Abstract
Streptococcus agalactiae (group B streptococcus [GBS]) causes neonatal sepsis, pneumonia, and meningitis, as well as infections of the bovine udder. The S. agalactiae hemolysin is regarded as an important virulence factor, and hemolysin expression is dependent on the cyl gene cluster. cylA and cylB encode the ATP binding and transmembrane domains of a typical ATP binding cassette (ABC) transporter. The deduced proteins contain the signature sequence of a multidrug resistance (MDR) transporter, and mutation of the genes results in a nonhemolytic and nonpigmented phenotype. To further elucidate the function of the putative transporter, nonpolar deletion mutants of cylA were constructed. These mutants are nonhemolytic and can be complemented by the transporter genes. Wild-type strain and nonhemolytic cylA and cylK deletion mutants were exposed to known substrates of MDR transporters. Mutation of cylA significantly impaired growth in the presence of daunorubicin, doxorubicin, and rhodamine 6G and resulted in a decreased export of doxorubicin from the cells. The mutation of cylK, a gene of unknown function located downstream from cylA, caused a loss of hemolysis but had no effect on the transport of MDR substrates. Furthermore, the hemolytic activity of the wild-type strain was inhibited by reserpine in a dose-dependent manner. We conclude that CylAB closely resembles an ABC-type MDR transporter and propose that the GBS hemolysin molecule represents a natural substrate of the transporter.
Multidrug resistance (MDR) transporters were first identified in eukaryotic organisms before bacterial homologues were found and characterized (22). In humans they are responsible for the failure of certain chemotherapeutics in cancer treatment. MDR transporters have the ability to extrude a wide variety of amphiphilic substrates with no apparent structural similarity. While the majority of eukaryotic MDR transporters are ATP binding cassette (ABC) transporters, most bacterial MDR transporters are proton-dependent multidrug efflux systems.
Bacterial ABC-type multidrug transporters displaying the typical structure of prokaryotic ABC proteins consist of two genes, encoding a nucleotide binding domain and a transmembrane domain. ABC transporters are one of the largest of all protein families, and almost 5% of the Escherichia coli genome codes for this type of transporter (12). In many bacterial genomes putative MDR transporters were found, as for example 34 in the Enterococcus faecalis genome (5). Despite this abundance, only a few bacterial ABC-type MDR transporters have been described. LmrA of Lactococcus lactis (3), HorA of Lactobacillus brevis (25), and MsbA of E. coli (13) belong to this group. Natural substrates of these transporters have not been identified in all cases. MsbA encodes the lipid A translocator of E. coli, and due to overlapping substrate specificities with MsbA, a role for LmrA in lipid trafficking has been suggested (23), while the natural substrate of HorA remains unknown.
Streptococcus agalactiae is a leading cause of sepsis, meningitis, and pneumonia in newborns (15, 32) and is an emerging pathogen in immunocompromised adult patients (9). The β-hemolysin represents an important virulence factor of S. agalactiae. Its ability to damage erythrocytes, lung epithelial cells (7), and brain microvascular endothelial cells (8) is regarded as an initial step in invasive disease.
In earlier studies we identified the cyl operon in S. agalactiae, which is essential for the production of the S. agalactiae hemolysin (28) and pigment (26). It contains 12 genes, and the deduced proteins of numerous genes are similar to fatty acid biosynthesis enzymes (cylD, cylG, acpC, cylZ, and cylI) (Fig. 1A). CylF shows similarities to an aminomethyltransferase, and CylJ carries the conserved domain of a glycosyltransferase. Two of the genes (cylX and cylK) display no significant similarities to genes of known function in the GenBank database. cylE coding for a 78-kDa deduced protein, has been proposed to encode the hemolysin molecule (21). The genes cylA and cylB display homology to typical ABC transporters, with CylA as the ATP-binding domain and CylB as the transmembrane protein. More precisely, this complex shows significant homology to DrrA, the daunorubicin transporter of Streptomyces peuceticus (11).
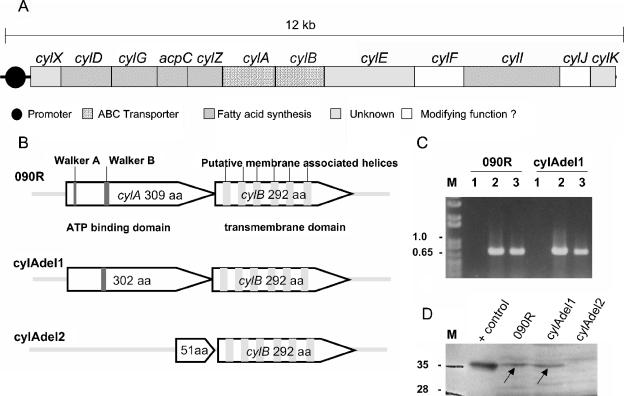
FIG. 1.
