Abstract
Pyoverdine-mediated iron uptake by the FpvA receptor in the outer membrane of Pseudomonas aeruginosa is dependent on the inner membrane protein TonB1. This energy transducer couples the proton-electrochemical potential of the inner membrane to the transport event. To shed more light upon this process, a recombinant TonB1 protein lacking the N-terminal inner membrane anchor (TonBpp) was constructed. This protein was, after expression in Escherichia coli, purified from the soluble fraction of lysed cells by means of an N-terminal hexahistidine or glutathione S-transferase (GST) tag. Purified GST-TonBpp was able to capture detergent-solubilized FpvA, regardless of the presence of pyoverdine or pyoverdine-Fe. Targeting of the TonB1 fragment to the periplasm of P. aeruginosa inhibited the transport of ferric pyoverdine by FpvA in vivo, indicating an interference with endogenous TonB1, presumably caused by competition for binding sites at the transporter or by formation of nonfunctional TonB heterodimers. Surface plasmon resonance experiments demonstrated that the FpvA-TonBpp interactions have apparent affinities in the micromolar range. The binding of pyoverdine or ferric pyoverdine to FpvA did not modulate this affinity. Apparently, the presence of either iron or pyoverdine is not essential for the formation of the FpvA-TonB complex in vitro.
Pseudomonas aeruginosa is a ubiquitous environmental gram-negative bacterium that has become one of the most prominent causative agents of opportunistic human infections (52). As it does for nearly all microorganisms, elemental iron plays an indispensable role in the growth and colonization of P. aeruginosa. In animal hosts, however, iron is usually bound to proteins such as transferrin and lactoferrin (53), and P. aeruginosa is therefore challenged with an environment that maintains a very small reservoir of free soluble iron. To overcome this problem, under these iron-limited conditions P. aeruginosa produces high-affinity iron chelators called siderophores (5). The major siderophore produced by P. aeruginosa is pyoverdine (Pvd), which sequesters iron from external sources and is concentrated as ferric Pvd on the outer membrane by the high-affinity transporter FpvA (38). Like all outer membrane siderophore receptors (51), this transporter is composed of two domains, a C-terminal β-barrel made of 22 β-strands and an N-terminal, globular cork or plug domain residing inside the barrel. FpvA belongs to a subclass of iron transporters which has an additional N-terminal domain that is involved in the regulation of the transcription of the fpvA operon (29, 47). In vivo, the normal state of FpvA under iron limitation is the FpvA-Pvd complex (45, 46). The dissociation of Pvd from FpvA, before binding of extracellular Pvd-Fe and the subsequent iron transport into the periplasm, is dependent on the inner membrane protein TonB and the proton motive force (PMF) (6, 45, 50).
In the P. aeruginosa genome (49), three tonB genes have been identified, i.e., tonB1, tonB2, and tonB3. Disruption of tonB1 inhibits siderophore-mediated iron uptake and heme uptake (39, 54). Inactivation of tonB2 has no adverse effect on iron or heme acquisition, but tonB1-tonB2 double mutants are more compromised with respect to growth in iron-restricted medium than is a single tonB1 knockout mutant (55). Inactivation of tonB3 appears to result in a defective twitching motility (19), and the gene product is most likely not involved in iron uptake.
Most knowledge about TonB function and structure has been obtained primarily from studies with Escherichia coli TonB (reviewed in reference 40). This protein comprises three domains: a hydrophobic amino-terminal helix that anchors the protein in the cytoplasmic membrane, followed by a proline-rich region and a carboxyl-terminal globular domain that protrudes in the periplasm. Although P. aeruginosa TonB displays a high homology with E. coli TonB, the protein is distinguished by an N-terminal extension, which is important for the TonB activity in P. aeruginosa (54). Furthermore, full TonB function is dependent on the ExbB and ExbD proteins, which are located in the cytoplasmic membrane (1, 14). The combined topologies of ExbB and ExbD mimic those of a signal transducer, with ExbD extending into the periplasmic space from its single transmembrane domain (21) and ExbB consisting of three transmembrane domains and a significant cytoplasmic domain (21, 23). Although P. aeruginosa homologues of ExbB and ExbD have been found, inactivation of these genes did not adversely affect the growth under iron limitation (55).
Structural determinations of carboxyl-terminal domains of different lengths of TonB have been performed, either by X-ray diffraction (4, 27, 28) or by spectroscopy (37). These different models show that the protein forms structurally different dimers but may also exist as a monomer. Further work suggests that FhuA, the E. coli iron-ferrichrome transporter, is able to bind two TonB proteins in vitro and that the TonB protein dimerizes in vivo (15, 24, 25, 44).
Whereas TonB is involved in the iron uptake by siderophore receptors, the way it exerts its mode of action for the exchange of siderophore on FpvA and the internalization of ferric siderophore is still unclear. Evidence for a direct physical interaction between TonB and TonB-dependent transporters was gathered from a range of in vivo and in vitro experiments (3, 31, 34, 35, 48). Located at the N termini of all TonB-dependent outer membrane transporters is the TonB box, a short stretch of amino acids shared by all the transporters (2, 20, 40). The nature of the change in the TonB box conformation after substrate binding is not well defined. The TonB box is disordered in the structures of FhuA and FecA (ferrichrome and ferric-dicitrate transporters, respectively, in E. coli), but a short helix to which the TonB box is attached is unfolded in the ferric siderophore-bound structures and moves across the periplasm (10, 33). However, unfolding of a switch helix is not a general feature, because it is absent in FepA and BtuB, the ferric enterobactin and vitamin B12 transporters, respectively, in E. coli (11). Consistently, removal of the switch helix in FhuA resulted in a decrease in iron transport but still conferred colicin M and phage sensitivity (9).
To date, two models for the activation of the outer membrane receptor by the TonB machinery have been postulated. In the propeller model, a TonB dimer undergoes rotary motion, similar to the mechanism described for the bacterial flagellar motor that is powered by MotA and MotB, which are homologous to ExbB and ExbD (41). In such a system, the energy for iron transport is delivered by the torque of the TonB-ExbB-ExbD system. Alternatively, energized TonB shuttles from the inner to the outer membrane, thereby releasing its energy to the iron transporter (30, 41). The shuttle model is supported by in vivo labeling experiments that demonstrate periplasmic accessibility of the extreme N terminus of TonB to a specific Cys marker.
