Abstract
We identified an N-terminal amphipathic helix (AH) in one of hepatitis C virus (HCV)’s nonstructural proteins, NS5A. This AH is necessary and sufficient for membrane localization and is conserved across isolates. Genetically disrupting the AH impairs HCV replication. Moreover, an AH peptide-mimic inhibits the membrane association of NS5A in a dose-dependent manner. These results have exciting implications for the HCV life cycle and novel antiviral strategies.
Hepatitis C virus (HCV) is a significant cause of morbidity and mortality, infecting over 100 million people worldwide (1, 9). Despite recent progress, current therapies remain inadequate for the majority of patients (27, 29, 45). HCV is a positive, single-stranded RNA virus. Its 9.6-kb genome encodes a single ∼3,000-amino-acid polyprotein which is proteolytically processed by cellular and viral proteinases into structural (components of the mature virus) and nonstructural (NS) (elements proposed to help replicate new virions) proteins (2, 6, 33). Like other plus-strand RNA viruses, HCV is thought to replicate its RNA in association with cytoplasmic membranes (4, 8, 12, 25, 34), although how the RNA replication complex is assembled and maintained remains unknown. That NS5A, one of the NS proteins of HCV, may play a key role in membrane-associated RNA replication is suggested by its apparent association with host cell membranes (17, 30, 37). NS5A has also been reported to interact with a variety of host cell proteins (13, 41, 44) and to determine the response to interferon therapy in some patients (11), although to date its precise role in HCV replication is not clear.
The study of HCV replication has been hampered by the lack of a convenient cell culture system. The recent advent of high-efficiency HCV subgenomic replicons (5), however, now opens the prospect of performing detailed molecular genetic studies. Such replicons, based on the original report by Lohmann et al. (28), contain all the cis and trans elements required for HCV RNA replication and should allow an analysis of structure-function relationships of engineered HCV mutants.
In the hope of further characterizing the role of NS5A in the HCV life cycle and identifying potential novel targets for antiviral therapy, we have been studying the cell biology of this NS protein. We were particularly interested in its mechanism of membrane association since it has no obvious transmembrane or endoplasmic reticulum (ER)-targeting domains (32). Here we report the identification of a key mechanism of the membrane association of NS5A and show that disrupting this mechanism abolishes HCV RNA replication.
Disruption of the N-terminal AH of NS5A abolishes membrane localization.
To study the determinants of NS5A intracellular localization, we first expressed NS5A in Huh-7 cells (a liver-derived cell line) by using the vaccinia virus expression system and a vector that encodes most of the HCV NS proteins (16). The cells were fixed with 4% formaldehyde and stained with a monoclonal antibody against NS5A (Virostat, Portland, Maine) and a Texas red-labeled anti-mouse secondary antibody (15). The staining pattern of NS5A in the transfected cells shows a cytoplasmic localization characteristic of membrane-associated proteins (Fig. 1A, middle panel). A perinuclear punctate vesicular staining suggestive of a Golgi-like intracellular distribution pattern was readily observed, as was, occasionally, a reticular chicken wire-like staining pattern, characteristic of the ER. Both patterns have been reported previously when NS5A is expressed either alone or in combination with other HCV NS proteins in a variety of expression systems (18, 21, 30, 37). As mentioned previously, although these findings strongly suggest that NS5A is associated with intracellular membrane compartments, no obvious transmembrane domains, ER retention domains, or evidence of lipid modifications—features commonly responsible for membrane association of proteins—have been reported.
FIG. 1.