(A) Genomic map of the cyl gene cluster. Depicted are the 12 genes belonging to the cluster. Homologies of the deduced proteins to genes with known function in the GenBank database and the putative promoter region are indicated. Genes that are indicated as open boxes display homologies to genes encoding enzymes such as an aminomethyltransferase (cylF annotated in genome sequencing project AAJS01000020 as belonging to protein family PF01571) and a glycosyl transferase (cylJ harbors the conserved motif COG1819 of glycosyl transferases), which may modify the structure of an assembled molecule. (B) Graphic representation of the deletion mutants of cylA. Indicated are the Walker A and Walker B sites of cylA and the six putative transmembrane helices of cylB in the wild-type strain. The truncated versions of CylA in both mutants are depicted. (C) Transcription analysis of cylE by RT-PCR. Strains 090R and cylAdel1 were grown to mid-logarithmic phase, and 50 ng of total RNA was used as a template for reverse transcription. The RT primer annealed at nucleotides 765 to 748 of cylE, and the subsequent PCR amplified nucleotides 39 to 765 of cylE (lane2). Chromosomal DNA served as a positive control (lane 3), and 50 ng of RNA that had not been subjected to an RT reaction (RT inactivated) was used to control for DNA contamination of RNA samples (lane 1). M, molecular mass marker (kb). (D) Western blot analysis of bacterial membrane fractions with anti-CylA antibodies. S. agalactiae wild-type strain 090R and the mutant strains cylAdel1 and cylAdel2 were grown to late logarithmic phase in THB supplemented with 1% proteose peptone. Subcellular fractions of all strains were prepared as detailed in Materials and Methods. Western blot analysis was carried out with polyclonal anti-cylA (rabbit) antibodies at a dilution of 1:1,000. M, molecular mass marker; lane 1, 2 μg of recombinant CylA (+control). For lanes 2 to 4, 40 μg of membrane fractions was applied to each lane, as follows: lane 2, 090R, lane 3, cylAdel1; lane 4, cylAdel2.
The aim of this study was to characterize the function of the genes cylA and cylB and to identify substrates of the putative transporter they encode.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are listed in Table 1. S. agalactiae strain 090R was used for growth curves, construction of mutants, and Western blotting experiments. Strain AC475 was used for reserpine inhibition experiments. E. coli strain DH5α served as a host for construction of recombinant pGhost5 and pAT28 plasmids. S. agalactiae strains were cultured on tryptic soy agar supplemented with 5% sheep blood, in Todd-Hewitt broth (THB) (Oxoid, Basingstoke, England), or in THB supplemented with 5% yeast extract (THY). To detect pigment production in S. agalactiae, strains were cultured on Granada medium (6). Streptococcal mutant strains with chromosomally integrated pGhost5 plasmids were cultured in medium with 1 mg/liter erythromycin, and strains harboring recombinant pAT28 plasmids were cultured on THY plates supplemented with 120 mg/liter spectinomycin. To analyze the influence of MDR substrates on the growth of the strains cylAdel1, cylKdel, and 090R, overnight cultures in THY broth were diluted 1:100 in fresh THY medium supplemented with various amounts of the indicated substrates. Doxorubicin, daunorubicin, rhodamine 6G, and benzalkonium chloride were obtained from Sigma (Deisenhofen, Germany). Growth was determined by measurement of the optical density at 600 nm (OD600).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | |
| ΒL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3); T7 polymerase gene under control of the lacUV5 promoter | Boehringer, Novagen |
| S. agalactiae strains | ||
| O90R (ATCC 12386) | R. Lancefield grouping strain | ATCC collection |
| AC475 | Clinical isolate serotype III | Aachen collection |
| cylAdel1 | O90R derivative; ΔWalker A motif of cylA | This study |
| cylAdel2 | O90R derivative; ΔcylA | This study |
| CylKdel | O90R derivative; ΔcylK | This study |
| cylAdel1 comp1 | O90R derivative; ΔWalker A motif of cylA carrying pBSU224 | This study |
| cylAdel2 comp1 | O90R derivative; ΔcylA carrying pBSU224 | This study |
| cylAdel1 comp2 | O90R derivative; ΔWalker A motif of cylA carrying pBSU362 | This study |
| cylAdel2 comp2 | O90R derivative; ΔcylA carrying pBSU301 | This study |
| Plasmids | ||
| pGhost5 | Eryr; ori pBR; ori Ts | Appligene |
| pAT28 | Specr; oripUC; ori pAmß1 | 30 |
| pBSU224 | pAT28 derivative carrying the complete coding region of cylA and cylB | This study |
| pBSU362 | pAT28 derivative carrying the complete coding region of cylA | This study |
| pBSU301 | pAT28 derivative carrying the complete coding region of cylE | This study |
| pBSU52 | pET21a vector carrying the coding region of cylA | This study |
General molecular biology techniques.
Standard recombinant DNA techniques were employed for nucleic acid preparation and analysis. PCR was carried out with Taq polymerase (Roche Diagnostics, Mannheim, Germany) at amplification steps of 94°C (1 min), 50 to 58°C (1 min), and 72°C (1 to 3 min, depending on product size) for 35 cycles. Genomic streptococcal DNA was isolated as previously described (20). Plasmid DNA was isolated and purified with a Qiaprep Spin Miniprep kit (QIAGEN, Hilden, Germany). PCR products were sequenced on an ABI 310 automated DNA sequencer using the ABI PRISM Big-Dye terminator cycle sequencing kit (Applied Biosystems, Weiterstadt, Germany). Transformation of S. agalactiae was carried out according to an established protocol (24). Southern blot hybridization analysis was performed as previously described (10).
Construction of mutants.
The plasmid pGhost5 (2) was used for construction of deletion mutants in strain 090R. For construction of cylAdel1, genomic regions adjacent to the deletion site were amplified with the following primers (restriction sites are underlined): 5′-GGCGGCGGATCCCTTGATGAATTAACAGCAGAG-3′ and 5′-GGCGGC CTCGAGAAGAAGGCCATAAAATTTTCC-3′ (region upstream of the deletion site); 5′-GGCGGCCTCGAGACAACTTTATTTAACCCTTTAATTC-3′ and 5′-GGCGGCGAATTCATTAGTACAACTGTCATCTGCG-3′ (region downstream of the deletion site). Amplified PCR products were subcloned into pGhost5 in E. coli. The resulting recombinant plasmid was transformed into strain 090R at 30°C. Integration into the chromosome was carried out via insertion duplication mutagenesis by a temperature shift to ≥37°C. To construct deletion mutants by mobilization of the integrated plasmid, the insertion mutant was passaged five times overnight at 30°C in THY without antibiotic pressure and subsequently plated on blood agar plates. Susceptibility to erythromycin was tested by subculturing single colonies on blood agar plates with or without erythromycin (1 mg/liter).
