Abstract
Ferric enterobactin utilization by Bordetella bronchiseptica and Bordetella pertussis requires the BfeA outer membrane receptor. Under iron-depleted growth conditions, transcription of bfeA is activated by the BfeR regulator by a mechanism requiring the siderophore enterobactin. In this study, enterobactin-inducible bfeA transcription was shown to be TonB independent. To determine whether other siderophores or nonsiderophore catechols could be utilized by the Bfe system, various compounds were tested for the abilities to promote the growth of iron-starved B. bronchiseptica and induce bfeA transcription. The BfeA receptor transported ferric salmochelin, corynebactin, and the synthetic siderophores TRENCAM and MECAM. Salmochelin and MECAM induced bfeA transcription in iron-starved Bordetella cells, but induction by corynebactin and TRENCAM was minimal. The neuroendocrine catecholamines epinephrine, norepinephrine, and dopamine exhibited a remarkable capacity to induce transcription of bfeA. Norepinephrine treatment of B. bronchiseptica resulted in BfeR-dependent bfeA transcription, elevated BfeA receptor production, and growth stimulation. Pyrocatechol, carbidopa, and isoproterenol were similarly strong inducers of bfeA transcription, whereas tyramine and 3,4-dihydroxymandelic acid demonstrated low inducing activity. The results indicate that the inducer structure requires a catechol group for function and that the ability to induce bfeA transcription does not necessarily correlate with the ability to stimulate bacterial growth. The expanded range of catechol siderophores transported by the BfeA receptor demonstrates the potential versatility of the Bordetella Bfe iron retrieval system. The finding that catecholamine neurotransmitters activate bfeA transcription and promote growth suggests that Bordetella cells can perceive and may benefit from neuroendocrine catecholamines on the respiratory epithelium.
The extremely limited availability of free iron in the host environment presents a major barrier to the growth of bacterial pathogens (18). One mechanism that microbes employ to retrieve iron is the production and utilization of siderophores that chelate ferric iron. In addition, some bacterial species have the transport machinery to utilize siderophores produced by other microbial species (xenosiderophores). In gram-negative bacteria, ferric siderophore complexes are internalized by outer membrane receptors in a process involving the TonB system, which transmits energy from the proton motive force to the receptors for high-affinity substrate transport to the periplasm (60).
Microbial iron transport is tightly controlled, in part because excess iron can participate in reactions leading to the production of toxic oxygen species (34). Under iron-replete growth conditions, bacterial iron uptake genes are transcriptionally repressed by regulators such as Fur that require iron as a corepressor (25, 35). Some bacterial species have evolved positive regulatory mechanisms to activate transcription of iron acquisition genes. In general, these positive regulators are repressed at the transcriptional level by Fur and iron, and their function often requires the presence of the iron source itself, acting as an inducer. Known positive regulators of iron transport genes belong to one of four mechanistic classes: classical two-component sensory transduction systems, extracytoplasmic function sigma factors, AraC/XylS family transcriptional regulators, and LysR-type regulators. For example, Pseudomonas aeruginosa uses the PfeR and PfeS two-component signal transduction proteins to elevate expression of the pfeA ferric enterobactin receptor gene when grown in the presence of enterobactin (23, 24). The Escherichia coli ferric citrate transport genes are transcriptionally activated by the FecI extracytoplasmic function sigma factor when ferric citrate becomes bound to its outer membrane receptor (73). Examples of the iron subfamily of the AraC family regulators include PchR and YbtA, which activate transcription of P. aeruginosa pyochelin (37, 38) and Yersinia pestis yersiniabactin (26) biosynthesis and transport genes, respectively, when the cognate siderophore is present. These regulators are hypothesized to require direct interaction with their iron source inducers, similar to AraC binding its arabinose effector (29). Lastly, the Vibrio cholerae IrgB protein is a LysR family transcriptional activator of the irgA enterobactin receptor gene; however, its function does not require induction by enterobactin (30, 49).
Bordetella bronchiseptica causes respiratory infections in a variety of mammalian hosts (31). Recent phylogenetic analyses suggest that a B. bronchiseptica-like organism was the progenitor of the human-adapted species Bordetella pertussis (57), the causative agent of the respiratory disease whooping cough (10). To date, there are three known Bordetella iron acquisition systems, and each is positively regulated at the transcriptional level in response to its cognate iron source. Transcription of the bhuRSTUV genes, encoding heme uptake and utilization functions, is heme inducible by a mechanism involving the extracytoplasmic function sigma factor HurI (72). These Bordetella species also produce the dihydroxamate siderophore alcaligin. The alcaligin biosynthesis and transport genes are transcriptionally activated in the presence of the siderophore by the AraC family regulator AlcR (12, 15). The third known Bordetella iron transport system utilizes the catechol xenosiderophore enterobactin (7). The AraC family regulator BfeR activates transcription of the bfeA enterobactin receptor gene in the presence of enterobactin (1). Although growth stimulation by ferric enterobactin is dependent on the BfeA receptor, transcriptional activation of the bfeA gene by BfeR and enterobactin does not require BfeA, suggesting that the inducing form of enterobactin enters the cell by another route.
The host upper respiratory tract is the natural colonization site for B. pertussis and B. bronchiseptica, yet both can use enterobactin, which is produced primarily by bacterial inhabitants of the intestinal tract such as E. coli. Enterobactin is also used as a xenosiderophore by a variety of other organisms, including Neisseria gonorrhoeae (20), Neisseria meningitidis (64), and respiratory Haemophilus species (76). Although E. coli and other enteric bacteria are not considered upper respiratory tract commensals of mammals, they can transiently colonize this anatomic site and, in the process, potentially excrete enterobactin (6, 71). Since the ferric ion stability constant of enterobactin (1049) is higher than that of virtually all other known siderophores (62), a microbe that can utilize this powerful siderophore may gain a significant growth advantage via iron piracy in the iron-restricted host environment.
It is also possible that respiratory tract commensals produce siderophores that are structurally similar to enterobactin, and these ferric siderophores may be transported by the ferric enterobactin uptake systems of other respiratory inhabitants or pathogens such as Bordetella species. In the present study, we identified other catechol siderophores transported by the B. bronchiseptica BfeA enterobactin receptor and assessed their capacity to induce bfeA transcription. In addition, several compounds, including the neuroendocrine catecholamines epinephrine, norepinephrine, and dopamine, were demonstrated to induce transcription of bfeA. These findings expand the functional classes of compounds that can activate the BfeR regulator and suggest that Bordetella cells may respond to these compounds while growing in the host.
MATERIALS AND METHODS
Bacterial strains and plasmid vectors.