In our laboratory we investigate the iron uptake mechanism of FpvA by in vivo and in vitro experiments (16, 45), and we recently solved the X-ray structure of FpvA (7). To gain more insight into the iron uptake mechanism in P. aeruginosa, we focused on the tonB1 gene product and its putative interaction with FpvA. In this study, we sought biochemical evidence for this interaction.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. The iron-deficient succinate minimal medium used in this study has been described previously (8). Luria-Bertani broth (Difco) and agar (Difco) were supplemented with 0.4% glucose and were employed as the rich media throughout the experiments. The antibiotics tetracycline (50 μg/ml), carbenicillin (150 μg/ml), kanamycin (25 μg/ml), and ampicillin (100 μg/ml) were included in growth media when appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| JM109(DE3) | F−traD36 lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV44 e14−gyrA96 recA1 relA1 endA1 thi hsdR17 | Promega |
| SG13009 | F−thi lac mtl | QIAGEN |
| P. aeruginosa strains | ||
| ATCC 15962 | Wild type | 17 |
| K691 | Wild type | 46 |
| CDC5 | PAO1 ΔPvd | 38 |
| PAD08 | PAO1 tonB::Tet | 50 |
| Plasmids | ||
| pCR4Blunt-TOPO | Ampr Kanr; T7 and T3 promoter | Invitrogen |
| pET41b | Kanr ColE1; T7 promoter, GST fusion | Novagen |
| pGST-TonBpp | Kanr; tonBpp gene in pET41b | This study |
| pHis-TonBpp | Ampr; tonBpp gene in pQE31 | This study |
| pMMB190 | Ampr; RSF replicon (IncQ), tac promoter | 36 |
| PMMB-TonBpp | Ampr; tonBpp gene in pMMB190 | This study |
| PMMB-TonB | Ampr; tonB gene in pMMB190 | This study |
| pPVR2 | CarbrfpvA+ | 38 |
| pREP4 | KanrlacIq | QIAGEN |
| pRK2013 | Kanr Tra+ Mob+ | 13 |
| pQE31 | Ampr ColE1; T5 promoter, His tag fusion | QIAGEN |
Abbreviations: Amp, ampicillin; Carb, carbenicillin; Kan, kanamycin; Tet, tetracycline.
Purification and preparation of FpvA, FpvA-Pvd, and FpvA-Pvd-Fe.
Unloaded FpvA or FpvA-Pvd was expressed, as described previously, in strain CDC5(pPVR2) or in strain K691(pPVR2), respectively (46). For the preparation of FpvA-Pvd-Fe, purified FpvA was incubated with a 10-fold excess of Pvd-Fe for 1 h.
PCR gene amplifications.
The plasmids used in this study are listed in Table 1. All enzymes for DNA manipulations were purchased from Fermentas and used according to the manufacturer's instructions. Strain TOP10 was used as the host strain for all plasmid constructions. Chromosomal DNA was isolated from strain PAO1 with a genomic DNA isolation kit (QIAGEN) and used as the template in the PCRs. The reaction mixture included 1 U of the proofreading DNA polymerase Pfx and 50 pmol (each) of primers that were designed to incorporate restriction sites into the amplified tonB1 gene. Primers AEH263 (5′-GGCCCGGGCCGACCCCGGCCGAACTC-3′) and AEH265 (5′-GAAAAGCTTCAGCGGCGCTTCTCGATCT-3′), containing a SmaI site and a HindIII site (underlined), respectively, were used for amplification of a 704-bp tonB1 gene product encoding the 234-amino-acid periplasmic domain of TonB1 (in this study called TonBpp). The combination of primers AEH266 (5′-GTCCCCGGGATGTCGCCACAGCCTTCACG-3′), containing a SmaI site (underlined), and AEH265 was used for the amplification of the full tonB1 gene (the gene product of which is called TonB in this study). The PCR was started with an annealing temperature of 55°C for 30 s. The annealing temperature was increased 0.5°C in 20 cycles to a final temperature of 65°C. Subsequently the PCR was performed at an annealing temperature of 65°C for another 15 cycles. The resulting DNA fragments were agarose gel purified (QIAGEN gel extraction kit) and were cloned into pCR4Blunt-TOPO (Invitrogen) according to the manufacturer's instructions, yielding pTOPO-TonBpp and pTOPO-TonB. The nucleotide sequences of the constructs resulting from the PCR amplifications were confirmed by sequence analysis on an ABI 377 sequencer with a dye terminator kit (Perkin-Elmer).
Expression plasmid constructions.
For the construction of a glutathione S-transferase (GST)-TonBpp fusion, plasmid pET41a was digested with NcoI and termini were filled in with Klenow fragment, followed by HindIII digestion and dephosphorylation. Plasmid pTOPO-TonBpp was digested with SmaI and HindIII. The DNA fragment was ligated into the digested pET41b vector, yielding pGST-TonBpp. Between the GST fusion protein and the TonBpp protein sequence (starting with the amino acids MetPro) (Fig. 1), the amino acid sequence SerProMetGly was inserted. For the construction of an N-terminally His-tagged TonBpp fusion, plasmid pQE31 was digested with SphI and blunted with T4 polymerase, followed by HindIII digestion and dephosphorylation. The SmaI and HindIII TonBpp fragment of pTOPO-TonBpp was ligated into the digested pQE31 vector, yielding pHis-TonBpp. Between the His tag and the TonBpp sequence, the amino acid sequence SerProMetGly was inserted.
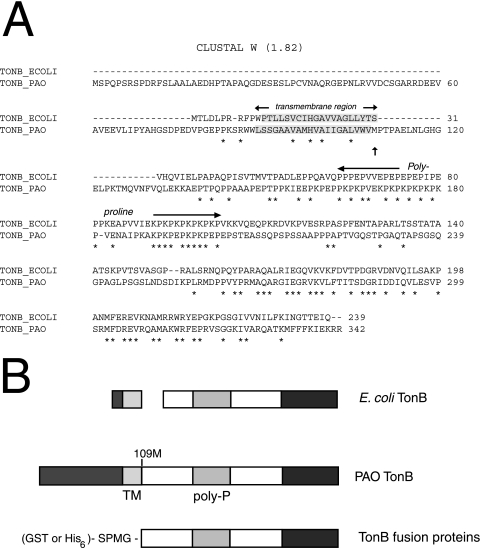
FIG. 1.
TonB fusion proteins. A. Clustal W alignment (http://www.ebi.ac.uk/clustalw/) of P. aeruginosa (PAO) and E. coli (ECOLI) TonB sequences. The putative transmembrane (shaded) and polyproline regions are depicted. Asterisks indicate residues conserved in the two sequences. B. Schematic diagram of E. coli and P. aeruginosa TonB proteins and the fusion protein constructs used. Homologous domains of P. aeruginosa TonB with E. coli TonB are depicted with the same gray shade. The 109 M (also indicated in panel A with an arrow) is the starting amino acid residue of the P. aeruginosa TonBpp sequence. Abbreviations: TM, transmembrane region; Poly-P, polyproline region. Note the different N-terminal extension.
P. aeruginosa expression plasmid constructions.
For proper expression of TonB or of TonBpp in P. aeruginosa, a synthetic RBS linker (5′-GAATTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATCCCGGGCAGCTGAAGCTT-3′), derived from the pET41b vector and encoding a ribosome binding site (indicated in boldface), was inserted into pCR4Blunt-TOPO. Furthermore, the RBS linker contains, in order, the restriction sites EcoRI, SmaI, and HindIII (all underlined). The resulting vector pTOPO-RBS was digested with SmaI and HindIII, and the SmaI-HindIII fragments from pTOPO-TonBpp or pTOPO-TonB, encoding either tonBpp or tonB, were inserted into the linearized vector, yielding pTOPO-RBS-TonBpp or pTOPO-RBS-TonB. For the construction of a TonBpp protein, which is targeted to the periplasm, vector pTOPO-RBS-TonBpp was digested with SmaI, and a synthetic linker, 5′-ATGAAGAAGGTTTCTACGCTTGACCTGTTGTTCGTTGCGATCATGGGTGTTTCGCCGGCCGCTTTTGCCAGTCCCATG-3′, encoding the LasB signal sequence (SS) (accession number DQ153386), was inserted. The vectors pTOPO-RBS-TonBpp-SS and pTOPO-RBS-TonB were EcoRI and HindIII digested, and the fragments, encoding the target genes, were cloned into the broad-host-range vector pMMB190 which had been digested with the same restriction enzymes. The resulting plasmids, pMMB-TonBpp and pMMB-TonB, were introduced into strain ATCC 15692 or strain PAD08 by triparental mating using the conjugative helper plasmid pRK2013 as described previously (22).