The strains cylAdel2 and cylKdel were created as described above. The fragment upstream of the deletion site in cylAdel2 was amplified with the primers 5′-GGAATTCCTAATGGTGGATAGAGTGC-3′ and 5′-CCCATCCACTAAACTTAAACAGAGATTAATGTCTCTCAATGC-3′; the downstream fragment of the deletion site was amplified with the primers 5′-TGTTTAAGTTTAGTGGATGGGATTCGAACTTAGAGGCGG-3′ and 5′-CCGCGGATCCCTGAATACTCTAGCAAGG-3′. In strain cylKdel, primers 5′-TGGCACAAGCTTTTAAACGAGATTGGGTGG-3′ and 5′-CCCATCCACTAAACTTAAACAATCTCATCAACCGATCGC-3′ were used for the fragment upstream of the deletion, and primers 5′-TGTTTAAGTTTAGTGGATGGGCAGTTAGTAGTGATATGG-3′ and 5′-CCGCGGATCCCTCACATTACTTAACCG G-3′ were used for the downstream region. For each strain, correct deletion of the targeted chromosomal region was confirmed by DNA sequencing of the deletion site and Southern blot analysis.
The genes cylA and cylE were amplified with the primers 5′-GGCGGCCTCGAGGAGGTTGCCTCAGGAAGGATG-3′ and 5′-GGCGGCGGATCCACGCTAACCGATCTCGCGTG-3′ and the primers 5′-GGAATTCTATTCATACTAGTTCAGC-3′ and 5′-CCGCGG ATCCTGCAGCATTTTATACTCG-3′, respectively. For complementation studies with both transporter genes, cylA and cylB were amplified with the primers 5′-GGCGGCGAGCTCGAGGTTGCCTCAGGAAGGATG-3′ and 5′-GGCGGCGGATCCTAATGTTGTAAGGAAGCTTCAG-3′ and subcloned in pAT28 (30). The recombinant plasmid was constructed in E. coli DH5α and subsequently transformed into strains cylAdel1 and cylAdel2.
For expression of cylA as a C-terminal histidine-tagged protein, cylA was amplified with the primers 5′-GGCGGCCATATGGAAATTAAACTCAAAAATATTGG-3′ and 5′-GGCGGCCTCGAGAACATTGTTTTTCCTTTCTTTTTCTC-3′ and subcloned into the vector pET21a in E. coli strain DH5α. For protein expression, the plasmid was transferred into E. coli strain BL21(DE3).
Detection of MDR substrate export.
Transport of the polyketide doxorubicin from S. agalactiae cells was measured by a method adapted from Nishino and Yamaguchi (19). To load the cells with the polyketides, S. agalactiae wild-type strain 090R and the cylAdel1 mutant strain were grown to equal optical densities at late exponential phase in medium supplemented with 0.5 mg/liter of doxorubicin, harvested by centrifugation, and washed in phosphate-buffered saline (PBS). Cells of a 30-ml culture were resuspended in 2 ml of PBS containing 100 mM glucose as an energy source. To measure release of the polyketides into the supernatant, aliquots of the suspension were removed at different time points (0, 2, 5, and 10 min) and subjected to centrifugation, and fluorescence of the supernatant was quantified in a fluorescence counter (Cytofluor II; Perseptive Biosystems, Inc.). Doxorubicin fluorescence was detected with an excitation at 485 and an emission at 590 nm.
Expression of recombinant CylA.
For the expression of histidine-tagged CylA protein, E. coli BL21(DE3) harboring the respective pET21a plasmid was grown to an OD600 of 0.6 at 30°C in Luria-Bertani broth. Protein expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 30°C. Bacterial cells were harvested by centrifugation, resuspended in lysis buffer B (8 M urca, 0.1 M NaH2PO4, 0.01 M Tris Cl [pH 8.0]), placed on ice for 30 min, and disrupted by sonication. Recombinant CylA was purified from the cell lysate by passage over a commercial nickel affinity matrix (Ni-NTA Spin kit; QIAGEN) and eluted under denaturing conditions.
Western immunoblot analysis.
Polyclonal rabbit antibodies directed against CylA were generated by Eurogentec (Brussels, Belgium). Bacterial cells were grown to late logarithmic phase in THB supplemented with 1% proteose peptone 3 (Beckton Dickinson, Heidelberg, Germany) and 5% fetal calf serum. Subcellular fractions were prepared from strains 090R, cylAdel1, and cylAdel2 as previously described (14). Samples of cell wall fractions, cytoplasmatic membrane fractions, and cytoplasma containing 40 μg of protein each were separated by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12% gel and transferred to an Immobilon P polyvinylidene difluoride membrane (Millipore, Eschborn, Germany). The blots were probed with the polyclonal anti-CylA antibody at a dilution of 1:1,000. Controls were probed with preimmune rabbit serum at a dilution of 1:1,000. An alkaline phosphatase-labeled anti-rabbit immunoglobulin G antibody was used as secondary antibody at a dilution of 1:5,000. Bound secondary antibody was visualized by the chemiluminescent CSPD (Roche, Mannheim, Germany).
RNA preparation and analysis.