B. bronchiseptica B013N has been described previously (3) and was used as the wild-type strain for this work. The isogenic tonB mutant strain BRM31 was constructed by insertional inactivation as described previously (55). The B. bronchiseptica bfeR deletion mutant strain BRM24 and the bfeA insertion mutant BRM25 are described elsewhere (1). B. bronchiseptica BRM26 is an alcaligin-deficient alcA mutant and has been described previously (13). E. coli DH5α (Invitrogen, Carlsbad, CA) was used for routine cloning purposes and as a donor in triparental matings. Bacillus subtilis strain ATCC 21332 was provided by Kenneth Raymond and was used for the production of corynebactin.
Plasmid pGEM3Z (Promega, Madison, WI) and the broad-host-range plasmid pBBR1MCS-1 (40) were used in the construction of recombinant plasmids. pMP220 carries a promoterless lacZ gene (67) and was used in the construction of bfeA-lacZ transcriptional reporter fusions. For conjugations, plasmid pRK2013 provided DNA mobilization functions to E. coli DH5α donor cells (27).
Culture conditions.
Luria-Bertani agar and broth (65) were used for routine cultivation of E. coli and B. bronchiseptica strains. B. subtilis was grown in a glucose mineral-salt medium that was depleted of iron by treatment with Chelex 100 (Bio-Rad, Richmond, CA) (70). Stainer-Scholte (SS) medium (69), modified as described previously (66), was used as a chemically defined growth medium for B. bronchiseptica. SS basal medium was treated with Chelex 100 prior to addition of nutritional supplements; iron-replete medium contained 36 μM iron, and iron-restricted medium contained no added iron (3). Media were supplemented with antibiotics at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; gentamicin, 10 μg/ml; tetracycline, 15 μg/ml. Bacterial growth in liquid culture was measured densitometrically at a wavelength of 600 nm.
Siderophores and catechol compounds.
Enterobactin was purified from culture supernatants of E. coli AN102 [araC14 leuB6(Am) secA206 (Azir) fhuA23 lacY1 proC14 tsx-67 fep-104 glnV44(As) λ− trpE38 rpsL109 (Strr) xylA5 mtl-1 thi-1] (22) by using a method derived from Neilands and Nakamura (54) as described previously (1). Corynebactin was purified from culture supernatants of B. subtilis 21332 by organic solvent extraction. Briefly, the B. subtilis biosurfactant surfactin was removed by adjusting the supernatant to pH 2 with concentrated HCl, according to the method of Arima et al. (2). The surfactin-containing precipitate was removed by centrifugation, and corynebactin was purified from the resulting supernatant by using the method for enterobactin purification (1). Corynebactin concentrations were determined using the Arnow assay (5) and 2,3-dihydroxybenzoic acid as the standard. Salmochelin S4 was provided by Klaus Hantke. N,N′,N"-[tris-(2,3-dihydroxybenzoyl)-2-aminoethyl]-amine (TRENCAM) (63) and 1,3,5-N,N′,N"-tris(2,3-dihydroxybenzoyl)triaminomethylbenzene (MECAM) (74, 75) were provided by Kenneth Raymond. Monomeric 2,3-dihydroxybenzoylserine (DHBS) was purchased from EMC Microcollections (Tuebingen, Germany). All other catechol compounds were purchased from Sigma-Aldrich (St. Louis, MO). Nonsiderophore solutions were prepared immediately prior to use.
Growth stimulation assays.
Iron-restricted agar bioassays were used to test the ability of catechol compounds to function as Bordetella iron sources. Luria-Bertani agar for bioassays contained the nonutilizable iron chelator ethylenediaminedi-(o-hydroxyphenyl)acetic acid (EDDA) and was seeded with B. bronchiseptica cells as described previously (14). Iron source test solutions were provided in 50-μl volumes placed in 6-mm-diameter wells cut into the agar; Bordetella growth zones around the wells were measured in millimeters. Results are means of triplicate assays and are representative of at least two experiments; the standard deviation from the mean was less than ±2 mm.
The effect of norepinephrine on the growth of B. bronchiseptica was assayed by measuring the bacterial growth yield in liquid cultures. The alcaligin-deficient strain BRM26 was grown in iron-replete SS medium, washed with SS basal medium lacking iron, and used to inoculate (1:2,000 dilution) parallel 10-ml cultures containing iron-depleted SS medium containing or lacking 30% (vol/vol) heat-treated fetal bovine serum (FBS) (Gibco Cell Culture, Carlsbad, CA). One culture from each of the two sets was supplemented with 6.5 μM enterobactin, 50 μM norepinephrine, or both compounds; control cultures were unsupplemented. After 20 h of growth at 37°C in a shaking incubator (250 rpm), the optical density at 600 nm (OD600) of each culture was measured. Results shown are representative of the growth trends from three independent experiments.
Genetic methods.
Recombinant plasmids were constructed by standard genetic methods (65). Plasmid pBB37 carries the B. pertussis bfeA gene and has been described previously (1). The 2.4-kb SmaI tonB+ exbBD+ fragment of B. bronchiseptica 19385 was subcloned from plasmid pRKTon (55) to pBBR1MCS-1 to create pBB41. Plasmid DNA was transferred to Bordetella cells by conjugation (11) or electroporation. B. bronchiseptica RB50 and B. pertussis Tohama I genomic DNA sequences were obtained from the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk). DNA sequences that were determined for this laboratory were provided by the Advanced Genetic Analysis Center at the University of Minnesota. Oligonucleotide primers were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Analysis of genetic sequences was aided by the use of the Lasergene suite of sequence analysis software (DNASTAR, Madison, WI).
To construct the lacZ fusion plasmid pMP4, primers bfeR-A′EcoRI (5′-GCGCGAATTCGCTGGAAGACATGGTCAAGG-3′) and bfe1Bam (5′-GGCCGGATCCTCTCCTCGGCGGTGATGAC-3′) were used to PCR amplify from the B. bronchiseptica B013N genomic DNA template a 1.5-kb product that contained the bfeR gene and the first 199 bp of the bfeA coding sequence. This primer pair was also used in the construction of pMP3 to PCR amplify the same genetic region from B. bronchiseptica bfeR mutant BRM24, resulting in a product containing the 594-bp in-frame ΔbfeR deletion allele. The bfeR+-bfeA′ and ΔbfeR-bfeA′ fragments were ligated to pGEM3Z to create p3Z129 and p3Z130, respectively. The DNA regions were then subcloned from those plasmids as EcoRI-SphI fragments into the promoterless lacZ plasmid pMP220, resulting in transcriptional reporter fusion plasmids pMP3 (ΔbfeR bfeA′-lacZ) and pMP4 (bfeR+ bfeA′-lacZ). The correct fusion junctions were verified by nucleotide sequencing.
β-Galactosidase assays.
