Abstract
Previous results have shown a correlation between the decrease in protease activity of several influenza A virus PA protein mutants and the capacity to replicate of the corresponding mutant ribonucleoproteins (RNPs) reconstituted in vivo. In this work we studied the phenotype of mutant viruses containing these mutations. Viruses with a T162A mutation, which showed a very moderate decrease both in protease and replication activities of reconstituted RNPs, showed a wild-type phenotype. Viruses with a T157A mutation, which presented a severe decrease in protease activity and replication of RNPs, showed a complex phenotype: (i) transport to the nucleus of PAT157A protein was delayed, (ii) virus multiplication was reduced at both low and high multiplicities, (iii) transcriptive synthesis was unaltered while replicative synthesis, especially cRNA, was diminished, and (iv) viral pathogenesis in mice was reduced, as measured by loss of body weight and virus titers in lungs. Finally, recombinant viruses with a T157E mutation in PA protein, which resulted in a drastic reduction of protease and replication activities of RNPs, were not viable. These results indicate that residue T157 in PA protein is important for the capacity of viral polymerase to synthesize cRNA.
The influenza virus RNA polymerase is a complex composed of three subunits, PA, PB1, and PB2. The polymerase, together with the nucleoprotein (NP) and the viral RNA template, form viral ribonucleoproteins (RNPs), which carry out viral transcription and replication. The polymerase complex synthesizes three different RNA classes: (i) mRNAs that contain a cap structure and 10 to 15 nucleotides derived from host cell mRNAs at their 5′ end and a poly(A) tail at their 3′ end; (ii) virion RNAs found in the viral particle (vRNAs); and (iii) cRNAs that are full-length complements of vRNA molecules and act as replicative intermediates (19, 24).
The PB1 subunit contains the polymerase active site, with sequence motifs characteristic of viral RNA-dependent RNA polymerases (34) which are essential for its activity (5). It also contains sites for sequence-specific binding to the conserved 5′- and 3′-terminal sequences of the vRNA and cRNA molecules (10, 11, 22). The PB2 protein binds cap1 structures (6, 15, 43). Although previous evidence indicated that the endonucleolytic activity responsible for the cleavage of host mRNA precursors could be contained on this subunit (7, 23), it has been proposed that the endonucleolytic activity lies on the PB1 subunit (21). Because it is responsible for cap-binding activity, PB2 is involved in transcription, but recently, we identified mutations in the PB2 subunit that specifically altered viral RNA replication (9a). The phenotype of viral temperature-sensitive mutants suggests that the PA subunit may be involved in the transition from mRNA transcription to cRNA and vRNA synthesis (reviewed in reference 24), but its precise role in this process is unknown at present. A recent report identified a mutation in the PA protein that shows a phenotype in cap-snatching activity but not in RNA replication of reconstituted RNPs (8).
The PA subunit induces a generalized proteolytic process when expressed from cloned cDNA (37). Hara et al. reported that the PA subunit is in fact a chymotrypsin-like protease having its catalytic site at serine 624 (14). However, by mutation analysis, we observed that the ability of PA to induce in vivo protein degradation lay within the 247 N-terminal amino acids of the protein, threonine 157 being involved in this activity (32, 39). The PA protein is phosphorylated in vivo and is a substrate of casein kinase II in vitro (38). Mutation of the N-terminal potential casein kinase II phosphorylation sites, especially positions T157 and T162, which are conserved among all strains of influenza type A viruses, led to protease-deficient PA mutants (32, 39). Viral RNPs reconstituted in vivo with these PA mutants showed reduced replication of model vRNAs that correlated with their protease phenotypes (32). Naffakh et al. (26) confirmed these results, although a mutation at cysteine 241 did not show such a correlation.
In this study, we tested the relevance of mutations at T157 and T162 during viral infection by rescuing the corresponding mutant viruses. The results showed that their capacity to replicate in tissue culture and their pathogenicity in mice correlated with their deficiency in protease activity.
MATERIALS AND METHODS
Biological materials.
HEK-293T cells and Madin-Darby canine kidney (MDCK) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum. Influenza virus A/Victoria/3/75 strain was propagated in MDCK cells. Plasmids pGPAT157A, pGPAT157E, and pGPAT162A with point mutations in the PA gene have been described (32).
Virus generation.