Total RNA was prepared from strains 090R and cylAdel1 grown to mid-logarithmic phase in THY broth and treated with RNAprotect bacteria reagent (QIAGEN) according to the manufacturer's instructions. Bacterial cells were lysed in a cell disrupter (Fastprep; QBiogene, Heidelberg, Germany) in the presence of RNeasy lysis buffer (QIAGEN). Subsequent purification of RNA was carried out with an RNeasy minikit (QIAGEN). For reverse transcription-PCR (RT-PCR), experiments 50 ng of DNase-treated RNA was used as a template in an RT reaction. RT-PCR experiments were carried out with the QIAGEN One Step RT-PCR kit (QIAGEN). The primer 5′-CAGTTTTAACCCGAACCC-3′ was used as the RT primer, and the primer combination 5′-CTTAGAAGATTATTCTGAAGTGG-3′ and 5′-CAGTTTTAACCCGAACCC-3′ was used for the subsequent PCR.
Nucleotide sequence of cylE.
The nucleotide sequence of cylE was amplified from genomic DNA of strain cylAdel1 with the primers 5′-GTCTTAATGAAGGGTTCTATTGC-3′ and 5′-GTAGCCACAAGTATC TGTATCTGC-3′. In addition to the amplification primers, the primers 5′-CTTAGAAGATTATTCTGA AGTGG-3′, 5′-AATGGCTTCAAAGAGTATG-3′, 5′-CCTAGCCTAACTTTAAGTCG-3′, and 5′-ATCATGACATTTACAAGTGACG-3′ were used to sequence the purified PCR product.
Hemolysin assay.
Bacterial cells from a fresh overnight culture were inoculated into 15 ml of fresh culture medium and grown at 37°C for 5 h. Cells were harvested by centrifugation and washed in PBS. The OD of each sample was measured at 600 nm and adjusted to equal values in order to compensate for minimal differences in growth kinetics between the samples. Hemolytic activity was extracted from bacterial cells by a modification of the method described by Marchlewicz and Duncan (17). After centrifugation, the bacterial pellet was resuspended in 600 μl of hemolysin extraction buffer (PBS, 1% starch, 1% glucose, 3% Tween 80) and incubated for 5 min at 37°C. After centrifugation, the supernatant was transferred to a new tube and stored on ice. For the determination of hemolytic activity, 15 to 100 μl of the extract was added to a 1% erythrocyte suspension in 1 ml of PBS and incubated for 10 min at 37°C. Hemolysis was detected by release of hemoglobin from erythrocytes. Hemoglobin was measured by the OD540 after centrifugation of the sample.
RESULTS
Transporter gene deletion mutants.
In our previous work we described several mutants of cylA and cylB that result in a nonhemolytic phenotype (27, 28). These mutants were targeted pGhost5 insertion mutants as well as naturally occurring nonhemolytic strains harboring insertion elements in cylA or cylB. To exclude the possibility of downstream polar effects on hemolysin expression, two different deletion mutants of cylA were constructed by insertion of the vector pGhost5 into cylA and subsequent mobilization of the integrated plasmid. One mutation comprises the Walker A motif generating a strain (cylAdel1) with a deletion of amino acids 35 to 42 of CylA. In the second strain (cylAdel2), amino acids 24 to 289 of the 309 residues of CylA are deleted (Fig. 1B). Both strains have a nonhemolytic and nonpigmented phenotype (Fig. 2).
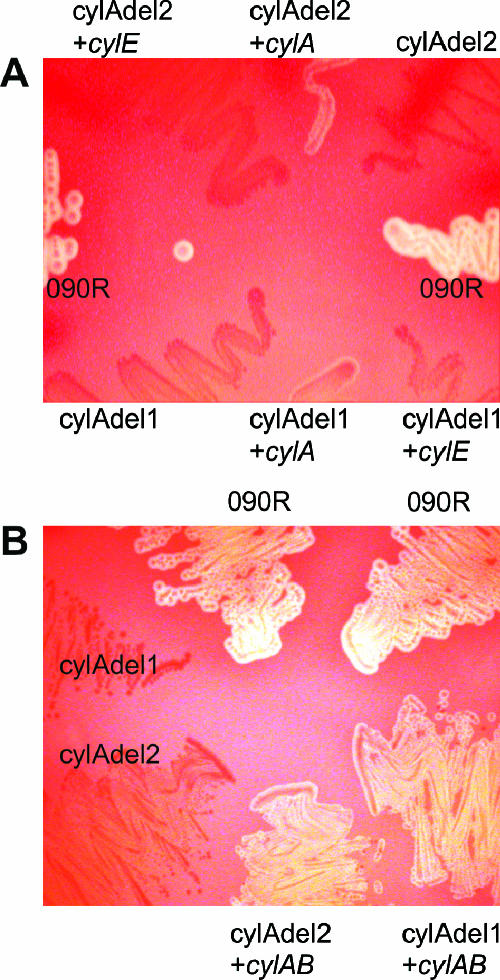
FIG. 2.
Phenotype of cylA deletion mutants and complemented strains on sheep blood agar plates. (A) Complementation studies with a plasmid harboring cylA or cylE in trans as indicated. (B) Strains complemented with a plasmid harboring cylAB in trans. Strain designations are indicated: 090R, wild-type strain; cylAdel1, cylA mutant with deleted Walker A motif; cylAdel2, cylA deletion as depicted in Fig. 1B.
cylE transcription and nucleotide sequence in the transporter deletion mutants.
cylE, the gene immediately downstream of cylB, has been proposed to encode the structural hemolysin of S. agalactiae (21). To ensure that a deletion of cylA does not impair the transcription of cylE, we performed RT-PCR experiments with strain cylAdel1. The transcription of cylE was unaltered compared to the wild-type strain (Fig. 1C). In addition, nonspecific mutations of cylE in the cylA mutant strain were excluded by sequencing of cylE.
Complementation studies.