Transcription of bfeA was analyzed using B. bronchiseptica strains carrying bfeA-lacZ reporter plasmids. Bacteria were grown for 24 h in iron-replete SS medium, washed with SS basal medium, and subcultured at a 1:50 dilution to iron-replete medium or iron-depleted medium in the presence or absence of an inducer. After 18 h of incubation at 37°C, bacteria were assayed for β-galactosidase activity using the method of Miller (51) as modified by Brickman et al. (16). Results are means for triplicate cultures ± 1 standard deviation and are representative of at least two experiments.
Immunoblot analysis.
B. bronchiseptica strains were grown in iron-replete SS medium for 24 h, washed, and used to inoculate (1:200 dilution) parallel iron-depleted cultures containing or lacking 50 μM norepinephrine. After 24 h of growth, bacteria were harvested and solubilized, and proteins from approximately 0.02 OD600 unit of cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (1). Proteins were transferred to nitrocellulose membranes and probed using a mouse antiserum raised to the FepA enterobactin receptor of E. coli (4) that cross-reacts with the BfeA protein (1).
RESULTS
Induction of bfeA expression is tonB independent.
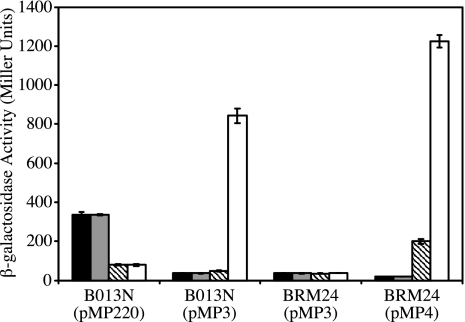
Since the Bordetella BfeR protein is an AraC family transcriptional regulator, it is predicted to interact with enterobactin in the cytoplasm to induce bfeA transcription. The BfeA receptor is not required for enterobactin-inducible transcriptional activation of bfeA (1). Thus, it was possible that another tonB-dependent outer membrane receptor could transport the enterobactin inducer. The B. bronchiseptica tonB strain BRM31, confirmed to be unable to use ferric enterobactin as an iron source (data not shown), was tested for its ability to activate transcription of bfeA-lacZ in response to enterobactin. Both BRM31(pMP3) and the wild-type parent strain B013N(pMP3) demonstrated iron-repressible, enterobactin-inducible expression of bfeA, producing similar levels of β-galactosidase activity when grown in iron-depleted SS medium supplemented with enterobactin (Fig. 1). Genetic complementation of BRM31(pMP3) using tonB exbBD provided in trans (pBB41) resulted in no change in bfeA-lacZ expression, although ferric enterobactin transport was restored by complementation (data not shown). Since Bordetella species have only one known tonB gene, these results demonstrate that neither tonB nor any other TonB-dependent receptor, including BfeA, is required for enterobactin-inducible bfeA transcription. It is possible that the enterobactin inducer may exert its effects from an extracellular location, or it may gain access to the BfeR regulator in the cytoplasm by a mechanism distinct from that used for uptake of the ferric enterobactin complex.
FIG. 1.
TonB-independent induction of bfeA transcription in B. bronchiseptica. Wild-type B013N and BRM31 (tonB::pSSt2) cells harboring the bfeA-lacZ reporter fusion plasmid pMP3 or the promoterless lacZ vector control plasmid pMP220 were grown in SS medium and assayed for β-galactosidase activity as described in Materials and Methods. Genetic complementation of BRM31 was accomplished using the tonB+ exbBD+ plasmid pBB41, and pBBR1MCS-1 served as the vector control. Error bars, ±1 standard deviation from the means of triplicate cultures. Culture conditions were as follows: iron replete (solid bars), iron depleted (shaded bars), and iron depleted and supplemented with 3.3 μM enterobactin (open bars).
BfeA-mediated transport of other ferric catechols.
The Bordetella BfeA receptor was hypothesized to transport siderophores or other compounds with structural similarity to enterobactin. Each compound shown in Fig. 2A was tested as a Bordetella iron source by assaying the growth of wild-type B013N cells in an iron-restricted agar bioassay. To determine if utilization of these catechols was dependent on the ferric enterobactin receptor, their ability to stimulate growth of the B. bronchiseptica bfeA mutant strain BRM25(pBBR1MCS-1) and the genetically complemented strain BRM25(pBB37) was tested. Salmochelin and corynebactin are catechol siderophores that have a high degree of structural similarity to enterobactin and were able to stimulate B. bronchiseptica growth in a concentration-dependent manner (Fig. 3). Salmochelin is a glucosylated form of enterobactin (8), and its utilization by B. bronchiseptica required the BfeA ferric enterobactin receptor (Table 1). Corynebactin and enterobactin have opposite chirality (9); corynebactin has l-threonine instead of the l-serine residues of enterobactin, and each of its side chains has a glycine spacer (17). The bfeA mutant exhibited a low level of growth stimulation by corynebactin. This result suggests that there may be another outer membrane receptor that is capable of transporting ferric corynebactin in addition to BfeA. In addition to these naturally occurring siderophores, two synthetic siderophores were tested. TRENCAM (63) and MECAM (74, 75) are enterobactin analogs, each of which has three iron-binding catechol groups but lacks the cleavable ester linkages of enterobactin (56, 59). Both analogs promoted the growth of strain B013N, with TRENCAM producing the largest growth stimulation zones of all of the compounds tested (Fig. 3). Growth stimulation by TRENCAM was dependent on BfeA (Table 1). MECAM promoted slight growth enhancement of the bfeA mutant, yielding results similar to those for corynebactin. The enterobactin breakdown product DHBS, as well as 3,4-dihydroxymandelic acid and pyrocatechol (Fig. 2A), was unable to stimulate growth of wild-type B. bronchiseptica cells in these assays. These results demonstrate that the Bordetella BfeA receptor can transport other catechol iron sources in addition to ferric enterobactin.
FIG. 2.
Molecular structures of catechol compounds (A) and of catecholamine synthesis pathway molecules and sympathetic nervous system agonists and antagonists (B).
FIG. 3.
Growth stimulation of B. bronchiseptica cells by natural and synthetic catechol siderophores. Wild-type B013N cells were seeded into iron-restricted agar as described in Materials and Methods and were provided with siderophores at 25 μM (solid bars), 12.5 μM (shaded bars), or 6.3 μM (open bars). Growth stimulation zones represent the means of triplicate bioassays and include the 6-mm diameter of the sample wells.
TABLE 1.