Recombinant viruses containing viral genes of strains A/Victoria/3/75 and A/WSN/33 in different combinations were generated by the method of Neumann et al. (29). To introduce PA point mutations in rescued viruses, EcoNI-StyI fragments from plasmid pGPAT157A or pGPAT157E were ligated to the PA genomic transfer plasmid digested with EcoNI and StyI. Likewise, fragment SfiI-StyI from plasmid pGPAT162A was ligated to the PA genomic transfer plasmid digested with SfiI and StyI. At different times posttransfection, aliquots of the supernatants were screened for rescued viruses by plaquing and amplification on MDCK cells. All rescued viruses were amplified from a single plaque, and the PA gene was sequenced after reverse transcription (RT)-PCR amplification.
35S in vivo labeling.
For protein synthesis analysis, cultures of MDCK cells were mock infected or infected with the recombinant viruses at 10 PFU/cell, starved for 60 min in methionine- and cysteine-free Dulbecco's modified Eagle's medium, and labeled for 1 h with 50 μCi of [35S]methionine-cysteine (Promix; Amersham) per ml at the indicated times. After the radiolabeling pulse, total cell extracts were prepared and analyzed by polyacrylamide gel electrophoresis and autoradiography.
Immunofluorescence.
Cultures of MDCK cells were infected with the recombinant viruses at a multiplicity of infection of 5 PFU per cell. At different times after infection, the cells were fixed with methanol at −20°C and stored in phosphate-buffered saline. Fixed cells were incubated with PA-specific monoclonal antibody 9 (1:3 dilution) (4), anti-RNP serum (1:500 dilution), anti-PB2 monoclonal antibody 22 (1:3 dilution) (4), or anti-PB1 serum (1:100 dilution) and processed as described elsewhere (30).
RNA assays.
Total RNAs were isolated at different times postinfection with the Ultraspec RNA isolation reagent from Biotex. The accumulation of vRNA, cRNA, and mRNA in infected cells was performed by real-time RT-PCR with oligonucleotide primers specific for the M or NP segment. Primer sequences are available upon request. To detect mRNA, the RT primer contained an oligo(dT) stretch of 16 to 18 residues fused to the sequence complementary to the 3′ terminus of the M or NP mRNA. These last sequences are also present in the corresponding cRNAs. Although the melting temperatures of the entire oligonucleotides are higher than if only those common sequences to cRNAs are considered, some cRNA could be amplified with these primers. Due to the small number of cRNA molecules present in infected cells compared to that of mRNA and keeping in mind the above considerations, RNA amplification with the oligo(dT) oligonucleotides should represent mainly mRNA molecules. The specificity of cRNA detection was obtained by using an RT primer containing the sequence of the 5′ terminus of M vRNA, not represented in mRNA.
Previous to RNA detection, the samples were boiled and chilled on ice. The RNA determination was done in two steps. First, the RT reaction was carried out, and after denaturation of the enzyme at 95°C for 15 min, the reverse oligonucleotide was added and PCR was continued. Quantitation was carried out with the Sybr Green RT-PCR kit from Applied Biosystems as recommended by the manufacturer. A standard curve was obtained with RNA isolated at late times after high-multiplicity infection with influenza virus. Only determinations that led to a standard curve with a fit better than 0.99 were considered. Denaturation curves of each reaction verified that it contained a single amplification product of the expected melting temperature. All RNA determinations were repeated at least seven times and assayed in duplicate.
Mice.
Mice were infected intranasally with 5 × 105 PFU of influenza virus A/Victoria/3/75 strain or rescued viruses containing wild-type PA (PAwt) or PAT157A or were mock infected. Their body weights were measured daily, and at different days after inoculation mice were sacrificed and their lungs were excised. Lungs were homogenized in phosphate-buffered saline-0.3% bovine serum albumin in a Dounce homogenizer, and viral titers were measured in the supernatants by plaque assay.
RESULTS
Rescue of recombinant mutant viruses.
The rescue of influenza viruses containing mutations in positions 157 and 162 of the PA subunit (32) was attempted with the procedures described by Neumann et al. (29). As we wanted to study the phenotype of the rescued viruses in a mouse model and the A/Victoria/3/75 strain is much less infectious in mice than the A/WSN/33 strain, we tried to rescue two different series of viruses. First we constructed the A series, having the RNP genes from the A/Victoria/3/75 strain and all other genes from the A/WSN/33 strain, and second, we constructed the B series, with all genes derived from the A/Victoria/3/75 strain except the WSN hemagglutinin (HA) gene, as a control to distinguish the recombinant viruses from other wild viruses used in the laboratory (Table 1). With this approach, we generated viruses containing wild-type PA and the T157A and T162A mutants in both genetic backgrounds. In contrast, recombinant viruses carrying the T157E mutant could not be rescued, although several trials were made. This result is in agreement with the low replication activity of recombinant RNPs with the T157E mutation (32). All studies in infected cultured cells were done with the rescued viruses of series A.