Expression and purification of soluble GST-tagged TonBpp.
The plasmid encoding GST-TonBpp was transferred to JM109(DE3) cells, and overnight cultures grown at 30°C were diluted 1:100 in 2 liters of rich medium supplemented with kanamycin and incubated at 30°C until an optical density at 600 nm (OD600) of 0.6 was reached. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM final concentration) was then added, and incubation was continued at 30°C for 4 h. The cells were chilled on ice and collected by centrifugation at 5,000 × g for 10 min at 4°C. The supernatant was discarded, and the pellet was either resuspended as described below or stored at −20°C.
The pellet was resuspended in 25 ml of 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol supplemented with Complete protease inhibitor (Roche). The cells were disrupted in a French press twice at 8,000 lb/in2. The intact cells and cell envelopes were removed by centrifugation at 9,600 × g for 10 min and at 113,600 × g for 90 min, respectively. The supernatant was loaded, at a flow rate of 0.5 ml/min, onto a 1-ml HiTrap GST affinity column (Amersham) equilibrated with phosphate-buffered saline (PBS). The column was washed with 10 ml PBS at a flow rate of 1 ml/min. The GST fusion proteins were eluted in 1-ml fractions with PBS containing 10 mM glutathione. GST fusion protein-containing fractions were pooled, and the buffer was exchanged for 125 mM HEPES (pH 7.3) and 5 mM NaCl. Subsequently, the GST fusion proteins were purified using a HiTrap SP column (Amersham). Proteins were eluted using a linear gradient from 5 mM to 1 M NaCl at a flow rate of 1 ml/min. Protein concentrations were estimated from the absorbance at 280 nm, assuming an extinction coefficient of 0.5 for a solution containing 1 mg of protein per ml.
Expression and purification of His-tagged TonBpp.
An overnight culture of strain SG13009(pREP4) carrying plasmid pHis-TonBpp was diluted 1:50 in 800 ml rich medium supplemented with kanamycin and ampicillin and was incubated at 37°C until an OD600 of 0.6 was reached. Synthesis of His-tagged TonBpp was induced by adding 500 μM IPTG to the medium, and incubation was continued for 3 h at 37°C. Cells were harvested by centrifugation as described in the previous paragraph, and the pellet was resuspended in 50 ml of 50 mM Tris-HCl (pH 7.3), 50 mM NaCl, and 1 mM EDTA. Complete protease inhibitor tablets (Roche) and 0.5 mM phenylmethylsulfonyl fluoride were added, and cells were lysed by sonication. Cell debris and membranes were removed as described in the previous paragraph, and the supernatant was loaded on a 10-ml SourceQ column (Amersham). His-tagged TonBpp eluted early in a gradient of 50 mM to 1 M NaCl. Fractions containing His-tagged TonBpp were pooled and concentrated, and simultaneously the buffer was changed to 50 mM Tris-HCl (pH 7.3). After loading of the His-tagged TonBpp on a SourceS column, proteins were eluted in a gradient of 0 to 1 M NaCl. As a final purification step, the His-tagged TonBpp was loaded (after a buffer change to 50 mM Tris-HCl [pH 8.0]) on a HiTrap Ni-nitrilotriacetic acid column that was preequilibrated with 50 mM Tris-Cl (pH 8) and 10 mM imidazole. Proteins specifically bound to the column material were eluted with 50 mM Tris-Cl (pH 8) buffer containing 300 mM imidazole. Sample purity was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue staining.
Small-scale interaction assays.
Interaction assays were conducted with purified FpvA and GST-TonBpp. All incubations were performed at room temperature. Purified FpvA, FpvA-Pvd, or FpvA-Pvd-Fe (10 μg) was mixed with 3 μg of GST-TonBpp and 50 μl 50% glutathione-Sepharose beads (Pharmacia) in PBS supplemented with 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) in a total volume of 100 μl and tumbled for 2 h at room temperature. The resin was pelleted by centrifugation (1 min at 5,000 × g) and washed with 1 ml PBS buffer with 1% CHAPS. Beads were mixed with an equal volume of denaturing electrophoresis sample buffer, boiled for 10 min, and resolved on 12% polyacrylamide gels, and proteins were visualized by Coomassie brilliant blue staining.
Characterization of the His-tagged-TonBpp-FpvA complex by size exclusion chromatography.
Proteins were applied to a Superose 6 HR 10/30 column (Amersham Pharmacia) equilibrated in TON buffer (50 mM Tris-HCl [pH 7.2], 0.8% octyl-polyoxyethylene (POE) and 50 mM NaCl). The flow rate was kept at 0.5 ml/min, and 0.5-ml fractions were collected. His-tagged-TonBpp (153 μg) and FpvA or FpvA-Pvd-Fe (100 μg each) were mixed (fivefold molar excess of TonB). All size exclusion chromatography was performed at room temperature.
Protein agarose gel electrophoresis.
Protein complexes were run in 0.9% agarose gels (20 mM Tris [pH 8.0], 50 mM glycine) supplemented with 1% octyl-POE. The gel was cast in a horizontal minigel apparatus typically used for electrophoresis of DNA. Gels were run in 20 mM Tris-HCl (pH 7.5)-50 mM glycine supplemented with 0.4% octyl-POE. The proteins were diluted in a 10-μl final volume in sample loading buffer (20 mM Tris-HCl [pH 7.5], 1% octyl-POE, and 20% glycerol). After loading of the gel, electrophoresis was performed at 120 V for 30 min and gels were stained with Coomassie brilliant blue solution. After destaining, the gel was extensively washed with water to sharpen the protein bands.
Iron uptake assays.
Pvd-55Fe (0.25 Ci/mmol) was prepared as described previously (45) with a fourfold excess of Pvd with respect to the concentration of iron. Cells of strain ATCC 15692 or strain PAD08, carrying plasmid pMMB-TonBpp or pMMB-TonB, respectively, were prepared at an OD600 of 0.6 in 50 mM Tris-HCl (pH 8.0) and incubated for 15 min. The transport assays were initiated by the addition of 200 nM Pvd-55Fe. Aliquots (100 μl) of the suspension were removed at different times and filtered (0.45-μm cellulose nitrate membrane filters; Whatman), and the filters were washed two times with 3 ml 50 mM Tris-HCl buffer (pH 8.0). The radioactivity retained was counted. The same experiment was repeated with ATCC 15692 cells pretreated with 200 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP).
Determination of TonB affinities for FpvA.
Anti-GST antibodies (5,100 resonance units [RU]) were immobilized on two channels of CM4 sensor surfaces (BR-1005-39), using standard amine-coupling chemistry (GST capture kit, BR-1002-23). One of the channels was used to capture the GST-TonBpp fragment (60 to 250 RU), and the other was used as a reference with a similar amount of GST.