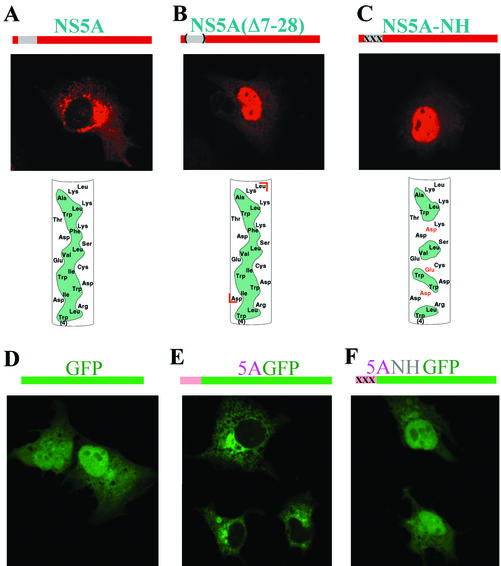
Disruption of the hydrophobic face of the N-terminal AH of NS5A abolishes the intracellular membrane localization pattern of NS5A. (A) (Top) The region of NS5A found to harbor an AH is shown in gray (above the picture) and in more detail in the bottom panel. (Middle) Huh-7 cells were infected with a vaccinia virus expressing T7 RNA polymerase and then transfected with plasmid pBRTM/HCV 827-3011 containing the HCV NS proteins downstream of a T7 promoter. Expression of NS5A was detected by indirect immunofluorescence using a monoclonal antibody against NS5A and a Texas red-labeled anti-mouse secondary antibody. (Bottom) α-Helix net diagram of the N terminus of NS5A wherein the cylindrical α-helical segment is “cut” longitudinally along one face and then “flattened” into the plane of the page. The amino acid sequence of NS5A from amino acids 4 to 27 in the N-terminal to C-terminal direction is shown. Hydrophobic amino acids in the amphipathic helix are shaded in green. Note the long, continuous stretch of such amino acids along one face of the helix, defining its amphipathic nature. (B) Same as for panel A except that plasmid 827/5AΔ7-28, which harbors a mutation of NS5A wherein the AH segment has been deleted (deleted amino acids shown between red brackets in bottom panel), was substituted for pBRTM/HCV 827-3011. (C) Same as for panel A except that plasmid 827/5ANH, wherein mutations which disrupt the hydrophobic nature of the AH (depicted in the bottom panel) were engineered into NS5A, was used for transfection. (D) Cells transfected with plasmid T7-GFP, which expresses wild-type GFP, a non-membrane-associated protein of 236 amino acids (schematically represented in the top panel). It is diffusely distributed throughout the cytoplasm and nucleus. (E) Cells transfected with plasmid T7-5AGFP, which expresses a fusion protein wherein the N-terminal segment of NS5A (containing the first 31 of the 448 amino acids of the full- length protein) which harbors the AH is grafted onto GFP (schematically represented in the top panel). The fusion protein displays an intracellular localization of a membrane-associated protein. (F) Cells transfected with plasmid T7-5ANHGFP, which is identical to T7-5AGFP except that the AH in the NS5A segment has been disrupted with the same mutations as in panel C (schematically represented in the top panel). This mutant displays a localization pattern similar to that of the native GFP.
Inspection of the extreme N-terminal amino acid sequence of NS5A revealed the presence of a potential amphipathic α-helix (36) (Fig. 1A, bottom panel). Preliminary deletion analysis suggested that the membrane localization is mediated by the N-terminal portion of NS5A (results not shown; consistent with results in references 20 and 39). We hypothesized that the N-terminal amphipathic helix (AH) might be responsible for mediating the membrane association of NS5A. As a first test of this hypothesis, we used PCR mutagenesis (14) to construct a mutant of pBRTM/HCV 827-3011, termed 827/5AΔ7-28, wherein the predicted AH of NS5A was deleted. This mutation resulted in a dramatic change in the intracellular distribution of NS5A (Fig. 1B, middle panel). Cytoplasmic membrane localization was completely abolished and replaced predominantly by a nuclear localization pattern.
We interpreted this result as being consistent with activation of a previously described cryptic nuclear localization signal (NLS) located within the C-terminal domain of NS5A (20). The NLS presumably became functional after the loss of the normal cytoplasmic membrane-targeting sequence of NS5A, which apparently lies within the deleted segment. These results are similar to those recently reported with an NS5A mutant harboring a larger N-terminal deletion consisting of the first 45 amino acids (7).