Transcriptional activation of bfeA and BfeA-dependent growth stimulation by catechol compounds
| Compound | β-Galactosidase activity (Miller units) of B013N(pMP3) (bfeA-lacZ):
|
Fold inductiona | BfeA-dependent growth stimulation | |||||
|---|---|---|---|---|---|---|---|---|
| Without Fe | Without Fe and with compound (50 μM) | |||||||
| Siderophores | ||||||||
| Enterobactin | 42.6 (±5.1)b | 1,531.5 (±123.9) | 36.0 | +c | ||||
| Salmochelin | 57.1 (±4.6) | 182.9 (±11.9) | 3.2 | + | ||||
| Corynebactin | 54.8 (±7.8) | 91.3 (±19.1) | 1.7 | +/−d | ||||
| DHBS | 17.2 (±10.7) | 15.5 (±10.9) | −e | NGf | ||||
| Synthetic analogs | ||||||||
| MECAM | 27.3 (±3.1) | 459.5 (±24.6) | 16.8 | +/− | ||||
| TRENCAM | 58.5 (±7.7) | 88.8 (±7.9) | 1.5 | + | ||||
| Catechols | ||||||||
| Pyrocatechol | 55.2 (±3.8) | 1,600.8 (±55.0) | 29.0 | NG | ||||
| 3,4-Dihydroxymandelic acid | 55.2 (±3.8) | 162.4 (±5.9) | 2.9 | NG | ||||
LacZ activity without Fe and with compound ÷ LacZ activity without Fe.
±1 standard deviation.
+, BfeA dependent.
+/−, partially BfeA dependent; growth stimulation of a bfeA strain was reduced compared to that of the wild-type strain.
−, no induction.
NG, no growth.
Inducers of bfeA transcription.
Since enterobactin stimulates transcriptional activation of bfeA, the ability of the catechol compounds (Fig. 2A) to induce bfeA transcription was tested by measuring the β-galactosidase activity of B013N(pMP3) cells (Table 1). Corynebactin and salmochelin yielded moderate levels of bfeA transcriptional activation compared with enterobactin. The DHBS enterobactin breakdown product failed to induce bfeA transcription; the enterobactin precursor, 2,3-dihydroxybenzoic acid, has already been shown to lack inducer capability (1). Although it elicited the largest Bordetella growth haloes among the compounds tested, TRENCAM exhibited low inducer activity, indicating that up-regulation of bfeA expression is not required for maximal growth stimulation by this compound. However, MECAM induced high levels of bfeA transcription. Pyrocatechol, which did not exhibit Bordetella growth-enhancing activity, induced bfeA expression 29-fold. 3,4-Dihydroxymandelic acid, which also lacked growth-promoting activity, elicited a 2.9-fold increase in bfeA transcription. Although enterobactin was the most potent inducer of the compounds tested, these results indicate that transcriptional activation of bfeA in Bordetella cells can occur in response to other catechol compounds.
Norepinephrine induction of bfeA transcription.
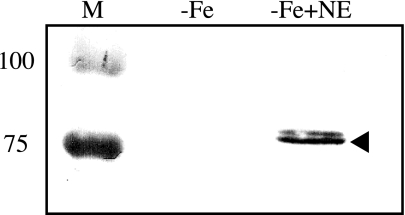
Burton and colleagues noted that exposure of a pathogenic strain of E. coli to the catecholamine neurotransmitter norepinephrine resulted in increased production of a protein reported to be the FepA enterobactin receptor (19). Given these experimental observations and the structural relatedness of norepinephrine to enterobactin, we hypothesized that norepinephrine may induce transcription of the Bordetella enterobactin receptor gene. The effect of norepinephrine on BfeA production was examined by immunoblot analysis using an anti-FepA antiserum that cross-reacts with the Bordetella BfeA protein (1). Iron-depleted B. bronchiseptica cells supplemented with norepinephrine produced an immunoreactive high-molecular-weight protein, presumed to be BfeA, and its putative precursor, whereas those grown in the absence of the catecholamine produced negligible levels of the protein (Fig. 4). Production of the norepinephrine-induced protein was absent in bfeA mutant strain BRM25 and was restored upon genetic complementation by bfeA in trans (data not shown), confirming its identity as BfeA.
FIG. 4.
Norepinephrine-induced production of BfeA in B. bronchiseptica. Bacterial lysates from wild-type B013N cells grown in iron-depleted SS medium (−Fe) or in iron-depleted medium supplemented with 50 μM norepinephrine (−Fe+NE) were subjected to immunoblot analysis using a FepA-specific antiserum. The sizes of molecular mass standards (in kilodaltons) are shown on the left. Arrowhead indicates the immunoreactive BfeA protein.
The ability of norepinephrine to induce transcription of bfeA-lacZ was investigated (Table 2). Iron-starved B013N(pMP3) demonstrated a ∼28-fold induction of bfeA transcription in response to 50 μM norepinephrine. To determine whether bfeA transcriptional activation by norepinephrine was dependent on the BfeR regulatory protein, bfeA-lacZ expression was assayed in the B. bronchiseptica bfeR mutant strain BRM24(pMP3). In contrast to bfeA transcription levels in the iron-starved wild-type strain exposed to norepinephrine, expression of bfeA in similarly cultured BRM24(pMP3) cells was negligible (Fig. 5). Norepinephrine-induced expression was restored to wild-type levels in BRM24 cells genetically complemented with the wild-type bfeR gene (pMP4). BRM24(pMP4) cultured under iron-depleted conditions exhibited norepinephrine-induced bfeA expression levels that were approximately 1.5-fold higher than those of the wild-type strain; increased transcription in the absence of norepinephrine was also observed in these cells, suggesting bfeR multicopy effects. Upstream of bfeA is a DNA region that contains predicted Fur-binding sites (7) and exhibits in vivo Fur binding activity (1). Norepinephrine-induced bfeA expression was absent in bacteria cultured under iron-replete conditions (Fig. 5), consistent with Fur-mediated repression. Norepinephrine-inducible transcription of bfeA requires BfeR and an iron-depleted physiologic state.
TABLE 2.
Transcriptional activation of bfeA by catecholamines
| Compound | β-Galactosidase activity (Miller units) of B013N(pMP3) (bfeA-lacZ):
|
Fold inductiona | |
|---|---|---|---|
| Without Fe | Without Fe and with compound (50 μM) | ||
| Norepinephrine | 32.1 (±27.8)b | 904.7 (±61.0) | 28.2 |
| Carbidopa | 55.2 (±3.8) | 1,388.1 (±257.4) | 25.1 |
| Isoproterenol | 55.2 (±3.8) | 1,375.5 (±209.7) | 24.6 |
| Phenylephrine | 38.4 (±9.2) | 24.1 (±15.2) | −c |
| Tyramine | 38.4 (±9.2) | 43.1 (±17.0) | 1.1 |
LacZ activity without Fe and with compound ÷ LacZ activity without Fe.
±1 standard deviation.
−, no induction.
FIG. 5.