To investigate whether the cylA deletion mutants can be complemented in trans, the coding region for CylA was amplified and subcloned into the plasmid pAT28. Transfer of this construct into the strains cylAdel1 and cylAdel2 resulted in a weak reconstitution of hemolysis for both strains (Fig. 2A). Further complementation studies were conducted with a recombinant pAT28 vector harboring the genes cylA as well as cylB in trans, resulting in a clearly visible hemolysis of both mutant strains (Fig. 2B). As a control, the strains were transformed with a plasmid harboring cylE, which failed to have any effect on the nonhemolytic phenotype (Fig. 2A). These results confirm the requirement of CylAB for β-hemolysis of S. agalactiae.
Hemolytic activity following cell lysis.
In E. coli intracellular hemolytic activity in nonhemolytic transporter mutants can be released by sonication (16). To investigate if the cylA mutant strains harbor intracellular hemolytic activity, strain cylAdel1 was grown to late logarithmic phase. Bacterial cells were disrupted with glass beads and subsequently subjected to hemolysin extraction. Hemolysin extracts of cellular fragments as well as hemolysin extracts from nondisrupted cylAdel1 cells were, however, nonhemolytic (data not shown).
Susceptibility to classical MDR substrates.
Analysis of the deduced protein sequence of cylA with the conserved domain search at the NCBI website reveals that it contains the sequence COG1131. COG1131 is a motif identified in the clusters of orthologous groups (COG) database (29). It is specific for the ATPase component of ABC-type multidrug transport systems. To investigate if classical MDR substrates can be transported via CylAB, we measured the growth of the wild-type strain O90R and the cylAdel1 mutant in the presence of daunorubicin, doxorubicin, rhodamine 6G, and benzalkonium chloride. Significant effects on the growth kinetics were observed for cultures containing daunorubicin, doxorubicin, and rhodamine 6G. In comparison to the 090R wild-type strain, growth of the mutant strain cylAdel1 was significantly impaired in the presence of 1 mg/liter doxorubicin (Fig. 3), 0.5 mg/liter and 1 mg/liter daunorubicin (Fig. 4), and 10 mg/liter rhodamine 6G (data not shown). The increased susceptibility of the strain was limited to specific MDR substrates. For benzalkonium chloride, no increased sensitivity of cylAdel1 could be observed at concentrations of 1, 3, and 5 mg/liter (data not shown). Growth of the wild-type and mutant strains was unaffected at 1 mg/liter and completely inhibited at 5 mg/liter benzalkonium chloride.
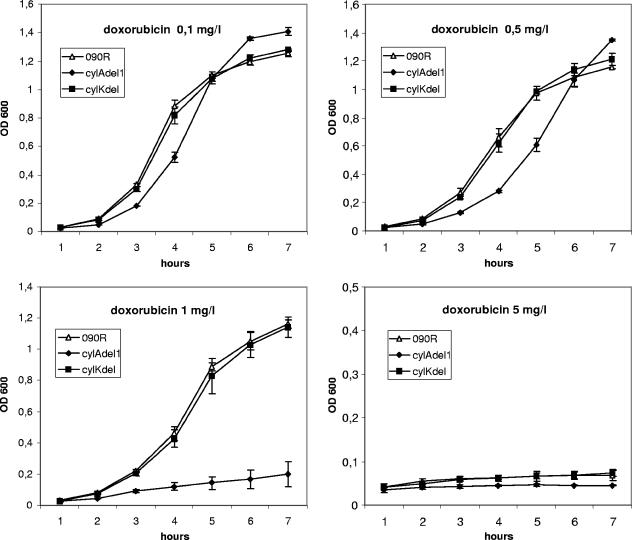
FIG. 3.
Growth curves of the 090R wild-type strain as well as the nonhemolytic strains cylAdel1 and cylKdel in the presence of the MDR substrate doxorubicin. Bacteria were inoculated into THB from a fresh overnight culture, and growth was determined by the measurement of the OD600 for 7 h. Depicted are mean values and standard deviations of three independent experiments.
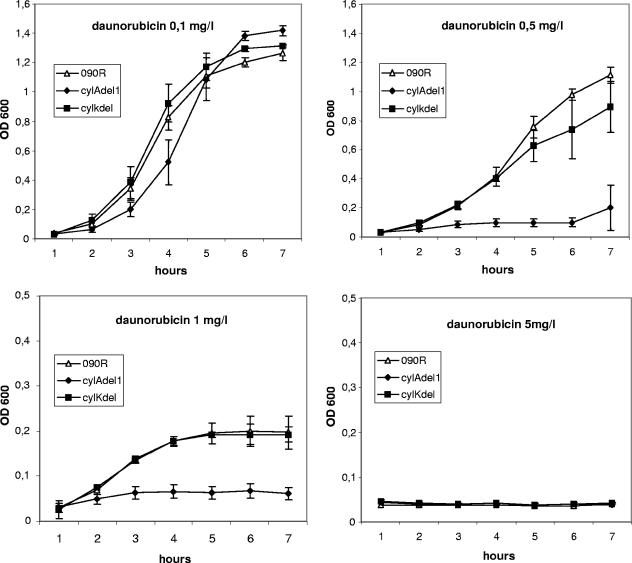
FIG. 4.
Growth curves of the 090R wild-type strain as well as the nonhemolytic strains cylAdel1 and cylKdel in the presence of the MDR substrate daunorubicin. Bacteria were inoculated into THB from a fresh overnight culture, and growth was determined by the measurement of the OD600 for 7 h. Depicted are mean values and standard deviations of three independent experiments.