To test the hypothesis that it is indeed the amphipathic nature of the deleted putative helix which mediates intracellular localization of NS5A, the hydrophobic face of the helix was disrupted by site-specific PCR mutagenesis. Our strategy was to introduce three charged amino acids spaced at intervals along the predicted N-terminal α-helix such that no sustained hydrophobic patch remained (Fig. 1C, bottom panel). The primer 5ANH (5′-TCCGGCTCCTGGCTAAGGGACGACTGGGACTGGGAATGCGAGGTGCTGAGCGACGATAAGACC-3′) was used to change the coding sequence for isoleucine-8, isoleucine-12, and phenylalanine-19 of NS5A to encode aspartate, glutamate, and aspartate, respectively. On expression of this mutant, termed 827/5ANH, in Huh-7 cells, complete loss of the characteristic membrane localization pattern of NS5A was observed (Fig. 1C, middle panel). Again, as seen for the mutant lacking the entire helix, nuclear localization was the predominant pattern observed. Interestingly, in both NS5A mutants, a few cells were occasionally noted in which the sharp nuclear localization pattern was replaced by diffuse staining throughout the cell (data not shown). Even then, however, no characteristic localization to cytoplasmic membrane structures was observed. Perhaps these cells represented a different phase of the cell cycle since no attempt had been made to synchronize cell division in any of our experiments. We conclude that the predicted N-terminal AH is necessary for the normal cytoplasmic membrane localization of NS5A.
We next sought to test the hypothesis that the N-terminal AH of NS5A is also sufficient to confer cytoplasmic membrane localization. For this, we first constructed a plasmid, T7GFP, in which the HCV-encoding sequences of pBRTM/HCV 827-3011 were replaced with the gene for the jellyfish Aequorea victoria green fluorescent protein (GFP), a non-membrane-associated protein (43). Next we modified plasmid T7GFP to construct plasmid T75AGFP, in which the first 31 amino acids of NS5A containing the AH are fused in frame to the N terminus of GFP. On expression in Huh-7 cells, GFP displays a diffuse cytoplasmic distribution pattern and also readily enters the nucleus (Fig. 1D). Grafting the N-terminal AH of NS5A onto GFP results in a dramatic change in intracellular distribution: the fusion protein can no longer enter the nucleus and appears restricted to cytoplasmic membrane structures (Fig. 1E). A similar finding has been recently reported (7). To determine the critical element within the grafted NS5A segment responsible for conferring the membrane association, we made plasmid T75ANHGFP, in which we introduced into the fusion protein the same mutations as in Fig. 1C, designed to destroy the hydrophobic face of the AH. As shown in Fig. 1F, disrupting the amphipathic nature of the NS5A helix portion of the fusion protein restored the distribution pattern characteristic of GFP. These results argue for the notion that the AH has an independent function—namely, membrane targeting— as opposed to being simply a part of NS5A crucial for proper folding.