Surface plasmon resonance (SPR) measurements were performed at 25°C using a Biacore 2000 device, with buffer A (10 mM Tris-HCl [pH 8], 150 mM NaCl, and 0.8% octyl-POE) as running buffer. Purified FpvA, purified FpvA preincubated with Pvd-Fe (complex formed in vitro with a 10-fold molar excess of Pvd-Fe), and iron-free FpvA-Pvd complex were tested for GST-TonBpp binding. The injection time for FpvA mixtures was 100 s, followed by injection of buffer for 300 s, with FpvA concentrations ranging from 90 nM to 10 μM. Bovine serum albumin (BSA) was used as a negative control. Regeneration of anti-GST antibody surfaces was achieved for each measurement by 1-min pulses of 10 mM glycine (pH 2.2), followed by injection of GST-TonBpp to regenerate a TonBpp-coated surface that had not previously interacted with FpvA, Pvd, or Pvd-Fe.
The use of a surface containing antibodies directed against GST to capture GST-TonBpp protein was found to be important in our measurements. We found that the presence of iron in the mixtures injected over the TonBpp-containing surface never allowed the baseline to reach its original value. To ensure reproducibility of the system, GST-TonBpp was always stripped from the antibody-containing surface after each experiment, and a clean surface was regenerated by injection of GST-TonBpp.
Data collected for the GST-TonBpp surface were processed using the double reference method (43). For each set of curves corresponding to the injection of various concentrations of FpvA and FpvA-Pvd-Fe complex (0 to 10 μM) over the same surface, global fitting was carried out using a simple 1:1 Langmuir model (BIAevaluation 3.1 software package). For the steady-state analysis, the equilibrium dissociation constants were determined by plotting the equilibrium plateau (Req) versus injected concentrations of solutes. Experimental saturation curves were adequately fit with the 1:1 model Req = Rmax × (C/(C + Kd), where Req is the equilibrium response, Rmax is the response at saturation, C is the concentration of solute, and Kd is the affinity constant.
RESULTS
Expression and purification of TonB fusion constructs.
To date there is no direct evidence for an interaction between P. aeruginosa TonB and the siderophore receptor FpvA. Previous experiments, however, demonstrated the necessity of an intact tonB1 gene for efficient iron uptake (39, 50, 54), which implies an interaction between TonB and FpvA. To investigate the physical interaction between TonB and FpvA, we constructed GST- and His-tagged TonB fusion proteins. The primers used for the PCR were designed to amplify the DNA sequence encoding amino acids 109 to 342 (Fig. 1). Vector-derived sequences appended either a stretch of six histidine residues or a GST fusion to the truncated TonB to facilitate downstream protein purification.
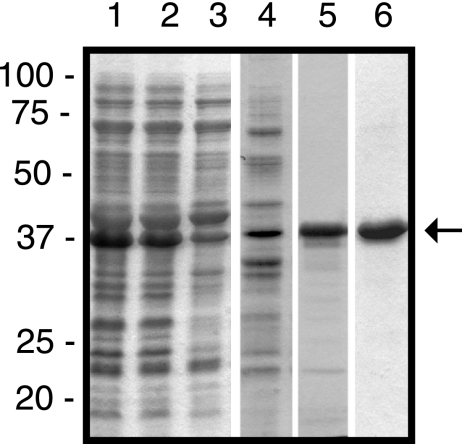
Cell fractionation studies showed that the majority of the GST- and His-tagged fusion constructs were present in the soluble fraction. Intriguingly, the fusion constructs GST-TonBpp (calculated molecular mass of 57 kDa) and His-TonBpp (calculated molecular mass of 23 kDa), both containing the polyproline stretch, migrated with apparent relative molecular weight (Mrs) of ∼75,000 (see Fig. 4) and ∼37,000 (Fig. 2), respectively, in SDS-polyacrylamide gels. This behavior has been reported for wild-type E. coli TonB (calculated molecular mass of 26.1 kDa; Mr of 36,000) (32, 42) and for a truncated version of TonB (35). All TonBpp fusion constructs were isolated with more than 95% purity, as judged from Coomassie blue-stained gels (Fig. 2, lane 6).
FIG. 4.
Pull-down assays with GST-TonBpp. GST-TonBpp-coated beads were incubated with FpvA, FpvA-Pvd, or FpvA-Pvd-Fe, followed by elution of the proteins and analysis by SDS-PAGE and staining with Coomassie brilliant blue. The positions of the molecular mass marker proteins (in kDa) are indicated at the right. The positions of FpvA and GST-TonBpp are marked with arrows.
FIG. 2.
Purification of His-tagged TonBpp. Samples from the purification steps were analyzed by SDS-PAGE. Lane 1, total cell lysate; lane 2, supernatant after sonication; lane 3, supernatant after ultracentrifugation; lane 4, pooled fractions after SourceQ column chromatography; lane 5, pooled fractions after SourceS column chromatography; lane 6. pooled fractions after Ni2+-nitrilotriacetic acid chromatography. The positions of the molecular mass marker proteins (in kDa) are shown at the left, and the position of His-TonBpp is marked with an arrow.
Iron transport through FpvA is inhibited by the periplasmic TonB fragment.
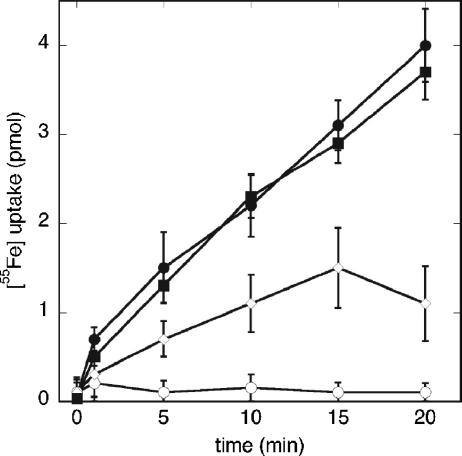
To evaluate the functionality of the cloned tonB constructs in P. aeruginosa, cells containing the plasmids encoding the periplasmic and full-length TonB were assayed for their ability to transport 55Fe via the Pvd pathway. Pvd-55Fe was transported with the same efficiency in the ΔtonB strain PAD08 carrying plasmid pMMB-TonB as in the wild-type strain (Fig. 3). This clearly demonstrates that the cloned TonB protein is fully functional. As shown in Fig. 3, the 55Fe uptake is completely inhibited by the uncoupler CCCP. When the TonBpp fragment was overexpressed, a 70% inhibition of the 55Fe uptake was observed (Fig. 3). Apparently, the function of FpvA is sensitive to the expression of the TonBpp fragment, suggesting an interaction of the fragment with FpvA. This is consistent with similar assays of ferrichrome and ferric citrate transport in E. coli, showing that expression of periplasmic TonB fragments in this bacterium inhibits transport of the ferric siderophore (18).
FIG. 3.