To exclude the possibility that the effects of MDR substrates on strain cylAdel1 are caused by the nonhemolytic phenotype of the strain, a cylK deletion strain (cylKdel1) served as a control. The strain is nonhemolytic due to a deletion of amino acids 22 to 115 of CylK. The gene is located more than 6.3 kb downstream of the stop codon of cylB, and the deduced protein consists of 178 amino acids with no significant homology to proteins of known function in the GenBank database. Growth of cylKdel1 was indistinguishable from the parent strain O90R (Fig. 3 and 4) for all substrates and concentrations tested. Our data indicate that CylAB can function as a transporter for the MDR substrates daunorubicin, doxorubicin, and rhodamine 6G.
Export of doxorubicin.
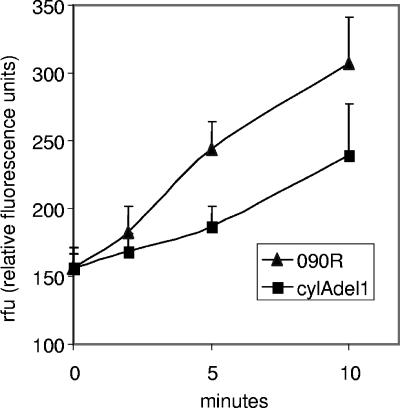
To further investigate the hypothesis that mutation of the transporter CylAB results in an inability to export toxic MDR substrates, strains 090R and cylAdel1 were loaded with subinhibitory concentrations of doxorubicin. Bacterial cells were resuspended in buffer containing glucose as an energy source, and the kinetics of doxorubicin export from bacterial cells into the supernatant was measured fluorometrically. Compared to the wild-type strain, the mutant strain displayed a markedly reduced capacity to export doxorubicin (Fig. 5).
FIG. 5.
Export studies of doxorubicin from S. agalactiae wild-type strain 090R and cylA mutant strain cylAdel1. Bacterial cells were grown in the presence of doxorubicin (0.5 mg/liter). Export of doxorubicin into the supernatant was determined fluorometrically as detailed in Materials and Methods. Mean values and the standard deviation of eight independent experiments are shown.
Hemolysis in the presence of reserpine.
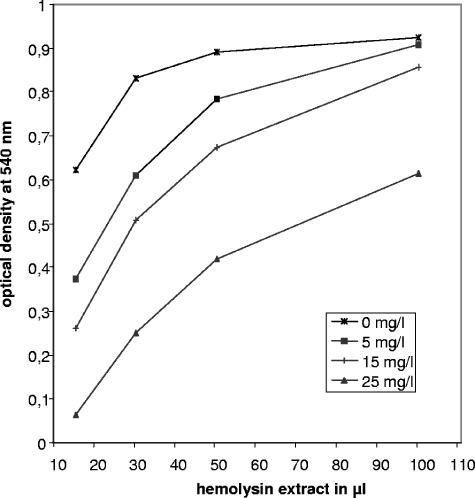
Based on the detection of the ATP-type MDR transporter motif COG1131 in the amino acid sequence of CylA, the increased sensitivity of the cylAdel1 mutant to MDR substrates, and the reduced transport of doxorubicin in cylAdel1, we hypothesized that cylA and cylB code for a transporter resembling ABC-type MDR transporters. To further substantiate our hypothesis, we analyzed the hemolytic activity of hemolysin extracts after growth of the wild-type strain AC475 in the presence of 0 to 25 mg/liter reserpine. Reserpine is a well-known inhibitor of MDR transporters and growth in the presence of reserpine resulted in a dose-dependent decrease of extractable hemolytic activity (Fig. 6).
FIG. 6.
Effect of reserpine on the hemolytic activity of the wild-type strain AC475. Bacteria were grown in the presence of 0 to 25 mg/liter reserpine to late logarithmic phase and subjected to hemolysin extraction as detailed in Materials and Methods. Hemolysin extracts were incubated with a 1% erythrocyte suspension in PBS for 15 min at 37°C. To detect hemolysis, hemoglobin release was measured as the OD540 of the supernatant. Depicted are mean values of five independent experiments.
Subcellular localization of the transporter.
Antibodies against recombinant CylA were used to analyze subcellular fractions of S. agalactiae strain O90R and the deletion mutants cylAdel1 and cylAdel2 by Western blotting. A positive reaction with the antibody could be observed for the membrane fraction of the wild-type strain and the cylAdel1 deletion mutant (Fig. 1D), which carries a cylA gene lacking the Walker A motif. No CylA protein was detectable in the membrane fraction of cylAdel2 (Fig. 1D). Analysis of the cytoplasma fraction, the cell wall fraction, and the culture supernatant did not reveal the presence of any CylA-specific bands in wild-type and mutant strains (data not shown).
DISCUSSION
Based on amino acid homologies, CylAB encodes the ATP binding and the transmembrane component of an ABC transporter. The genes are located in the cyl gene cluster that is required for β-hemolysis expression in S. agalactiae. Involvement of both genes in the production of β-hemolysin has been suggested by the analysis of various nonhemolytic insertion mutants of cylA and cylB (27, 28). To further investigate the role of these genes for the expression of β-hemolysis and to rule out polar effects we constructed two nonpolar deletion mutants of the putative transporter. The mutants display a nonhemolytic phenotype which can be restored by a plasmid harboring cylAB in trans. Partial reconstitution of the beta-hemolytic phenotype was observed after complementation with cylA (Fig. 2). The partial reconstitution that was observed for complementation experiments with cylA alone may be explained by translational coupling of the genes cylA and cylB. Polar effects on genes further downstream could be ruled out by a plasmid harboring cylE which failed to complement the nonhemolytic mutant strains. Furthermore, RT-PCR analysis and sequencing of cylE did not give any indication that other genes of the cyl cluster are responsible for the observed effects. However, conflicting results have been obtained regarding the phenotype of insertion mutants of cyl transporter genes by other groups. Pritzlaff et al. reported that cylB insertion mutants display a regular hemolytic phenotype. This observation seems to contradict our finding that naturally occurring cylB insertion mutants (27) as well as a targeted disruption of the cylB gene (28) result in a loss of hemolytic activity. The hemolytic phenotype of the transporter mutant Pritzlaff described might be explained by the fact that it harbors an insertion at codon 265 of the 292-amino-acid-long CylB protein (21). It is possible that the expression of a slightly truncated CylB protein does not interfere with the function of the transporter.