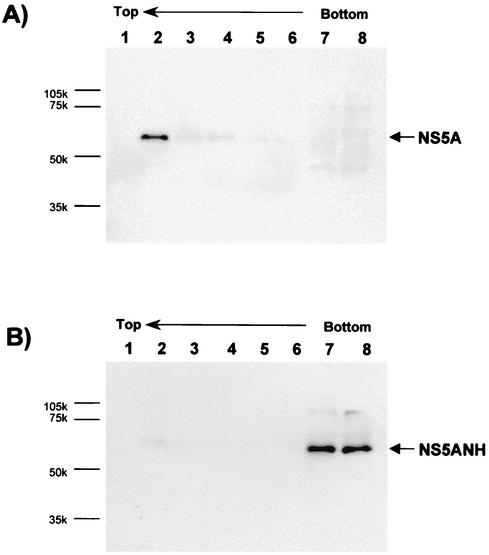
We next wished to complement the above morphological data with biochemical evidence. To determine the effect of disrupting the amphipathic nature of the N-terminal α-helix of NS5A on the membrane association of NS5A, we performed membrane flotation assays (26) on cells expressing wild-type and AH mutant NS5A. Whole-cell extracts from Huh-7 cells transfected with pBRTM/HCV 827-3011 or 827/5ANH were overlaid with a 5 to 40% OptiPrep (Sigma) step density gradient and subjected to ultracentrifugation for 4 h at 40,000 × g in an SW60 rotor. Fractions were collected from the top, and the proteins in each gradient fraction were precipitated with methanol-chloroform and analyzed by Western blotting for NS5A using a monoclonal antibody against NS5A and a horseradish peroxidase-labeled anti-mouse secondary antibody. In such assays, non-membrane-associated proteins remain at the bottom of the gradient whereas membranes—and associated proteins—“float” toward less dense gradient fractions present at the top. As shown in Fig. 2, wild-type NS5A quantitatively floated with the membrane fraction (Fig. 2A) whereas NS5A with a genetically disrupted AH did not (Fig. 2B). Moreover, these Western blots show that this is not simply a function of the mutant NS5A being expressed at a lower level than the wild-type protein. Finally, both NS5B and NS4B have been reported to each be independently targeted to membranes (19, 35) and are thought to interact with NS5A as part of a multiprotein complex (23, 38). However, as shown in Fig. 1C and 2B, neither of these potential interactions appears to be sufficient to keep AH-mutated NS5A on the membranes.
FIG. 2.
An intact AH is required for biochemical association of NS5A with membranes. Huh-7 cells were infected with a vaccinia virus expressing T7 RNA polymerase and then transfected with plasmid pBRTM/HCV 827-3011 (A) or 827/5ANH (B), which expresses wild-type or an AH mutant of NS5A, respectively. Cell extracts were analyzed by membrane flotation assays for membrane association of NS5A by overlaying the extracts with an OptiPrep gradient (5 to 40%) and subjecting them to ultracentrifugation. Non-membrane-associated proteins remain at the bottom (right side) of the gradient, whereas membranes and associated proteins float toward the less dense gradient fractions present at the top (left side). Numbers correspond to OptiPrep gradient fractions, which were analyzed by Western blotting using a monoclonal antibody to NS5A as the probe.
Genetic disruption of the NS5A AH impairs HCV RNA replication.
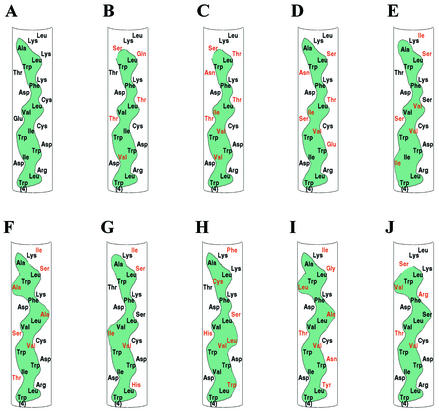
The above experiments identified an essential structural element capable of mediating the membrane association of NS5A when expressed in cultured cells. To assess the possible importance of this element in natural infections of HCV, we examined the predicted amino acid sequences of the N terminus of NS5A contained in isolates obtained from a variety of patients and representing multiple genotypes. As shown in Fig. 3, while there was considerable variability in the specific amino acid sequence of this region among isolates, the amphipathic nature of the predicted α-helix was preserved. Moreover, when a similar analysis was extended to all sequenced isolates currently available in public databases, the same result was observed (7; Y. Yang and J. Glass, personal communication). This suggests that such a structural motif is indeed important for some essential aspect of the HCV life cycle.
FIG. 3.
The AH of NS5A is conserved across HCV isolates. Amino acid sequences of the AH of NS5A obtained from sequenced isolates of HCV from around the world and representing a variety of genotypes are shown. Note that although there are differences in particular amino acids (shown in red) compared to a reference sequence (A), the amphipathic nature of the helix (indicated by the green hydrophobic continuous band) is conserved. (A) Genotype 1a (accession no. AF009606), (B) genotype 1b (accession no. P26663), (C) genotype 2a (accession no. P26660), (D) genotype 2b (accession no. AB030907), (E) genotype 3a-K (accession no. D28917), (F) genotype 3a-NZL (accession no. D17763), (G) genotype 3b (accession no. D49374), (H) genotype 4a (accession no. Y11604), (I) genotype 10a (accession no. D63821), (J) genotype 11a (accession no. D63822).
