further work has to be done to clarify the exact role of high urease activity for pH balance and/or nitrogen metabolism in ClpP-deficient cells.
Concluding remarks.
Global DNA expression analysis using DNA microarray technology revealed a broad impact of the S. aureus ClpP protease on several regulons involved in virulence, heat shock response, oxidative stress response, DNA repair, autolysis, and anaerobic growth. Targets of proteolytic ClpP activity in S. aureus are presently not known; however, the clustering of deregulated genes suggests that the expression of genes within specific regulons is controlled by ClpP-dependent proteolysis. In E. coli many proteins cleaved by ClpXP are involved in the oxidative stress response and a shift between aerobic and anaerobic growth. It has been suggested that ClpXP degrades proteins whose Fe-S clusters have been damaged by oxidation (19). Many results presented in this study are consistent with this idea. Possibly, ClpP plays a major role in the maintenance of reducing conditions within the cell by degradation of oxidized proteins. In consequence, oxidation-susceptible proteins like Spx may accumulate in the ΔclpP strain (77). An important challenge for the future will be to identify substrates of ClpP proteolytic activity in S. aureus and to clarify the role of functional ClpP for the infection process. In addition, ClpP may serve as an attractive new target for anti-infective agents. Interestingly, acyldepsipeptides, a new class of antibiotics that targets ClpP protease, has been recently identified (8). Surprisingly, the antimicrobial activity of the compound was not due to an inhibition of the target ClpP, but bacterial cells were killed by uncontrolled ClpP-dependent proteolysis. These observations impressively stress the importance of controlled ClpP-mediated proteolysis for protein homeostasis in bacterial cells.
Supplementary Material
Acknowledgments
This study was supported by Intervet Innovation GmbH, Schwabenheim (Germany), by a grant of the Bundesministerium für Bildung und Forschung Network of Competence “Pathogenomics” Alliance “Gram-Positive Cocci,” and by the Deutsche Forschungsgemeinschaft (SFB630).
We thank Karin Hilgert for technical assistance and Indranil Chatterjee for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agerer, F., A. Michel, K. Ohlsen, and C. R. Hauck. 2003. Integrin-mediated invasion of Staphylococcus aureus into human cells requires Src family protein-tyrosine kinases. J. Biol. Chem. 278:42524-42531. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. A. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology, vol. 4. John Wiley and Sons, Inc., New York, N.Y.
- 3.Bandow, J. E., H. Brotz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckers, G., A. K. Bendt, R. Kramer, and A. Burkovski. 2004. Molecular identification of the urea uptake system and transcriptional analysis of urea transporter- and urease-encoding genes in Corynebacterium glutamicum. J. Bacteriol. 186:7645-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisognano, C., W. L. Kelley, T. Estoppey, P. Francois, J. Schrenzel, D. Li, D. P. Lew, D. C. Hooper, A. L. Cheung, and P. Vaudaux. 2004. A RecA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 279:9064-9071. [DOI] [PubMed] [Google Scholar]
- 7.Brandenburg, J. L., L. V. Wray, Jr., L. Beier, H. Jarmer, H. H. Saxild, and S. H. Fisher. 2002. Roles of PucR, GlnR, and TnrA in regulating expression of the Bacillus subtilis ure P3 promoter. J. Bacteriol. 184:6060-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brotz-Oesterhelt, H., D. Beyer, H. P. Kroll, R. Endermann, C. Ladel, W. Schroeder, B. Hinzen, S. Raddatz, H. Paulsen, K. Henninger, J. E. Bandow, H. G. Sahl, and H. Labischinski. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11:1082-1087. [DOI] [PubMed] [Google Scholar]
- 9.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 10.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee, I., P. Becker, M. Grundmeier, M. Bischoff, G. A. Somerville, G. Peters, B. Sinha, N. Harraghy, R. A. Proctor, and M. Herrmann. 2005. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 187:4488-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 15.Dong, Y., Y. Y. Chen, and R. A. Burne. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton, K. A., J. V. Gilbert, E. A. Joyce, A. E. Wanken, T. Thevenot, P. Baker, A. Plaut, and A. Wright. 2002. In vivo complementation of ureB restores the ability of Helicobacter pylori to colonize. Infect. Immun. 70:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedtke, I., A. Kamps, B. Krismer, and F. Gotz. 2002. The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J. Bacteriol. 184:6624-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flynn, J. M., S. B. Neher, Y. I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671-683. [DOI] [PubMed] [Google Scholar]
- 20.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournier, B., and A. Klier. 2003. Response to the criticisms of Richard P. Novick in his review “Autoinduction and signal transduction in the regulation of staphylococcal virulence” (Novick, 2003, Mol. Microbiol. 48:1429-1449). Mol. Microbiol. 50:1085-1086. [DOI] [PubMed] [Google Scholar]
- 22.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 23.Frank, E. G., D. G. Ennis, M. Gonzalez, A. S. Levine, and R. Woodgate. 1996. Regulation of SOS mutagenesis by proteolysis. Proc. Natl. Acad. Sci. USA 93:10291-10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franks, F. 1995. Protein destabilization at low temperatures. Adv. Protein Chem. 46:105-139. [DOI] [PubMed] [Google Scholar]
- 25.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 26.Frees, D., S. N. Qazi, P. J. Hill, and H. Ingmer. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565-1578. [DOI] [PubMed] [Google Scholar]