TABLE 1.
Rescue with PA point mutantsa
| Series | Virus | Gene origin
|
Rescue | |
|---|---|---|---|---|
| WSN strain | Victoria strain | |||
| A | PAwt | HA, NA, M, NS | PB1, PB2, PA, NP | + |
| PAT157A | HA, NA, M, NS | PB1, PB2, PA, NP | + | |
| PAT157E | HA, NA, M, NS | PB1, PB2, PA, NP | − | |
| PAT162A | HA, NA, M, NS | PB1, PB2, PA, NP | + | |
| B | PAwt | HA | All except HA | + |
| PAT157A | HA | All except HA | + | |
| PAT157E | HA | All except HA | − | |
| PAT162A | HA | All except HA | + | |
Recombinant viruses containing the full length of viral RNAs of strains A/Victoria/3/75 and A/WSN/33 in different combinations with PAwt and PA point mutants were generated. The recombinant viruses that could be rescued are indicated by +, while those that could not are indicated by −.
PAT157A mutant virus is affected in PA nuclear transport and replication.
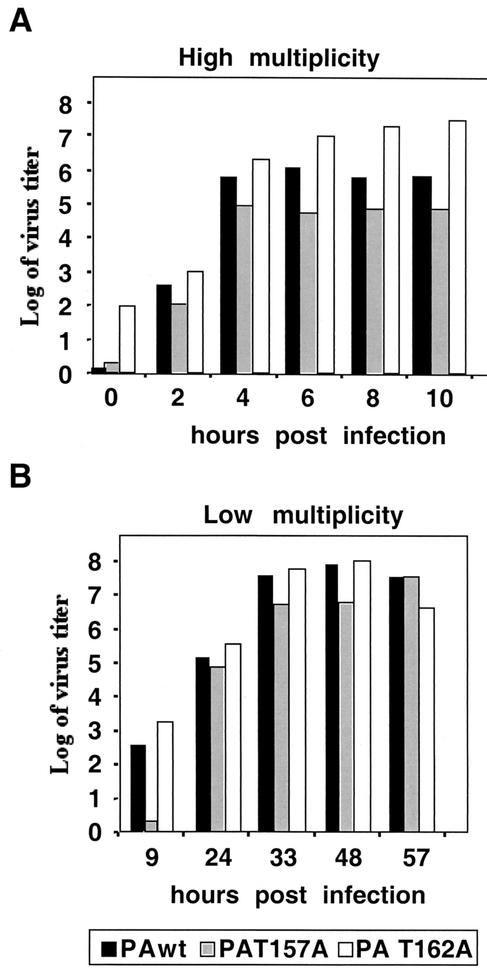
We first investigated the effect of PA mutations on viral multiplication in tissue culture. MDCK cells were infected with mutant or wild-type viruses at 5 PFU/cell, and the production of infectious virus was evaluated by plaque assay. The results are presented in Fig. 1A. Recombinant virus with the PAT162A mutation produced a higher virus yield than the wild-type virus, while recombinant PAT157A mutant virus showed a final titer 10 to 100 times lower than that obtained with wild-type recombinant virus. The production of infectious viruses was also studied in a multiple-cycle infection at a multiplicity of 10−3 PFU/cell. The results are presented in Fig. 1B and show that under these conditions there was a delay in virus production for the PAT157A virus, although the final titers were similar for all viruses. These results indicate that viruses with the PAT157A mutation have a defect in virus amplification, while the PAT162A mutant virus is similar to the wild-type virus.
FIG. 1.
Multiplication of recombinant viruses. MDCK cells were infected at either high (A) or low (B) multiplicity. At the indicated times, aliquots were taken, and virus infectivity was evaluated by plaque assay.