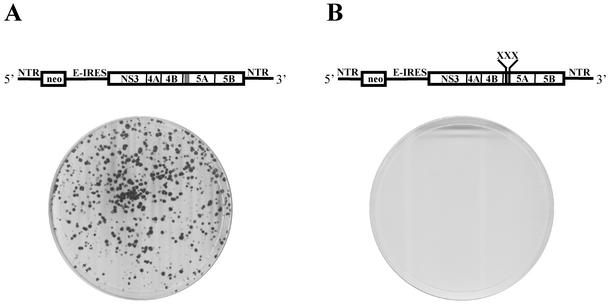
The dramatic relocalization of NS5A to the nucleus after disruption of the amphipathic nature of its N-terminal helix and the strict preservation of this motif in all known HCV isolates suggest that disruption of the AH may have significant consequences for HCV RNA replication. To test this hypothesis, we constructed high-efficiency second-generation bicistronic subgenomic RNA replicons of HCV (5) harboring a neomycin resistance gene along with an HCV polyprotein containing NS5A with either a wild-type or mutated N-terminal AH (Fig. 4, top panels). The RNA replication efficiency of these replicons was then assayed on the basis of their ability to establish G418-resistant colonies after transfection of Huh-7 cells. As shown in Fig. 4, while a replicon with a wild-type NS5A N terminus gave rise to numerous colonies, as previously observed (5), disruption of the amphipathic nature of the N-terminal helix of NS5A resulted in a dramatic inhibition of HCV genome replication. It is unlikely that these results are due to an increased cytotoxicity associated with the mutant NS5A, since we observed no decreased transfection efficiency or ability to establish colonies by using plasmids encoding a drug resistance marker along with just the wild-type or mutant NS5A proteins (data not shown). It is difficult to suggest that NS5A simply cannot tolerate mutations, since a much larger alteration (47 deleted amino acids) than our three point mutations was tolerated without any effect on RNA replication (5). Nor are these results likely to be due to disruption of a critical RNA structural motif, since multiple mutations appear to be tolerated within the mutated segment of NS5A (including wobble mutations at the amino acid codons mutated in NS5ANH [Fig. 3]). Rather, it is the altered intracellular targeting of NS5A that is most clearly associated with the profound inhibition of HCV RNA replication.
FIG. 4.
Disrupting the amphipathic nature of the 5A helix impairs replication of HCV subgenomic replicons. (Top) Schematic representation of HCV high-efficiency subgenomic replicons used for HCV replication assays. (A) Bart79I, a high-efficiency subgenomic replicon of HCV harboring the neomycin resistance gene (neo) and the HCV NS proteins. (5′NTR and 3′NTR represent the nontranslated regions at the end of the HCV genomic RNA, which contain presumed recognition sequences for the viral replication machinery. E-IRES, encephalomyocarditis internal ribosome entry site.) (B) The amphipathic helix-disrupting mutations of Fig. 1C were introduced into the Bart79I replicon, yielding the depicted Bart5X79I. (Bottom) The above replicons were electroporated into Huh-7 cells, and neomycin-resistant colonies were selected and stained with crystal violet. Each dot represents a colony of Huh-7 cells that was able to grow in the presence of neomycin due to the presence of efficiently replicating intracellular replicons.
Because the three mutations completely disrupting the amphipathic nature of the NS5A AH contained in Bart5X79I resulted in no HCV RNA replication, we hypothesized that a partial disruption of the hydrophobic face of the AH might be associated with an intermediate level of replication. Indeed, while Bart79I with a wild-type AH had a ∼3% G418 transduction efficiency and Bart5X79I gave rise to no colonies, when just one of the mutations illustrated in Fig. 1C (I12E) was introduced into the Bart79I replicon, a G418 transduction efficiency of ∼0.0003% was observed (data not shown). This 4-log-unit-reduced level of replication is comparable to that observed with the first-generation HCV replicons without adaptive mutations (5, 28). A more complete description of a larger collection of such intermediate mutants will be presented elsewhere.