The presence of a conserved protein motif for an ABC-type MDR transporter (COG1131) in the deduced protein sequence of cylA and the homology of CylB to the daunorubicin transporter of Streptomyces peuceticus prompted us to investigate the effect of a CylAB transporter mutation on the extrusion of classical MDR substrates. MDR proteins transport multiple, structurally dissimilar compounds that are characterized by common physicochemical characteristics such as high hydrophobicity, an amphiphilic nature, and a positive or neutral charge. Deletion of the Walker A motif in cylA resulted in an increased sensitivity of the strain to the MDR substrates daunorubicin, doxorubicin, and rhodamine 6G (Fig. 3 and 4). The increased sensitivity to MDR substrates is probably due to a failure of the mutant strain to export the toxic compounds to the same extent as the wild-type strain. Further experiments showed a decreased export of doxorubicin from the bacterial cells and support this interpretation of our results (Fig. 5). Genome sequencing projects of various streptococcal species demonstrated multiple putative MDR transporters in each streptococcal genome, a fact that may explain why the mutation of one transporter does not lead to a complete loss of MDR substrate transport. To test our hypothesis that CylAB closely resembles MDR transporters, we investigated if the hemolytic activity of an S. agalactiae wild-type strain can be inhibited by reserpine and found a dose-dependent decrease of extractable hemolytic activity (Fig. 6). Reserpine is a well-known inhibitor of bacterial MDR transporters. It inhibits the MDR transporter Bmr of Bacillus subtilis (1) and NorA of Staphylococcus aureus (18) at concentrations we tested in our hemolysin assay. Based on these data, we conclude that the hemolysin transporter of S. agalactiae is capable of transporting MDR substrates.
Although MDR transporters extrude a wide variety of structurally dissimilar compounds and CylE of S. agalactiae has been proposed to encode the structural hemolysin (21), the transport of a 78-kDa protein by this type of transporter seems unlikely. However, the hypothesis that CylE represents the hemolysin was only one of the explanations offered in the paper by Pritzlaff et al. The failure to induce antibodies against the S. agalactiae hemolysin in repeated attempts (4) would also argue against a large protein. Moreover, the presence of CylE has not been demonstrated in either hemolytic S. agalactiae extracts or the membrane of erythrocytes, nor does the deduced protein itself display any similarities to other characterized hemolysin molecules. Taken together, the data that were generated by the different groups could better be explained by a model in which cylE participates in the synthesis of the hemolysin rather than representing the hemolysin itself. Based on the fact that the transporter displays characteristic features of MDR transporters, it is tempting to speculate that the natural substrate of CylAB possesses certain structural similarities with classical MDR substrates. While the genes present in the cyl gene cluster do not represent a classical polyketide synthesis cluster, many genes resemble fatty acid synthesis genes that are also essential for polyketide synthesis. The data of the different S. agalactiae genome projects show that S. agalactiae has regular fatty acid synthesis genes apart from the cyl genes. This might explain why a mutation of cyl genes is not lethal for S. agalactiae and supports the theory that these genes participate in another biosynthetic pathway.
The S. agalactiae hemolysin is associated with the bacterial cell surface, from where it can be extracted by mild detergents. It is not released into the culture supernatant under regular growth conditions (17). To further investigate the transporter function of CylAB, we tried to extract hemolytic activity from the transporter mutants cylAdel1 and cylAdel2 after cellular disruption. In E. coli, sonication of nonhemolytic transporter mutants leading to a disruption of bacterial cells results in the recovery of intracellular hemolytic activity (16). Cell disruption of our transporter mutants, however, did not result in the detection of any extractable hemolytic activity. A possible explanation is provided by the finding that MDR transporters mediate the extrusion of lipophilic compounds from the inner leaflet of the plasma membrane to the outside of the cell rather than just representing an outlet for cytosolic compounds to the extracellular environment. For LmrA of L. lactis, it has been shown that hydrolysis of ATP results in the transfer of the MDR substrate from a high-affinity drug binding site at the inner membrane surface to a low-affinity drug binding site on the outer membrane surface (31). We think that a simple destruction of the cellular integrity of S. agalactiae cannot substitute for this complex extrusion mechanism. Another possible explanation could be that modifications of the hemolysin molecule are made during or after export to the bacterial cell surface and that no functional hemolysin molecule is present in the cytosolic compartment.
CylA deletion mutants of S. agalactiae are nonhemolytic and can be complemented by the transporter genes. Mutations of the transporter lead to an increased sensitivity to typical MDR substrates and a decreased export of doxorubicin. Furthermore, hemolytic activity of S. agalactiae is decreased in the presence of a classical MDR-transport inhibitor. Based on these data, we conclude that CylAB closely resembles ABC-type MDR transporters and propose that the S. agalactiae hemolysin molecule represents a natural substrate of the transporter.
Acknowledgments
The work of B.S. and M.K. was supported by DFG grant Sp511/4-1.