Pvd-55Fe uptake. Cells of strain ATCC 15692 carrying no plasmid (•) or plasmid pMMB-TonBpp (⋄) and of the ΔTonB strain PAD08 carrying plasmid pMMB-TonB (▪), were grown to an OD600 of 0.6 and were incubated for 15 min in 50 mM Tris-HCl (pH 8.0) before initiation of the transport assays by addition of 200 nM of Pvd-55Fe. Aliquots (100 μl) were removed at different time points and filtered, and the retained radioactivity was counted. As a control, the experiment was repeated with ATCC 15692 cells pretreated with 200 μM CCCP (○). The data shown are the averages and standard deviations from three experiments.
Interaction of GST-TonBpp with FpvA.
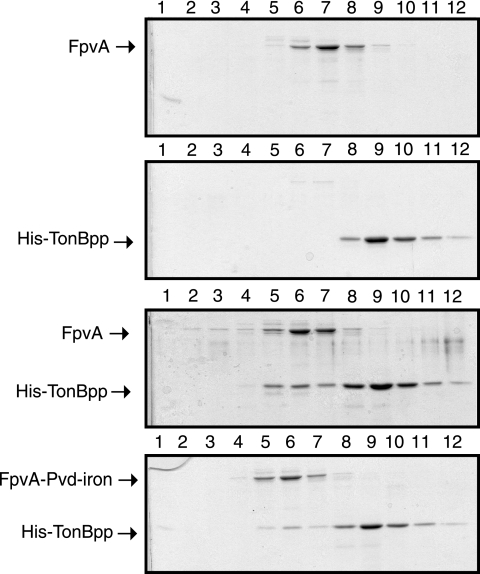
To investigate the possible interaction of the TonB protein with FpvA, we employed small-scale pull-down assays with the GST-TonBpp fusion construct. After incubation of purified FpvA-Pvd-Fe, FpvA-Pvd, or FpvA with glutathione-agarose beads coated with GST-TonBpp, complexes were analyzed by SDS-PAGE (Fig. 4). Unloaded or Pvd-Fe- or Pvd-loaded FpvA incubated under the same conditions but in the absence of GST-TonBpp showed no affinity for the glutathione resin (Fig. 4), demonstrating the specificity of the capture. Taken together, these data demonstrate that purified GST-TonBpp forms a specific complex with unloaded FpvA, as well with Pvd-Fe- or Pvd-loaded FpvA.
Characterization of His-tagged-TonBpp-FpvA complexes by gel filtration.
Since results from the small-scale pull-down experiments demonstrated an interaction between immobilized TonBpp and FpvA, size exclusion chromatography was used to further characterize the TonBpp-FpvA complex. Size exclusion chromatography experiments performed on FpvA or His-TonBpp alone showed that FpvA eluted as a single peak centered at fraction 7, while the peak of His-TonBpp was centered at fraction 9 (Fig. 5). The same experiment performed with FpvA mixed with His-TonBpp resulted in the shift of the FpvA elution peak to fraction 6, i.e., towards higher-molecular-weight complexes. SDS-PAGE analysis of the eluted fraction showed that His-TonBpp coeluted with FpvA, suggesting the formation of a stable complex between these two species. Apparently, FpvA loaded with or without Pvd-iron interacts with His-TonBpp.
FIG. 5.
Analysis of His-tagged TonBpp-FpvA complex by gel filtration. Proteins were incubated together and then applied to a Superose 6 column. In all experiments, the amount of FpvA applied to the column was 100 μg and the amount of His-tagged TonBpp was 153 μg. Samples from the fractions passing through the column were analyzed by SDS-PAGE (lanes 1 to 12).
Characterization of His-tagged TonBpp-FpvA complexes by agarose gel electrophoresis.
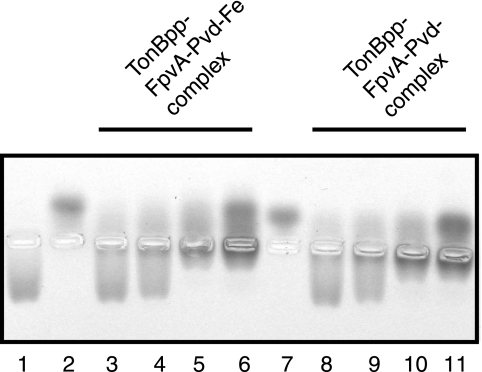
To substantiate the evidence for an interaction between TonB and FpvA, native agarose gel electrophoresis was performed. We took advantage of the marked difference of charge between FpvA and TonBpp at neutral pH. The theoretical isoelectric points of FpvA and TonBpp are 5.08 and 9.93, respectively, and the proteins should thus migrate in opposite directions at neutral pH in an electric field. As shown in Fig. 6, TonBpp and FpvA loaded either with Pvd or with Pvd-iron migrated in opposite directions at pH 7.5. Formation of TonBpp-FpvA complexes is clearly evidenced with this experimental method, since the band corresponding to FpvA alone disappeared when TonBpp was present in excess (Fig. 6, lanes 3, 4, 8, and 9), while the band corresponding to TonBpp disappeared in the presence of a stoichiometric amount or excess of FpvA (lanes 5, 6, 10, and 11). Under all conditions, in samples containing both FpvA and TonBpp, a band of low mobility appeared close to the wells, which likely corresponds to the FpvA-TonBpp complex. In conclusion, it appears again that the presence of iron does not influence the interaction between TonBpp and FpvA.
FIG. 6.
Native electrophoresis of His-tagged TonBpp-FpvA complex in agarose gels. Agarose gels of FpvA and His-TonBpp were stained with Coomassie brilliant blue. Lanes 1, 4 pmol TonBpp; 2, 2 pmol FpvA-Pvd-Fe; 3 to 6, 4 pmol TonBpp mixed with, respectively, 1, 2, 4, and 8 pmol FpvA-Pvd-Fe; 7, 2 pmol FpvA-Pvd; 8 to 11, 4 pmol TonBpp mixed with, respectively, 1, 2, 4, and 8 pmol FpvA-Pvd.
Determination of affinities of TonBpp for the FpvA receptor in the presence or absence of Pvd-Fe or Pvd.
If the interaction of TonBpp with FpvA is not influenced by the presence of iron-loaded or unloaded Pvd, the affinity of TonBpp for FpvA should be the same for the three different complexes. To obtain quantitative data about the TonB binding to FpvA, SPR experiments were conducted. In a typical SPR experiment, one of the binding partners is immobilized on a sensor surface and the other interactant is injected over the surface. The interactions are recorded in RU, which are proportional to the mass accumulation on the surface.