Norepinephrine-induced bfeA transcription. Strains B013N (wild type) and BRM24 (ΔbfeR) carried the bfeA-lacZ reporter plasmid pMP3 or the vector control plasmid pMP220. Strain BRM24 was complemented by using the bfeR+ bfeA-lacZ plasmid pMP4. Bacteria were assayed for β-galactosidase activity after growth in SS medium under the following conditions: iron replete (solid bars), iron replete and supplemented with 50 μM norepinephrine (shaded bars), iron depleted (striped bars), or iron depleted and supplemented with 50 μM norepinephrine (open bars). Error bars, ±1 standard deviation from the means of triplicate cultures.
Bordetella growth stimulation by norepinephrine.
To determine whether norepinephrine could provide a nutritional benefit to Bordetella cells, its ability to promote growth in an iron-restricted medium was investigated. Agar bioassays did not demonstrate a role for norepinephrine in growth stimulation of iron-starved B. bronchiseptica cells (data not shown). It was hypothesized that since B. bronchiseptica produces the siderophore alcaligin, any growth-stimulatory effects that may be provided by norepinephrine may be masked by endogenous siderophore activity. Therefore, the alcaligin-deficient mutant strain BRM26 was used to assess the ability of norepinephrine to stimulate Bordetella growth in a defined liquid medium. Norepinephrine alone or in combination with enterobactin failed to stimulate growth of BRM26 cells in iron-depleted SS medium (Fig. 6). This lack of growth even in the presence of a siderophore was consistently observed and was likely due to the extremely limited availability of iron in the SS medium in this closed batch culture system. In previous work by others, addition of norepinephrine to a defined minimal medium containing bovine serum resulted in increased growth of E. coli cells (28, 45). To determine if serum could similarly potentiate norepinephrine-mediated growth stimulation of Bordetella cells, strain BRM26 was cultured in iron-depleted SS medium containing 30% heat-treated FBS (SS+FBS) that either lacked or contained enterobactin, norepinephrine, or both compounds. Addition of serum to iron-depleted SS medium resulted in enhanced growth of B. bronchiseptica strain BRM26, and supplementation of the SS+FBS medium with either norepinephrine or enterobactin caused a further increase in the growth yield (Fig. 6). The bacterial growth yield in SS+FBS containing both norepinephrine and enterobactin was greater than that with either compound alone. These results demonstrate that norepinephrine, as well as enterobactin, stimulates the growth of Bordetella cells in the presence of serum. Furthermore, Bordetella cells apparently do not require the function of a siderophore for serum-enhanced growth in SS medium or for norepinephrine-mediated growth stimulation in SS+FBS.
FIG. 6.
Growth stimulation by norepinephrine. B. bronchiseptica cells were cultured in iron-depleted SS medium (SS) or iron-depleted SS medium containing 30% FBS (SS+FBS), and the growth yield after 20 h was measured densitometrically at a wavelength of 600 nm. Cultures either had no supplementation (solid bars) or were supplemented with either enterobactin (shaded bars), norepinephrine (striped bars), or both enterobactin and norepinephrine (open bars).
Catecholamine activators of bfeA transcription.
Norepinephrine is a precursor of epinephrine in the major mammalian catecholamine biosynthesis pathway (78) (Fig. 2B). To determine whether compounds in the pathway could act as inducers of bfeA transcription, each was subjected to a titration analysis using iron-depleted B013N(pMP3) cells (Fig. 7). Norepinephrine, dopamine, and epinephrine stimulated high levels of bfeA transcription, exhibiting similar inducer activity over the range of concentrations tested. These results contrast with those obtained using l-DOPA and tyrosine, which had little, if any, ability to activate bfeA transcription. Enterobactin elicited the highest bfeA transcriptional response compared to the other compounds tested.
FIG. 7.
Analysis of bfeA transcriptional activation by catecholamines. B. bronchiseptica B013N carrying the bfeA-lacZ reporter plasmid pMP3 was assayed for β-galactosidase activity as described in Materials and Methods. Bacteria were grown in iron-depleted SS medium and supplemented with one of the following catechol compounds: enterobactin (solid squares), norepinephrine (open squares), epinephrine (solid triangles), dopamine (open circles), l-DOPA (open triangles), and tyrosine (solid circles). Values represent means of triplicate cultures ± 1 standard deviation. Error bars representing less than ±50 Miller units are not shown.
Several adrenergic activators and inhibitors have structural features similar to those of the known inducers dopamine, norepinephrine, and epinephrine (Fig. 2B) and were tested to determine if they could activate transcription of bfeA (Table 2). Carbidopa, an inhibitor of aromatic l-amino acid decarboxylase, closely resembles l-DOPA (which did not induce bfeA transcription) except that it has a methyl group on carbon 2 of the propanoic acid and a second amine that forms a hydrazinyl group. Carbidopa strongly influenced bfeA expression, resulting in a 25-fold increase in transcription compared to uninduced control cultures. Isoproterenol is a β-adrenergic receptor agonist that resembles norepinephrine and epinephrine but contains a propyl group on the terminal amine. This 3-carbon difference did not alter the ability of this molecule to induce bfeA transcription, since high levels of β-galactosidase activity were observed when B013N(pMP3) cells were exposed to isoproterenol (Table 2). The α-adrenergic receptor antagonist phenylephrine differs from epinephrine due to the lack of a hydroxyl substituent at carbon 4 of its aromatic ring. This difference accounted for the inability of phenylephrine to induce bfeA transcription (Table 2), since epinephrine, which contains a complete catechol structure, was a potent inducer (Fig. 7). Tyramine was also unable to induce bfeA transcription due to the absence of a hydroxyl on carbon 3 of the benzene ring, compared to dopamine (dihydroxylated), which acts as an inducer. Together, these results indicate that a complete catechol structure is required for induction of bfeA transcription.
DISCUSSION
Enterobactin-producing organisms can inhabit environments such as soil and water (41), and host activities in those environments may allow transient colonization of the respiratory mucosa. In humans, microbes known to produce enterobactin can be isolated from the upper respiratory tract and oral cavity (6, 71). Since mucosal inhabitants such as B. bronchiseptica, B. pertussis (7), Haemophilus parainfluenzae (76), and Neisseria species (20, 64) possess transport systems for the enterobactin xenosiderophore, their ability to use this potent iron chelator may provide an in vivo growth advantage. We postulated that the Bordetella Bfe ferric enterobactin system could function in the utilization of other, structurally similar siderophores. In Salmonella enterica, for example, both the FepA and IroN receptors transport ferric enterobactin and DHBS; in addition, FepA can transport myxochelin C (61), and IroN is a receptor for ferric corynebactin (61) and salmochelin (36). Similarly, the Bordetella BfeA enterobactin receptor was shown in the present study to transport other ferric catechols including corynebactin, salmochelin, and the two synthetic siderophores TRENCAM and MECAM. Of the naturally occurring siderophores tested, enterobactin exhibited the greatest ability to enhance the growth of iron-starved B. bronchiseptica.