We thank Simone Martin and Claudia Reichardt for expert technical assistance.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, A. A. Neyfakh, and S. Schuldiner. 1993. Mutants of the Bacillus subtilis multidrug transporter Bmr with altered sensitivity to the antihypertensive alkaloid reserpine. J. Biol. Chem. 268:11086-11089. [PubMed] [Google Scholar]
- 2.Biswas, I., A. Gruss, D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolhuis, H., D. Molenaar, G. Poelarends, H. W. van Veen, B. Poolman, A. J. Driessen, and W. N. Konings. 1994. Proton motive force-driven and ATP-dependent drug extrusion systems in multidrug-resistant Lactococcus lactis. J. Bacteriol. 176:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dal, M. C., and H. Monteil. 1983. Hemolysin produced by group B Streptococcus agalactiae. FEMS Microbiol. Lett. 16:89-94. [Google Scholar]
- 5.Davis, D. R., J. B. McAlpine, C. J. Pazoles, M. K. Talbot, E. A. Alder, C. White, B. M. Jonas, B. E. Murray, G. M. Weinstock, and B. L. Rogers. 2001. Enterococcus faecalis multi-drug resistance transporters: application for antibiotic discovery. J. Mol. Microbiol. Biotechnol. 3:179-184. [PubMed] [Google Scholar]
- 6.De la Rosa, M., R. Villareal, D. Vega, C. Miranda, and A. Martinzbrocal. 1983. Granada medium for the detection and identification of group B streptococci. J. Clin. Microbiol. 18:779-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doran, K. S., J. C. Chang, V. M. Benoit, L. Eckmann, and V. Nizet. 2002. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J. Infect. Dis. 185:196-203. [DOI] [PubMed] [Google Scholar]
- 8.Doran, K. S., G. Y. Liu, and V. Nizet. 2003. Group B streptococcal beta-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J. Clin. Investig. 112:736-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farley, M. 1995. Group B streptococcal infection in older patients. Spectrum of disease and management strategies. Drugs Aging 6:293-300. [DOI] [PubMed] [Google Scholar]
- 10.Franken, C., G. Haase, C. Brandt, J. Weber-Heynemann, S. Martin, C. Lammler, A. Podbielski, R. Lutticken, and B. Spellerberg. 2001. Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb. Mol. Microbiol. 41:925-935. [DOI] [PubMed] [Google Scholar]
- 11.Guilfoile, P. G., and C. R. Hutchinson. 1991. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc. Natl. Acad. Sci. USA 88:8553-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holland, B., S. Cole, K. Kuchler, and C. Higgins. 2003. ABC proteins from bacteria to man. Academic Press, San Diego, Calif.
- 13.Karow, M., and C. Georgopoulos. 1993. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 7:69-79. [DOI] [PubMed] [Google Scholar]
- 14.Kling, D. E., L. C. Madoff, and J. L. Michel. 1999. Subcellular fractionation of group B Streptococcus. BioTechniques 27:24-. 6:28. [DOI] [PubMed] [Google Scholar]
- 15.Luck, S., M. Torny, K. d'Agapeyeff, A. Pitt, P. Heath, A. Breathnach, and A. B. Russell. 2003. Estimated early-onset group B streptococcal neonatal disease. Lancet 361:1953-1954. [DOI] [PubMed] [Google Scholar]
- 16.Mackman, N., and I. B. Holland. 1984. Functional characterization of a cloned haemolysin determinant from E. coli of human origin, encoding information for the secretion of a 107K polypeptide. Mol. Gen. Genet. 196:129-134. [DOI] [PubMed] [Google Scholar]
- 17.Marchlewicz, B. A., and J. L. Duncan. 1980. Properties of a hemolysin produced by group B streptococci. Infect. Immun. 30:805-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neyfakh, A. A., C. M. Borsch, and G. W. Kaatz. 1993. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 37:128-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino, K., and A. Yamaguchi. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 21.Pritzlaff, C. A., J. C. Chang, S. P. Kuo, G. S. Tamura, C. E. Rubens, and V. Nizet. 2001. Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol. Microbiol. 39:236-247. [DOI] [PubMed] [Google Scholar]
- 22.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuter, G., T. Janvilisri, H. Venter, S. Shahi, L. Balakrishnan, and H. W. van Veen. 2003. The ATP binding cassette multidrug transporter LmrA and lipid transporter MsbA have overlapping substrate specificities. J. Biol. Chem. 278:35193-35198. [DOI] [PubMed] [Google Scholar]
- 24.Ricci, M. L., R. Manganelli, C. Berneri, G. Orefici, and G. Pozzi. 1994. Electrotransformation of Streptococcus agalactiae with plasmid DNA. FEMS Microbiol. Lett. 119:47-52. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto, K., A. Margolles, H. W. van Veen, and W. N. Konings. 2001. Hop resistance in the beer spoilage bacterium Lactobacillus brevis is mediated by the ATP-binding cassette multidrug transporter HorA. J. Bacteriol. 183:5371-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spellerberg, B., S. Martin, C. Brandt, and R. Lutticken. 2000. The cyl genes of Streptococcus agalactiae are involved in the production of pigment. FEMS Microbiol. Lett. 188:125-128. [DOI] [PubMed] [Google Scholar]
- 27.Spellerberg, B., S. Martin, C. Franken, R. Berner, and R. Lutticken. 2000. Identification of a novel insertion sequence element in Streptococcus agalactiae. Gene 241:51-56. [DOI] [PubMed] [Google Scholar]
- 28.Spellerberg, B., B. Pohl, G. Haase, S. Martin, J. Weber-Heynemann, and R. Lütticken. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 181:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatusov, R. L., R. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 30.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Veen, H. W., A. Margolles, M. Muller, C. F. Higgins, and W. N. Konings. 2000. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J. 19:2503-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zangwill, K., A. Schuchat, and J. D. Wenger. 1992. Group B streptococcal disease in the United States, 1990: report from a multistate active surveillance system. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 41:25-32. [PubMed] [Google Scholar]