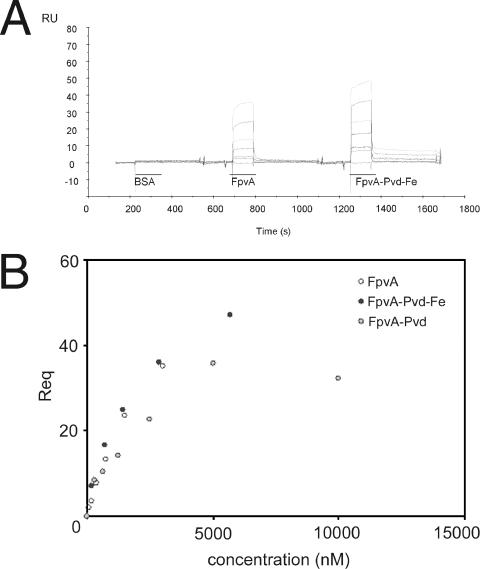
For the determination of the kinetics and equilibrium constants of the FpvA-TonBpp binding, GST-TonBpp (60 to 200 RU) was captured by anti-GST antibodies bound to the sensor surface. FpvA, FpvA-Pvd-Fe, iron free FpvA-Pvd, and BSA as a control were injected at concentrations ranging from 90 to 10,000 nM. The sensorgrams corresponding to the successive injections of BSA, FpvA, FpvA-Pvd, and FpvA-Pvd-Fe on a GST-TonBpp (80 RU) surface are shown in Fig. 7A. As shown, we did not observe any binding to the GST-TonBpp-coated surface with BSA. A concentration-dependent equilibrium plateau (Req) is observed for FpvA and FpvA-Pvd-Fe (Fig. 7A) and for FpvA-Pvd (data not shown). The equilibrium dissociation constants were determined from saturation curves by plotting the equilibrium plateau (Req) versus injected concentrations of solutes. Saturation curves obtained on an 80-RU GST-TonBpp surface for FpvA and FpvA-Pvd-Fe and on a 60-RU GST-TonBpp surface for FpvA-Pvd are shown in Fig. 7B. The experiments were performed over different GST-TonBpp surfaces (200 RU, 80 RU, 60 RU, or 70 RU). Table 2 summarizes the evaluated affinity parameters determined on the three different TonB surfaces. As can be concluded from Table 2, FpvA, FpvA-Pvd-Fe, and FpvA-Pvd show similar affinities for TonBpp in the micromolar range, suggesting that the interaction between FpvA and TonBpp in vitro is not influenced by the presence of Pvd or Pvd-Fe. The dissociation rate is high and dissociation is complete for FpvA and FpvA-Pvd.
FIG. 7.
SPR experiments with the GST-TonBpp-FpvA complex. A. Control corrected sensorgrams corresponding to the successive injections of BSA (312 to 10,000 nM), FpvA (90 to 3,000 nM), and FpvA-Pvd-Fe (180 to 5,700 nM) over 80 RU of GST-TonBpp. B. Saturation curves obtained on an 80-RU GST-TonBpp surface for FpvA and FpvA-Pvd-Fe and on a 60-RU GST-TonBpp surface for FpvA-Pvd are shown.
TABLE 2.
Equilibrium constants (Kd) and maximal binding capacities (Rmax) as evaluated for the interactions of FpvA, FpvA-Pvd-Fe, and FpvA-Pvd with immobilized GST-TonBpp
| Immobilized GST-TonBpp (RU) | Solute | Kd (μM) | Rmax | χ2 | Mean Kd ± SE (μM) |
|---|---|---|---|---|---|
| 60 | FpvA | 1.96 | 27.6 | 2.9 | 2.9 ± 0.8 |
| 80 | FpvA | 3.30 | 73.9 | 0.13 | |
| 200 | FpvA | 3.45 | 237 | 3.3 | |
| 70 | FpvA-Pvd-Fe | 4.62 | 36.5 | 0.77 | 2.8 ± 1.6 |
| 80 | FpvA-Pvd-Fe | 2.11 | 63.7 | 1.54 | |
| 200 | FpvA-Pvd-Fe | 1.68 | 175 | 20.1 | |
| 60 | FpvA-Pvd | 1.84 | 41.6 | 13.1 | 2.4 ± 0.8 |
| 70 | FpvA-Pvd | 2.96 | 68.7 | 23 |
In the case of FpvA and Pvd-Fe mixtures, the dissociation from TonBpp was found not to be complete, as some remaining complexes were observed during the late postinjection phase, which could be interpreted as a more stable population of TonBpp binding molecules. However, this was also observed during the postinjection phase with Pvd-Fe alone, while no binding was found for Pvd only (data not shown). These stable complexes observed for FpvA incubated with a 10-fold molar excess of Pvd-Fe are thus due to the excess of Pvd-Fe molecules, and a dose-dependent response could be observed on the TonB surface by varying the injected Pvd-Fe concentration (data not shown). Taken this into account, analysis of binding curves shows that dissociation rates are similar for FpvA, FpvA-Pvd, and FpvA-Pvd-Fe.
DISCUSSION
TonB is a key protein in the ferric siderophore uptake via outer membrane transporters. It couples the PMF of the inner membrane to the active transport of ferric siderophores across the outer membrane in gram-negative bacteria. How TonB acts is still a matter of conjecture. The P. aeruginosa and E. coli TonB proteins are highly homologous, except for an N-terminal extension which is present only in P. aeruginosa TonB1 (Fig. 1) and has been found to be essential for its activity in P. aeruginosa (54). To investigate the TonB interaction with the outer membrane siderophore receptor FpvA in P. aeruginosa, we cloned and expressed the full-length tonB1 gene and its C-terminal domain (Fig. 2). The TonBpp fragment encompasses the entire periplasmic domain but does not contain the membrane anchor domain that is essential for TonB activity. Iron transport assays using Pvd-55Fe demonstrate that the cloned TonB protein is fully functional (Fig. 3). The ferric siderophore was transported with the same efficiency in the ΔtonB strain PAD08 carrying plasmid pMMB-TonB as in the wild-type strain. On the other hand, the transport of Pvd-55Fe was markedly inhibited when the periplasmic TonBpp fragment was overproduced. This inhibition may be due to competition of TonBpp with the endogenous TonB for FpvA. Alternatively, if TonBpp is able to dimerize upon binding to its cognate receptor, analogous to the case for E. coli TonB, then TonBpp forms heterodimers with the endogenous encoded TonB, leading to a nonfunctional FpvA-TonB complex.
Different techniques were used to assess FpvA-TonBpp interactions. All of the results presented convincingly demonstrate that FpvA interacts in vitro with the periplasmic fragment of TonB. Binding of GST-TonBpp and FpvA on glutathione-coated agarose beads (Fig. 4), size exclusion chromatography of mixtures of FpvA and TonBpp (Fig. 5), and electrophoresis in native agarose gels (Fig. 6) all suggest the formation of a stable TonBpp-FpvA complex. In vivo, FpvA can be found in the outer membrane under three different loading statuses: FpvA, FpvA-Pvd, and FpvA-Pvd-Fe (47). All these FpvA forms are able to interact with TonBpp (Fig. 4, 5, 6, and 7). Since TonB activates the dissociation of Pvd from FpvA and the uptake of ferric-Pvd, it is not surprising that TonBpp interacts with FpvA-Pvd and FpvA-Pvd-Fe, but it is intriguing that TonBpp interacts with unloaded FpvA. However, this behavior has also been observed for the E. coli TonBpp-FhuA complex, in which the two proteins interact regardless of the presence of iron siderophore (25).