In iron-depleted Bordetella cells, transcription of bfeA is enterobactin inducible by a process requiring BfeR, an AraC-type regulator. Based on the mechanism of transcriptional activation by the AraC protein of E. coli, the AraC/XylS family regulators are generally predicted to bind small molecule inducers (29). Within this regulator family exists a subset of proteins that control transcription of genes that encode siderophore biosynthesis and utilization functions. The B. pertussis and B. bronchiseptica BfeR (1) and AlcR (12, 15) proteins, as well as the P. aeruginosa PchR (37, 38), Yersinia pestis YbtA (26), and Sinorhizobium meliloti RhrA (44) proteins, are among the members of this iron acquisition subclass of AraC-type regulators. With the exception of RhrA, which has not yet been fully characterized, all of these regulators are activated by their cognate siderophores when the bacteria are starved for iron. BfeR (1) (Fig. 1), AlcR (T. J. Brickman and S. K. Armstrong, unpublished data), and YbtA (26, 58) can perceive their siderophore inducers in the absence of their cognate outer membrane siderophore receptors and TonB. The mechanism by which these inducers are internalized in the absence of TonB-dependent receptor function remains unknown. However in P. aeruginosa, PchR requires the FptA pyochelin receptor for maximal fptA induction by pyochelin (38). PchR was recently demonstrated to bind target gene promoter sequences in a pyochelin-dependent manner (50), strongly supporting the hypothesis that the regulators of this iron acquisition subclass interact directly with their cognate siderophore inducers to activate transcription.
The results from this study showed that transcription of bfeA was induced significantly by the MECAM synthetic siderophore and by pyrocatechol, whereas salmochelin had moderate inducing activity. TRENCAM, which elicited the best growth enhancement response in B. bronchiseptica, showed negligible induction ability, as did the siderophore corynebactin. Pyrocatechol was a strong inducer but lacked growth-promoting function. These results indicate that the ability to promote Bordetella growth does not necessarily correlate with the ability to induce bfeA transcription, and they support the concept of distinct cell entry pathways for inducers and iron sources. Although nonutilizable monomeric catecholates could potentially form bidentate ferric iron complexes and thus cause iron restriction, increased bfeA expression resulted from induction rather than Fur derepression, since Bordetella cells in iron-depleted SS medium are iron starved in the absence of exogenous chelators.
Since B. bronchiseptica is an obligate pathogen, it is possible that catechols of host origin may be perceived and potentially used by the Bfe system. The ability of norepinephrine to enhance in vitro growth has been reported for several bacterial species including E. coli (45), Listeria species (21), Staphylococcus epidermidis (53), Aeromonas hydrophila, and Klebsiella pneumoniae (39). Internalization of norepinephrine has also been observed in some bacteria (28, 39). Previous studies reported that growth of pathogenic E. coli strains was stimulated by catecholamine neurotransmitters in serum-containing growth medium (46), and increased production of the ferric enterobactin receptor in response to norepinephrine was observed (19). Our studies demonstrated that Bordetella growth was stimulated in serum-supplemented SS medium by addition of either enterobactin or norepinephrine. The addition of serum to the SS medium also caused enhanced Bordetella growth; this effect may be due to the reported presence of epinephrine in FBS (68). The fact that growth stimulation by norepinephrine occurred in the absence of the alcaligin or enterobactin siderophores is intriguing, since norepinephrine is not known to have iron-chelating activity similar to that of siderophores. One previous report implicated transferrin as the component of serum responsible for norepinephrine-mediated growth stimulation of E. coli, and norepinephrine was demonstrated to cause the release of transferrin- and lactoferrin-bound iron in vitro (28). For B. bronchiseptica, acquisition of the iron released from transferrin by norepinephrine may occur by a siderophore-independent mechanism, possibly involving putative transporters such as annotated asATP-dependent permease complexes having iron binding components (locus tags BB2946 and BB4774). Ongoing studies in our laboratory are aimed at identifying other Bordetella genes whose expression is inducible by catecholamines and determining whether transferrin is involved in norepinephrine-mediated growth stimulation of Bordetella cells.
In enterohemorrhagic E. coli, expression of the locus of enterocyte effacement pathogenicity island and flagellar genes is activated by a signaling system that uses the autoinducer AI-3, epinephrine, and norepinephrine (68). In this way, pathogenic E. coli strains can intercept and respond to host hormone signals. In mammals, the main adrenergic catecholamines are epinephrine, norepinephrine, and dopamine, all of which strongly induced Bordetella bfeA transcription at biologically relevant concentrations. A metabolite of norepinephrine, 3,4-dihydroxymandelic acid, exhibited low inducer activity and did not promote the growth of B. bronchiseptica (Table 1). Nanomolar concentrations of epinephrine and norepinephrine are found in the plasma of mammals, and in tissues the concentration of norepinephrine is dependent on the degree of sympathetic innervation at that site (78). Tissue injury is known to result in the release of norepinephrine from neurons (77), and high levels of circulating catecholamines and their metabolites have been correlated with severe bacterial infection (32) and onset of acute respiratory illness (33).
Bordetella responsiveness to catecholamine hormones may fulfill a nutritional need, such as iron acquisition, or impart some other benefit to the organism that improves its fitness in the host environment. It is also possible that these neuroendocrine hormones do not function in iron acquisition per se but may serve as a host environmental cue. Surprisingly little is known of the catecholamine concentrations that may be found on healthy respiratory mucosal surfaces. Airway tone and reflexes such as coughing, bronchodilation, mucociliary function, and plasma leakage are regulated primarily by the sympathetic (adrenergic) and parasympathetic (cholinergic) branches of the autonomic nervous system (47, 52). Human tracheal gland cells (48) and rabbit tracheal epithelial cells (42) respond to exogenous epinephrine and norepinephrine, and inhaled epinephrine is an effective bronchodilator that has been widely used clinically (52). Rat nasal mucus contains nanomolar concentrations of catecholamines, including norepinephrine and dopamine (43). Therefore, catecholamines may be present on the host respiratory epithelium and detectable by Bordetella cells.
Bordetella species may not only have the ability to sense and respond to the iron-depleted host environment to obtain nutritional iron but may also be able to perceive important host signaling molecules. Transiently colonizing bacteria or commensals of the respiratory tract may produce catechol xenosiderophores that can be used by Bordetella via the Bfe system. Host catecholamines may be present at the initial site of mucosal infection or liberated by transudation in significant concentrations later during infection, after Bordetella-mediated damage to the epithelium has occurred. The ability of the Bordetella BfeR protein to activate transcription in response to a variety of catechol compounds, including those of microbial and host origin, illustrates the potential flexibility of this iron acquisition system.