Most, if not all, plus-strand RNA viruses appear to replicate in association with cytoplasmic membranes (4, 8, 12, 25, 34). The detailed mechanisms of RNA replication complex assembly, however, are not known. The AH of NS5A might play a role in localizing, inducing, or maintaining such membrane-associated replication complexes and may be an example of a more general theme. For example, the NS protein 2C of both poliovirus and hepatitis A virus also contains a membrane-associating AH (24, 42), which, at least for poliovirus, appears to play a role in RNA synthesis (31) and the induction of membrane rearrangements derived from the host ER and Golgi (40) on which replication complexes are localized (3). In this regard, the HCV NS4B protein has also recently been associated with the induction of novel intracellular membrane structures hypothesized to be the site of HCV RNA replication (10). It is interesting that we also have found an AH within NS4B, which will be presented elsewhere.
Pharmacologic disruption of NS5A AH-mediated membrane association.
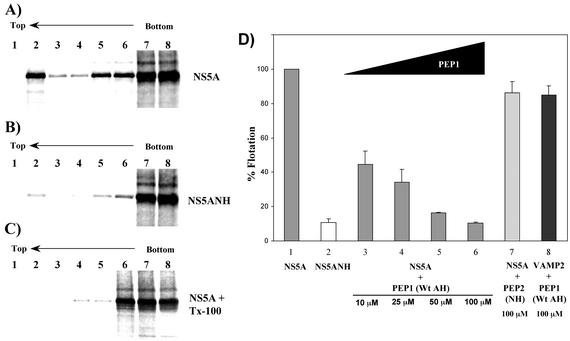
Finally, because genetically mutating the ability of the AH to associate with membranes disrupts RNA replication efficiently, we hypothesized that the AH-mediated membrane association of NS5A might also be amenable to pharmacologic disruption and thereby form the basis for a novel approach to anti-HCV therapy. To test this hypothesis, we used a modified version of the membrane flotation assay to evaluate the ability of a first-generation inhibitor to competitively block the association of NS5A with host cell membranes. Because NS5A associates with membranes in a posttranslational manner (7), aliquots of in vitro-translated and 35S-labeled NS5A proteins prepared using the TNT reticulocyte lysate kit (Promega, Madison, Wis.) were combined with a membrane fraction derived from Huh-7 cells, with or without synthetic peptides, and overlaid with a 5 to 40% OptiPrep step gradient. Following ultracentrifugation, the proteins in each gradient fraction were precipitated with methanol-chloroform and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The percentage of protein floating to the top of the gradient with the membrane fraction (fraction 2 from the top) was then quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Similar to the results shown in Fig. 2, wild-type NS5A specifically floated with the low-density gradient membrane fraction (Fig. 5A). In contrast, when NS5A contained the mutations (depicted in Fig. 1C) that disrupt the amphipathic nature of the N-terminal helix, minimal flotation was observed (Fig. 5B). As expected, if the cell extracts were treated with the detergent Triton X-100 to dissolve the target membranes prior to gradient assembly, no flotation of NS5A was observed (Fig. 5C). These results are in good agreement with the morphologic and biochemical evidence for AH-mediated membrane association (Fig. 1 and 2), especially considering the potential limitations of this type of in vitro assay (22). Moreover, as shown in Fig. 5D, a synthetic peptide (PEP1) designed to mimic the wild-type AH not only inhibited membrane association of NS5A but also did so in a dose-dependent manner. A control peptide (PEP2) mimicking the mutated AH had no such effect on NS5A membrane association. An additional measure of specificity against NS5A was provided by testing the effect of PEP1 on the membrane association of VAMP2, a control host cell protein whose membrane association is mediated by its own AH in a posttranslational fashion (22). The precise