To obtain quantitative data about the FpvA-TonBpp interaction, the affinity constants for the interaction between TonBpp and FpvA were determined using SPR technology (Fig. 7). The results show that FpvA, FpvA-Pvd-Fe, and FpvA-Pvd, have similar 2 to 3 μM affinities for GST-TonBpp. Apparently, the binding of TonBpp to FpvA is independent of the presence of the Pvd siderophore. For the E. coli ferrichrome-FhuA system, ferrichrome enhances the total amount of complex but is not essential for its formation (25, 27). A Kd in the nanomolar range has been determined (25). Numerous different C-terminal fragments of E. coli TonB have been engineered, and the structures of the constructs containing amino acids (aa) 154 to 239, aa 164 to 239, and aa 103 to 239 have been solved by X-ray diffraction or by nuclear magnetic resonance (4, 27, 37). Structural and biochemical analyses of these different constructs show that the length of the construct has a large influence on its oligomerization state (aa 154 to 239 and aa 164 to 239 form two structurally different dimers, while the longer fragment stays monomeric) and how it binds to FhuA or other outer membrane receptors. The short C-terminal fragments form homodimers in solution and interact weakly with FhuA (25, 27). The longer fragments remain monomeric in solution and form only dimers when they interact with FhuA (25, 27). The experimental design to measure association between TonB and TonB-dependent receptors might thus be important, as illustrated with the different results obtained by Khursigara et al. (25, 26). Using different chip surfaces and TonB constructs for SPR experiments, it was shown that FhuA is able to bind two TonB molecules only if the immobilized TonB molecules are able to interact spatially and form a 2:1 TonB-FhuA complex. Furthermore, analytical ultracentrifugation studies showed that FhuA interacts in solution with TonB to form a 2:1 TonB-FhuA complex (25). The kinetics of binding of FpvA to immobilized TonBpp were best fit with a 1:1 Langmuir model. However, it was not possible to determine whether FpvA binds to one or two immobilized GST-TonBpp molecules based solely on the SPR data. Examination of the maximum binding capacities of FpvA on the different TonBpp-coated surfaces (Rmax) (Table 2) showed that the values are lower than would be expected for a 1:1 FpvA-TonBpp stoichiometry. Although this can be a consequence of a 1:2 stoichiometry, it could also be due to an inactive population of immobilized TonBpp. Further experiments are needed to precisely determine the stoichiometry of the FpvA-TonBpp complex.
It has been postulated that E. coli TonBpp undergoes some rearrangement leading to a higher-affinity state for FhuA: FhuA would bind a first TonB molecule with reduced affinity (Kd in the micromolar range) (25), followed by a rearrangement of this initial complex and then binding of a second TonB with a Kd in the nanomolar range (Kd,app = 25.7 and 11.5 nM in the absence or presence of ferrichrome iron, respectively) (26) Such a rearrangement is not evidenced with P. aeruginosa TonBpp, as we could not discriminate two different affinity states. The observed Kds for FpvA and TonBpp are all in the micromolar range (2.9, 2.4, and 2.8 μM without Pvd, with Pvd, and with Pvd-iron, respectively). Compared with the FhuA-TonB system, the binding of FpvA to TonBpp would reflect the first step of binding of FhuA to TonB but would lack the proposed rearrangement leading to the formation of a 2:1 complex with enhanced stability. A possible explanation would be that the fusion of GST with TonBpp affects the interaction with FpvA in such a way that this hypothetical rearrangement could not take place. It is clear that further experiments are needed for characterizing the interactions between FpvA and P. aeruginosa TonBpp. As seen from the structural similarities between FhuA (12, 33) and FpvA (7) and the high sequence identity between E. coli TonB and P. aeruginosa TonB1, it is expected that the same molecular events take place for the transport of iron from the extracellular side of the receptor to the periplasmic compartment.
The analysis of interaction between different fragments of E. coli TonB and FhuA, using different surfaces for TonB immobilization in SPR experiments, led to the conclusion that TonB possesses two distinct binding regions: one in the C terminus of the protein, for which binding to FhuA is ferrichrome independent, and a higher-affinity region outside the C terminus, for which ferrichrome enhances interactions with FhuA (25, 26). By using an N-terminal deletion mutant of FhuA, it was further shown that FhuA contains multiple TonB binding sites (26). Such deletion mutants of FpvA and/or P. aeruginosa TonB are not yet available, and the FpvA-TonB interaction cannot be described as detailed. The main difference observed between P. aeruginosa GST-TonBpp and FpvA and E. coli TonBpp and FhuA reside in their ability to form a stable complex: as evidenced with SPR experiments, FhuA and E. coli TonBpp interact to form a stable complex, with stability enhanced by the presence of ferrichrome, while FpvA and P. aeruginosa TonBpp dissociate rapidly and completely, independently of the presence of Pvd-iron. Our results do not show evidence for the second binding site of higher affinity that is described for FhuA and E. coli TonBpp.
In conclusion we have shown that the soluble periplasmic C-terminal construct of P. aeruginosa TonB1 is able to bind to the receptor FpvA in vitro. SPR experiments show that FpvA binds to TonBpp with micromolar affinities and that this binding is independent of the presence of Pvd, loaded or not with iron. However, all these experiments have been performed in vitro, using fragments of TonB which are unable to promote the active transport of siderophore. The fact that Pvd-iron does not influence the interaction between FpvA and TonBpp could be due to the absence of the ExbB and ExbD proteins and PMF. The next experimental challenge would be to isolate a functional TonB machinery and then assess the binding of outer membrane receptors in the presence and absence of the PMF.
Acknowledgments
We thank Renaud Wagner and Juliette Kempf for their helpful suggestions regarding the cloning procedures.
This work was supported by EU grant HPRN-CT-2000-00075 from the European Community.