Acknowledgments
We thank Timothy Brickman for helpful discussions and critical review of the manuscript and Ryan Suhadolc for the construction of the tonB mutant strain BRM31. We thank Bernard Beall for B. bronchiseptica strains and for plasmids pRKTon and pSSt2. We are also grateful to Klaus Hantke and Kenneth Raymond for providing siderophores.
Support for this study was provided by University of Minnesota grant-in-aid 19473 and Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases. M.A. was supported by Public Health Service grant T32 AI-07421 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Anderson, M. T., and S. K. Armstrong. 2004. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J. Bacteriol. 186:7302-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arima, K., A. Kakinuma, and G. Tamura. 1968. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 31:488-494. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, S. K., and M. O. Clements. 1993. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J. Bacteriol. 175:1144-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong, S. K., C. L. Francis, and M. A. McIntosh. 1990. Molecular analysis of the Escherichia coli ferric enterobactin receptor FepA. J. Biol. Chem. 265:14536-14543. [PubMed] [Google Scholar]
- 5.Arnow, L. E. 1937. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixtures. J. Biol. Chem. 118:531-537. [Google Scholar]
- 6.Barrie, J. D., and J. B. Gallacher. 1975. The significance of Escherichia coli in the upper respiratory tract of children under 2 years of age. Postgrad. Med. J. 51:373-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 8.Bister, B., D. Bischoff, G. J. Nicholson, M. Valdebenito, K. Schneider, G. Winkelmann, K. Hantke, and R. D. Sussmuth. 2004. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17:471-481. [DOI] [PubMed] [Google Scholar]
- 9.Bluhm, M. E., S. S. Kim, E. A. Dertz, and K. N. Raymond. 2002. Corynebactin and enterobactin: related siderophores of opposite chirality. J. Am. Chem. Soc. 124:2436-2437. [DOI] [PubMed] [Google Scholar]
- 10.Bordet, J., and O. Gengou. 1906. Le microbe de la coqueluche. Ann. Inst. Pasteur (Paris) 20:731-741. [Google Scholar]
- 11.Brickman, T. J., and S. K. Armstrong. 1996. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J. Bacteriol. 178:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brickman, T. J., and S. K. Armstrong. 2005. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 187:3650-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brickman, T. J., J. G. Hansel, M. J. Miller, and S. K. Armstrong. 1996. Purification, spectroscopic analysis and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. Biometals 9:191-203. [DOI] [PubMed] [Google Scholar]
- 15.Brickman, T. J., H. Y. Kang, and S. K. Armstrong. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brickman, T. J., B. A. Ozenberger, and M. A. McIntosh. 1990. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212:669-682. [DOI] [PubMed] [Google Scholar]
- 17.Budzikiewicz, H., A. Bössenkamp, K. Taraz, A. Pandey, and J. M. Meyer. 1997. Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC 14067 (Brevibacterium sp. DSM 20411). Z. Naturforsch. 52:551-554. [Google Scholar]
- 18.Bullen, J. J., H. J. Rogers, and E. Griffiths. 1978. Role of iron in bacterial infection. Curr. Top. Microbiol. Immunol. 80:1-35. [DOI] [PubMed] [Google Scholar]
- 19.Burton, C. L., S. R. Chhabra, S. Swift, T. J. Baldwin, H. Withers, S. J. Hill, and P. Williams. 2002. The growth response of Escherichia coli to neurotransmitters and related catecholamine drugs requires a functional enterobactin biosynthesis and uptake system. Infect. Immun. 70:5913-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carson, S. D. B., P. E. Klebba, S. M. Newton, and P. F. Sparling. 1999. Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J. Bacteriol. 181:2895-2901.10217784 [Google Scholar]
- 21.Coulanges, V., P. Andre, and D. J. Vidon. 1998. Effect of siderophores, catecholamines, and catechol compounds on Listeria spp. Growth in iron-complexed medium. Biochem. Biophys. Res. Commun. 249:526-530. [DOI] [PubMed] [Google Scholar]
- 22.Cox, G. B., F. Gibson, R. K. Luke, N. A. Newton, I. G. O'Brien, and H. Rosenberg. 1970. Mutations affecting iron transport in Escherichia coli. J. Bacteriol. 104:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean, C. R., S. Neshat, and K. Poole. 1996. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 178:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean, C. R., and K. Poole. 1993. Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol. Microbiol. 8:1095-1103. [DOI] [PubMed] [Google Scholar]
- 25.Ernst, J. F., R. L. Bennett, and L. I. Rothfield. 1978. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J. Bacteriol. 135:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fetherston, J. D., S. W. Bearden, and R. D. Perry. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol. Microbiol. 22:315-325. [DOI] [PubMed] [Google Scholar]
- 27.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freestone, P. P., M. Lyte, C. P. Neal, A. F. Maggs, R. D. Haigh, and P. H. Williams. 2000. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J. Bacteriol. 182:6091-6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg, M. B., S. A. Boyko, and S. B. Calderwood. 1991. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:1125-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodnow, R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44:722-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groves, A. C., J. Griffiths, F. Leung, and R. N. Meek. 1973. Plasma catecholamines in patients with serious postoperative infection. Ann. Surg. 178:102-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruchow, H. W. 1979. Catecholamine activity and infectious disease episodes. J. Hum. Stress. 5:11-17. [DOI] [PubMed] [Google Scholar]
- 34.Halliwell, B., and J. M. Gutteridge. 1984. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 36.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 100:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinrichs, D. E., and K. Poole. 1993. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J. Bacteriol. 175:5882-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinrichs, D. E., and K. Poole. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinney, K. S., C. E. Austin, D. S. Morton, and G. Sonnenfeld. 2000. Norepinephrine as a growth stimulating factor in bacteria—mechanistic studies. Life Sci. 67:3075-3085. [DOI] [PubMed] [Google Scholar]
- 40.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 41.Leclerc, H., D. A. Mossel, S. C. Edberg, and C. B. Struijk. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55:201-234. [DOI] [PubMed] [Google Scholar]
- 42.Liedtke, C. M. 1988. Differentiated properties of rabbit tracheal epithelial cells in primary culture. Am. J. Physiol. 255:C760-C770. [DOI] [PubMed] [Google Scholar]
- 43.Lucero, M. T., and A. Squires. 1998. Catecholamine concentrations in rat nasal mucus are modulated by trigeminal stimulation of the nasal cavity. Brain Res. 807:234-236. [DOI] [PubMed] [Google Scholar]
- 44.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. O. Cuiv, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyte, M., and S. Ernst. 1992. Catecholamine induced growth of gram negative bacteria. Life Sci. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 46.Lyte, M., C. D. Frank, and B. T. Green. 1996. Production of an autoinducer of growth by norepinephrine cultured Escherichia coli O157:H7. FEMS Microbiol. Lett. 139:155-159. [DOI] [PubMed] [Google Scholar]
- 47.Mathé, A. A., and P. Hedqvist. 1988. Catecholamines and pulmonary function, p. 497-503. In A. Dahlström, R. H. Belmaker, and M. Sandler (ed.), Progress in catecholamine research, part A: basic aspects and peripheral mechanisms. Alan R. Liss, Inc., New York, N.Y.