mechanistic details of how the peptide exerts its inhibition remains to be determined, although a variety of interesting possibilities can be envisaged. For example, the host cell membrane receptor for NS5A may be composed of a specific protein—presumably different from that used by VAMP2—which can be specifically blocked by PEP1 but not by the control peptide, PEP2. Alternatively, PEP1 may somehow perturb the lipid membrane itself so as to alter efficient binding by NS5A. Finally, PEP1 might directly bind to NS5A and thereby prevent the latter from efficient membrane binding. Experiments are being performed to help distinguish between these models. Whatever the precise mechanism, however, the maximal extent of NS5A inhibition achieved pharmacologically is comparable to that obtained by genetic mutation of the AH in NS5A. In addition, unlike current treatments for HCV, the synthetic peptide appears equally effective against NS5A derived from different genotypes (data not shown), including those most refractory to current therapies. This convenient membrane flotation assay has also proven well suited for current efforts focused on studying the mechanistic details of the AH membrane-targeting domain and ideal for guiding ongoing development of peptidomimetic compounds designed to resemble key elements, or bind to specific features, of the AH. Such compounds represent an exciting potential addition to current anti-HCV combination therapy regimens.
FIG. 5.
Pharmacologic disruption of AH-mediated membrane association of NS5A. (A and B) Membrane association of NS5A (containing a wild-type AH) (A) and NS5ANH (NS5A with a genetically mutated AH) (B) proteins was analyzed by in vitro membrane flotation assays. Numbers correspond to OptiPrep gradient (5 to 40%) fractions. Non-membrane-associated proteins remain at the bottom (right side) of the gradient, whereas membranes and associated proteins float toward the less dense gradient fractions present at the top (left side). (C) Same as panel A except that the reaction mixture was treated with the detergent Triton X-100 to dissolve the membranes prior to gradient assembly. (D) Pharmacologic inhibition of NS5A membrane association. Huh-7 cell-derived membranes were treated with the indicated concentration of PEP1 (SGSWLRDVWDWICTVLTDFKTWLQSKLDYKD), a peptide mimicking the wild-type amphipathic helix of NS5A, or PEP2 (SGSWLRDDWDWECTVLTDDKTWLQSKLDYKD), a control peptide mimicking the NS5ANH mutant amphipathic helix, or mock treated with water and incubated with in vitro-translated NS5A, NS5ANH, or VAMP2 before being analyzed by membrane flotation assays. The percentage of NS5A, NS5ANH, or VAMP2 floating to the top of the gradient with the membrane fraction (fraction 2 from the top) under the indicated conditions was quantified using a PhosphorImager and expressed relative to the mock-treated control. Error bars represent standard error of the mean.
Acknowledgments
M.E. and K.H.C. contributed equally to this work.
We thank Ellie Ehrenfeld and Karla Kirkegaard for helpful discussions and critical reading of the manuscript and David Andrews for providing the VAMP2 clone.
M.E. is the recipient of the ALF Postdoctoral Research Fellowship Award and Stanford University Dean's Fellowship. J.S.G. is supported by an Amgen/AASLD American Liver Foundation Award and a Burroughs Wellcome Fund Career Award. This work was also supported in part by the Eli Lilly-Stanford University HCV initiative, a pilot study award from the Stanford Digestive Disease Center, a Veterans Administration REAP Award, and an NIH U19 award.