REFERENCES
- 1.Ahmer, B. M., M. G. Thomas, R. A. Larsen, and K. Postle. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 177:4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 3.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96:10673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, C., A. Mooser, A. Plückthun, and A. Wlodawer. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276:27535-27540. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, T. E., L. W. Tari, and H. J. Vogel. 2001. Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem. 1:7-30. [DOI] [PubMed] [Google Scholar]
- 6.Clement, E., P. J. Mesini, F. Pattus, and I. J. Schalk. 2004. The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry 43:7954-7965. [DOI] [PubMed] [Google Scholar]
- 7.Cobessi, D., H. Celia, N. Folschweiller, I. J. Schalk, M. A. Abdallah, and F. Pattus. 2005. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. J. Mol. Biol. 347:121-134. [DOI] [PubMed] [Google Scholar]
- 8.Demange, P., A. Bateman, C. Mertz, A. Dell, Y. Piemont, and M. A. Abdallah. 1990. Bacterial siderophores: structures of pyoverdins Pt, siderophores of Pseudomonas tolaasii NCPPB 2192, and pyoverdins Pf, siderophores of Pseudomonas fluorescens CCM 2798. Identification of an unusual natural amino acid. Biochemistry 29:11041-11051. [DOI] [PubMed] [Google Scholar]
- 9.Endriss, F., M. Braun, H. Killmann, and V. Braun. 2003. Mutant analysis of the Escherichia coli FhuA protein reveals sites of FhuA activity. J. Bacteriol. 185:4683-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson, A. D., and J. Deisenhofer. 2002. TonB-dependent receptors—structural perspectives. Biochim. Biophys. Acta 1565:318-332. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 13.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, E., K. Günter, and V. Braun. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 171:5127-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, J., and K. Postle. 2005. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol. Microbiol. 55:276-288. [DOI] [PubMed] [Google Scholar]
- 16.Hoegy, F., H. Celia, G. L. Mislin, M. Vincent, J. Gallay, and I. J. Schalk. 2005. Binding of iron-free siderophore, a common feature of siderophore outer membrane transporters of Escherichia coli and Pseudomonas aeruginosa. J. Biol. Chem. 280:20222-20230. [DOI] [PubMed] [Google Scholar]
- 17.Holloway, B. W. 1969. Genetics of Pseudomonas. Bacteriol. Rev. 33:419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard, S. P., C. Herrmann, C. W. Stratilo, and V. Braun. 2001. In vivo synthesis of the periplasmic domain of TonB inhibits transport through the FecA and FhuA iron siderophore transporters of Escherichia coli. J. Bacteriol. 183:5885-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, B., K. Ru, Z. Yuan, C. B. Whitchurch, and J. S. Mattick. 2004. tonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J. Bacteriol. 186:4387-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadner, R. J. 1990. Vitamin B12 transport in. Escherichia coli: energy coupling between membranes. Mol. Microbiol. 4:2027-2033. [DOI] [PubMed] [Google Scholar]
- 21.Kampfenkel, K., and V. Braun. 1992. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 174:5485-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 109:137-141. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8:389-396. [DOI] [PubMed] [Google Scholar]
- 24.Khursigara, C. M., G. De Crescenzo, P. D. Pawelek, and J. W. Coulton. 2005. Deletion of the proline-rich region of TonB disrupts formation of a 2:1 complex with FhuA, an outer membrane receptor of Escherichia coli. Protein Sci. 14:1266-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khursigara, C. M., G. De Crescenzo, P. D. Pawelek, and J. W. Coulton. 2004. Enhanced binding of TonB to a ligand-loaded outer membrane receptor: role of the oligomeric state of TonB in formation of a functional FhuA. TonB complex. J. Biol. Chem. 279:7405-7412. [DOI] [PubMed] [Google Scholar]
- 26.Khursigara, C. M., G. De Crescenzo, P. D. Pawelek, and J. W. Coulton. 2005. Kinetic analyses reveal multiple steps in forming TonB-FhuA complexes from Escherichia coli. Biochemistry 44:3441-3453. [DOI] [PubMed] [Google Scholar]
- 27.Ködding, J., P. Howard, L. Kaufmann, P. Polzer, A. Lustig, and W. Welte. 2004. Dimerization of TonB is not essential for its binding to the outer membrane siderophore receptor FhuA of Escherichia coli. J. Biol. Chem. 279:9978-9986. [DOI] [PubMed] [Google Scholar]
- 28.Ködding, J., F. Killig, P. Polzer, S. P. Howard, K. Diederichs, and W. Welte. 2005. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J. Biol. Chem. 280:3022-3028. [DOI] [PubMed] [Google Scholar]
- 29.Koebnik, R. 2005. TonB-dependent trans-envelope signaling: the exception or the rule? Trends Microbiol. 13:343-347. [DOI] [PubMed] [Google Scholar]
- 30.Larsen, R. A., T. E. Letain, and K. Postle. 2003. In vivo evidence of TonB shuttling between the cytoplasmic and outer membrane in Escherichia coli. Mol. Microbiol. 49:211-218. [DOI] [PubMed] [Google Scholar]
- 31.Larsen, R. A., P. S. Myers, J. T. Skare, C. L. Seachord, R. P. Darveau, and K. Postle. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178:1363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809-1824. [DOI] [PubMed] [Google Scholar]
- 33.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 34.Moeck, G. S., J. W. Coulton, and K. Postle. 1997. Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor fhua promotes interaction with the energy-transducing protein TonB. J. Biol. Chem. 272:28391-28397. [DOI] [PubMed] [Google Scholar]
- 35.Moeck, G. S., and L. Letellier. 2001. Characterization of in vitro interactions between a truncated TonB protein from Escherichia coli and the outer membrane receptors FhuA and FepA. J. Bacteriol. 183:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 37.Peacock, S. R., A. M. Weljie, S. Peter Howard, F. D. Price, and H. J. Vogel. 2005. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J. Mol. Biol. 345:1185-1197. [DOI] [PubMed] [Google Scholar]
- 38.Poole, K., S. Neshat, K. Krebes, and D. E. Heinrichs. 1993. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J. Bacteriol. 175:4597-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole, K., Q. Zhao, S. Neshat, D. E. Heinrichs, and C. R. Dean. 1996. The Pseudomonas aeruginosa tonB gene encodes a novel TonB protein. Microbiology 142:1449-1458. [DOI] [PubMed] [Google Scholar]
- 40.Postle, K. 1993. TonB protein and energy transduction between membranes. J. Bioenerg. Biomembr. 25:591-601. [DOI] [PubMed] [Google Scholar]
- 41.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 42.Postle, K., and J. T. Skare. 1988. Escherichia coli TonB protein is exported from the cytoplasm without proteolytic cleavage of its amino terminus. J. Biol. Chem. 263:11000-11007. [PubMed] [Google Scholar]
- 43.Rich, R. L., and D. G. Myszka. 2000. Advances in surface plasmon resonance biosensor analysis. Curr. Opin. Biotechnol. 11:54-61. [DOI] [PubMed] [Google Scholar]
- 44.Sauter, A., S. P. Howard, and V. Braun. 2003. In vivo evidence for TonB dimerization. J. Bacteriol. 185:5747-5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schalk, I. J., C. Hennard, C. Dugave, K. Poole, M. A. Abdallah, and F. Pattus. 2001. Iron-free pyoverdin binds to its outer membrane receptor FpvA in Pseudomonas aeruginosa: a new mechanism for membrane iron transport. Mol. Microbiol. 39:351-360. [DOI] [PubMed] [Google Scholar]
- 46.Schalk, I. J., P. Kyslik, D. Prome, A. van Dorsselaer, K. Poole, M. A. Abdallah, and F. Pattus. 1999. Copurification of the FpvA ferric pyoverdin receptor of Pseudomonas aeruginosa with its iron-free ligand: implications for siderophore-mediated iron transport. Biochemistry 38:9357-9365. [DOI] [PubMed] [Google Scholar]
- 47.Schalk, I. J., W. W. Yue, and S. K. Buchanan. 2004. Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol. Microbiol. 54:14-22. [DOI] [PubMed] [Google Scholar]
- 48.Skare, J. T., B. M. Ahmer, C. L. Seachord, R. P. Darveau, and K. Postle. 1993. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302-16308. [PubMed] [Google Scholar]
- 49.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 50.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Requirement of the Pseudomonas aeruginosa tonB gene for high-affinity iron acquisition and infection. Infect. Immun. 68:4498-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiener, M. C. 2005. TonB-dependent outer membrane transport: going for Baroque? Curr. Opin. Struct. Biol. 15:394-400. [DOI] [PubMed] [Google Scholar]
- 52.Wilson, R., and R. B. Dowling. 1998. Lung infections. 3. Pseudomonas aeruginosa and other related species. Thorax 53:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wooldridge, K. G., and P. H. Williams. 1993. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 12:325-348. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, Q., and K. Poole. 2002. Mutational analysis of the TonB1 energy coupler of Pseudomonas aeruginosa. J. Bacteriol. 184:1503-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, Q., and K. Poole. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol. Lett. 184:127-132. [DOI] [PubMed] [Google Scholar]