- 27.Frees, D., K. Sorensen, and H. Ingmer. 2005. Global virulence regulation in Staphylococcus aureus: pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect. Immun. 73:8100-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaillot, O., S. Bregenholt, F. Jaubert, J. P. Di Santo, and P. Berche. 2001. Stress-induced ClpP serine protease of Listeria monocytogenes is essential for induction of listeriolysin O-dependent protective immunity. Infect. Immun. 69:4938-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 30.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 31.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 32.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565-587. [DOI] [PubMed] [Google Scholar]
- 33.Gottesman, S., and M. R. Maurizi. 1992. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 56:592-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haima, P., D. van Sinderen, H. Schotting, S. Bron, and G. Venema. 1990. Development of a beta-galactosidase alpha-complementation system for molecular cloning in Bacillus subtilis. Gene 86:63-69. [DOI] [PubMed] [Google Scholar]
- 36.Hengge, R., and B. Bukau. 2003. Proteolysis in prokaryotes: protein quality control and regulatory principles. Mol. Microbiol. 49:1451-1462. [DOI] [PubMed] [Google Scholar]
- 37.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 38.Horsburgh, M. J., S. J. Wharton, A. G. Cox, E. Ingham, S. Peacock, and S. J. Foster. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44:1269-1286. [DOI] [PubMed] [Google Scholar]
- 39.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howell, A., S. Dubrac, K. K. Andersen, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639-1655. [DOI] [PubMed] [Google Scholar]
- 42.Hussain, M., K. Becker, C. von Eiff, J. Schrenzel, G. Peters, and M. Herrmann. 2001. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 183:6778-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingavale, S. S., W. Van Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 44.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenal, U., and R. Hengge-Aronis. 2003. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6:163-172. [DOI] [PubMed] [Google Scholar]
- 46.Kallipolitis, B. H., and H. Ingmer. 2001. Listeria monocytogenes response regulators important for stress tolerance and pathogenesis. FEMS Microbiol. Lett. 204:111-115. [DOI] [PubMed] [Google Scholar]
- 47.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kock, H., U. Gerth, and M. Hecker. 2004. MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol. Microbiol. 51:1087-1102. [DOI] [PubMed] [Google Scholar]
- 49.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 50.Krüger, E., D. Zuhlke, E. Witt, H. Ludwig, and M. Hecker. 2001. Clp-mediated proteolysis in gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20:852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene. 237:223-234. [DOI] [PubMed] [Google Scholar]
- 52.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang, X., L. Zheng, C. Landwehr, D. Lunsford, D. Holmes, and Y. Ji. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187:5486-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maghnouj, A., A. A. Abu-Bakr, S. Baumberg, V. Stalon, and C. Vander Wauven. 2000. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 191:227-234. [DOI] [PubMed] [Google Scholar]
- 56.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 175:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 59.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 61.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 62.Palma, M., and A. L. Cheung. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Porankiewicz, J., and A. K. Clarke. 1997. Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 179:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reeve, C. A., A. T. Bockman, and A. Matin. 1984. Role of protein degradation in the survival of carbon-starved Escherichia coli and Salmonella typhimurium. J. Bacteriol. 157:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 69.Savijoki, K., H. Ingmer, D. Frees, F. K. Vogensen, A. Palva, and P. Varmanen. 2003. Heat and DNA damage induction of the LexA-like regulator HdiR from Lactococcus lactis is mediated by RecA and ClpP. Mol. Microbiol. 50:609-621. [DOI] [PubMed] [Google Scholar]
- 70.Scherl, A., P. François, M. Bento, J. M. Deshusses, Y. Charbonnier, V. Converset, A. Huyghe, N. Walter, C. Hoogland, R. D. Appel, J.-C. Sanchez, C. G. Zimmermann-Ivol, G. L. Corthals, D. F. Hochstrasser, and J. Schrenzel. 2005. Correlation of proteomic and transcriptomic profiles of Staphylococcus aureus during the post-exponential phase of growth. J. Microbiol. Methods 60:247-257. [DOI] [PubMed] [Google Scholar]
- 71.Sinha, B., P. P. Francois, O. Nusse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell Microbiol. 1:101-117. [DOI] [PubMed] [Google Scholar]
- 72.Streker, K., C. Freiberg, H. Labischinski, J. Hacker, and K. Ohlsen. 2005. Staphylococcus aureus NfrA (SA0367) is a flavin mononucleotide-dependent NADPH oxidase involved in oxidative stress response. J. Bacteriol. 187:2249-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomoyasu, T., A. Takaya, E. Isogai, and T. Yamamoto. 2003. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 48:443-452. [DOI] [PubMed] [Google Scholar]
- 74.Viala, J., and P. Mazodier. 2002. ClpP-dependent degradation of PopR allows tightly regulated expression of the clpP3 clpP4 operon in Streptomyces lividans. Mol. Microbiol. 44:633-643. [DOI] [PubMed] [Google Scholar]
- 75.Webb, C., M. Moreno, M. Wilmes-Riesenberg, R. Curtiss III, and J. W. Foster. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34:112-123. [DOI] [PubMed] [Google Scholar]
- 76.Winterling, K. W., D. Chafin, J. J. Hayes, J. Sun, A. S. Levine, R. E. Yasbin, and R. Woodgate. 1998. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 180:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zuber, P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zuniga, M., M. C. Miralles, and G. Perez-Martinez. 2002. The product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl. Environ. Microbiol. 68:6051-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.