REFERENCES
- 1.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1980. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology 100:390-399. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Branch, A. D. 2000. Hepatitis C virus RNA codes for proteins and replicates: does it also trigger the interferon response? Semin. Liver Dis. 20:57-68. [DOI] [PubMed] [Google Scholar]
- 7.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 8.Chu, P. W., and E. G. Westaway. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177-191. [DOI] [PubMed] [Google Scholar]
- 9.Di Bisceglie, A. M. 2000. Natural history of hepatitis C: its impact on clinical management. Hepatology 31:1014-1018. [DOI] [PubMed] [Google Scholar]
- 10.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. [DOI] [PubMed] [Google Scholar]
- 12.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale, M. J. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 14.Glenn, J. S., J. C. Marsters, Jr., and H. B. Greenberg. 1998. Use of a prenylation inhibitor as a novel antiviral agent. J. Virol. 72:9303-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glenn, J. S., J. M. Taylor, and J. M. White. 1990. In vitro-synthesized hepatitis delta virus RNA initiates genome replication in cultured cells. J. Virol. 64:3104-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. USA 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, Y., Y. Uchiyama, T. Fujimura, H. Kanamori, T. Doi, A. Takamizawa, T. Hamakubo, and T. Kodama. 2001. A human hepatoma cell line expressing hepatitis C virus nonstructural proteins tightly regulated by tetracycline. Biochem. Biophys. Res. Commun. 281:732-740. [DOI] [PubMed] [Google Scholar]
- 19.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 20.Ide, Y., L. Zhang, M. Chen, G. Inchauspe, C. Bahl, Y. Sasaguri, and R. Padmanabhan. 1996. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene 182:203-211. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. E., W. K. Song, K. M. Chung, S. H. Back, and S. K. Jang. 1999. Subcellular localization of hepatitis C viral proteins in mammalian cells. Arch. Virol. 144:329-343. [DOI] [PubMed] [Google Scholar]
- 22.Kim, P. K., C. Hollerbach, W. S. Trimble, B. Leber, and D. W. Andrews. 1999. Identification of the endoplasmic reticulum targeting signal in vesicle- associated membrane proteins. J. Biol. Chem. 274:36876-36882. [DOI] [PubMed] [Google Scholar]
- 23.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusov, Y. Y., C. Probst, M. Jecht, P. D. Jost, and V. Gauss-Muller. 1998. Membrane association and RNA binding of recombinant hepatitis A virus protein 2C. Arch. Virol. 143:931-944. [DOI] [PubMed] [Google Scholar]
- 25.Lazarus, L. H., and R. Barzilai. 1974. Association of foot-and-mouth disease virus replicase with RNA template and cytoplasmic membranes. J. Gen. Virol. 23:213-218. [DOI] [PubMed] [Google Scholar]
- 26.Lecat, S., P. Verkade, C. Thiele, K. Fiedler, K. Simons, and F. Lafont. 2000. Different properties of two isoforms of annexin XIII in MDCK cells. J. Cell Sci. 113:2607-2618. [DOI] [PubMed] [Google Scholar]
- 27.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 29.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, and the Hepatitis Interventional Therapy Group. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 30.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 31.Paul, A. V., A. Molla, and E. Wimmer. 1994. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology 199:188-199. [DOI] [PubMed] [Google Scholar]
- 32.Pelham, H. R. B. 1995. Sorting and retrieval between the endoplasmic reticulum and Golgi apparatus. Curr. Opin. Cell Biol. 7:530-535. [DOI] [PubMed] [Google Scholar]
- 33.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 34.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-959. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publications, Philadelphia, Pa.
- 35.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 36.Segrest, J. P., H. De Loof, J. G. Dohlman, C. G. Brouillette, and G. M. Anantharamaiah. 1990. Amphipathic helix motif: classes and properties. Proteins 8:103-117. [DOI] [PubMed] [Google Scholar]
- 37.Selby, M. J., Q.-L. Choo, K. Berger, G. Kuo, E. Glazer, M. Eckart, C. Lee, D. Chien, C. Kuo, and M. Houghton. 1993. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J. Gen. Virol. 74:1103-1113. [DOI] [PubMed] [Google Scholar]
- 38.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 39.Song, J., M. Nagano-Fujii, F. Wang, R. Florese, T. Fujita, S. Ishido, and H. Hotta. 2000. Nuclear localization and intramolecular cleavage of N-terminally deleted NS5A protein of hepatitis C virus. Virus Res. 69:109-117. [DOI] [PubMed] [Google Scholar]
- 40.Suhy, D. A., T. H. Giddings, and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan, S.-L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 42.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 44.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 45.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]