- 48.Merten, M. D., and C. Figarella. 1993. Constitutive hypersecretion and insensitivity to neurotransmitters by cystic fibrosis tracheal gland cells. Am. J. Physiol. 264:L93-L99. [DOI] [PubMed] [Google Scholar]
- 49.Mey, A. R., E. E. Wyckoff, A. G. Oglesby, E. Rab, R. K. Taylor, and S. M. Payne. 2002. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect. Immun. 70:3419-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michel, L., N. Gonzalez, S. Jagdeep, T. Nguyen-Ngoc, and C. Reimmann. 2005. PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol. Microbiol. 58:495-509. [DOI] [PubMed] [Google Scholar]
- 51.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 52.Nadel, J. A., and P. J. Barnes. 1984. Autonomic regulation of the airways. Annu. Rev. Med. 35:451-467. [DOI] [PubMed] [Google Scholar]
- 53.Neal, C. P., P. P. Freestone, A. F. Maggs, R. D. Haigh, P. H. Williams, and M. Lyte. 2001. Catecholamine inotropes as growth factors for Staphylococcus epidermidis and other coagulase-negative staphylococci. FEMS Microbiol. Lett. 194:163-169. [DOI] [PubMed] [Google Scholar]
- 54.Neilands, J. B., and K. Nakamura. 1991. Detection, determination, isolation, characterization and regulation of microbial iron chelates, p. 1-14. In G. Winkelmann (ed.), Handbook of microbial iron chelates. CRC Press, Inc., Boca Raton, Fla.
- 55.Nicholson, M. L., and B. Beall. 1999. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussis prevents utilization of ferric siderophores, haemin and haemoglobin as iron sources. Microbiology 145:2453-2461. [DOI] [PubMed] [Google Scholar]
- 56.O'Brien, I. G., G. B. Cox, and F. Gibson. 1970. Biologically active compounds containing 2,3-dihydroxybenzoic acid and serine formed by Escherichia coli. Biochim. Biophys. Acta 201:453-460. [DOI] [PubMed] [Google Scholar]
- 57.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 58.Perry, R. D., J. Abney, I. Mier, Jr., Y. Lee, S. W. Bearden, and J. D. Fetherston. 2003. Regulation of the Yersinia pestis Yfe and Ybt iron transport systems. Adv. Exp. Med. Biol. 529:275-283. [DOI] [PubMed] [Google Scholar]
- 59.Porra, R. J., L. Langman, I. G. Young, and F. Gibson. 1972. The role of ferric enterochelin esterase in enterochelin-mediated iron transport and ferrochelatase activity in Escherichia coli. Arch. Biochem. Biophys. 153:74-78. [DOI] [PubMed] [Google Scholar]
- 60.Postle, K. 1993. TonB protein and energy transduction between membranes. J. Bioenerg. Biomembr. 25:591-601. [DOI] [PubMed] [Google Scholar]
- 61.Rabsch, W., W. Voigt, R. Reissbrodt, R. M. Tsolis, and A. J. Baumler. 1999. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J. Bacteriol. 181:3610-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reid, R. T., D. H. Live, D. J. Faulkner, and A. Butler. 1993. A siderophore from a marine bacterium with an exceptional ferric ion affinity constant. Nature 366:455-458. [DOI] [PubMed] [Google Scholar]
- 63.Rodgers, S. J., C. Lee, C. Y. Ng, and K. N. Raymond. 1987. Ferric ion sequestering agents. 15. Synthesis, solution chemistry, and electrochemistry of a new cationic analogue of enterobactin. Inorg. Chem. 26:1622-1625. [Google Scholar]
- 64.Rutz, J. M., T. Abdullah, S. P. Singh, V. I. Kalve, and P. E. Klebba. 1991. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J. Bacteriol. 173:5964-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 66.Schneider, D. R., and C. D. Parker. 1982. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect. Immun. 38:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 68.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stainer, D. W., and M. J. Scholte. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63:211-220. [DOI] [PubMed] [Google Scholar]
- 70.Stintzi, A., and K. N. Raymond. 2000. Amonabactin-mediated iron acquisition from transferrin and lactoferrin by Aeromonas hydrophila: direct measurement of individual microscopic rate constants. J. Biol. Inorg. Chem. 5:57-66. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka, I., K. Suzuki, E. Tanaka, and S. Baba. 1996. Investigation of normal bacterial flora in the upper respiratory tract. Acta Otolaryngol. Suppl. 525:44-50. [PubMed] [Google Scholar]
- 72.Vanderpool, C. K., and S. K. Armstrong. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Hove, B., H. Staudenmaier, and V. Braun. 1990. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K-12. J. Bacteriol. 172:6749-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venuti, M. C., W. H. Rastetter, and J. B. Neilands. 1979. 1,3,5-Tris(N,N′,N"-2,3-dihydroxybenzoyl)amino-methylbenzene, a synthetic iron chelator related to enterobactin. J. Med. Chem. 22:123-124. [DOI] [PubMed] [Google Scholar]
- 75.Weitl, F. L., and K. N. Raymond. 1979. Ferric ion sequestering agents. 1. Hexadentate O-bonding N,N′,N"-tris(2,3-dihydroxybenzoyl) derivatives of 1,5,9-triazacyclotridecane and 1,3,5-triaminomethylbenzene. J. Am. Chem. Soc. 101:2728-2731. [Google Scholar]
- 76.Williams, P., D. J. Morton, K. J. Towner, P. Stevenson, and E. Griffiths. 1990. Utilization of enterobactin and other exogenous iron sources by Haemophilus influenzae, H. parainfluenzae and H. paraphrophilus. J. Gen. Microbiol. 136:2343-2350. [DOI] [PubMed] [Google Scholar]
- 77.Woolf, P. D., J. V. McDonald, D. V. Feliciano, M. M. Kelly, D. Nichols, and C. Cox. 1992. The catecholamine response to multisystem trauma. Arch. Surg. 127:899-903. [DOI] [PubMed] [Google Scholar]
- 78.Young, J. B., and L. Landsberg. 1998. Catecholamines and the adrenal medulla, p. 665-728. In J. D. Wilson, D. W. Foster, H. M. Kronenberg, and P. R. Larsen (ed.), Williams textbook of endocrinology, 9th ed. W. B. Saunders Co., Philadelphia, Pa.