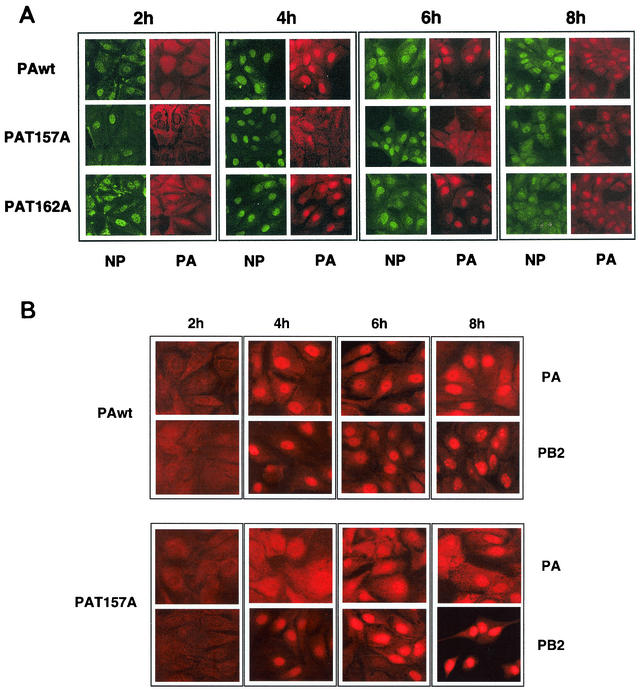
We have reported previously that the PA polymerase subunit is a phosphorylated protein (38). Moreover, when the phosphorylation state of mutant PAT157A was studied with a vaccinia virus PAT157A recombinant, we observed that this mutant protein was underphosphorylated (32). Two regions of PA are involved in its nuclear transport, a region located between amino acids 124 and 139 and another region between amino acids 186 and 247 (30). As position 157 is very close to the PA nuclear localization signal, we tested whether the T157A mutation had any effect on PA transport in virus-infected cells.
MDCK cells were infected with either wild-type, PAT157A, or PAT162A virus. At various times after infection, the cells were fixed and processed for immunofluorescence with antibodies specific for PA and NP proteins. The results are shown in Fig. 2A. At 4 h postinfection, NP was located in the nucleus in all three infections. At that time, PA was also nuclear in wild-type- and PAT162A-infected cells but was mainly in the cytosol of PAT157A-infected cells. At 6 h postinfection, the PAT157A mutant was located in both the nucleus and the cytosol, and at 8 h postinfection it showed mainly a nuclear localization. The localization of PB2 protein in PAwt- and PAT157A-infected cells was also studied, and the results are presented in Fig. 2B. As can be observed, no differences in PB2 localization were observed between cells infected with the two viruses, and its distribution paralleled that of NP protein. These results indicate that mutation T157A in PA protein dissociates its nuclear transport from the rest of the components of the RNP and suggest that phosphorylation of PA protein modulates its nuclear transport.
FIG. 2.
Kinetics of nuclear transport of wild-type and mutant PA proteins in infected cells. Cultures of MDCK cells were infected with wild-type and PA mutant viruses. At the indicated times, the cultures were fixed and processed for immunofluorescence with PA- and NP-specific antibodies as indicated in Materials and Methods.
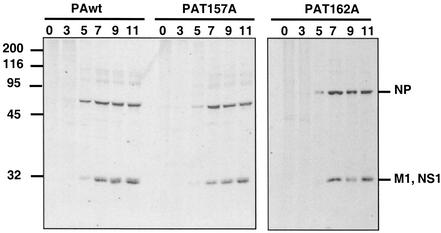
To study the molecular basis for the defect in viral amplification of the PAT157A mutant virus, we first analyzed the kinetics of viral protein synthesis. MDCK cells were infected with either wild-type, PAT157A, or PAT162A virus and labeled in vivo with [35S]methionine-cysteine for 60 min at various times after infection. Total cell extracts were analyzed by polyacrylamide gel electrophoresis and autoradiography. The results are presented in Fig. 3. Not remarkable differences were observed among the three viruses, although the amounts of protein synthesized were slightly smaller in PAT157A virus-infected cells than in wild-type- or PAT162A-infected cells.
FIG. 3.
Kinetics of protein synthesis in PA mutant virus-infected cells. Cultures of MDCK cells were infected at 10 PFU/cell and radiolabeled in vivo with [35S]methionine-cysteine for 60 min at the times indicated (hours postinfection). After the radiolabeling pulse, total cell extracts were prepared and analyzed by polyacrylamide gel electrophoresis and autoradiography. Sizes are shown to the left in kilodaltons.
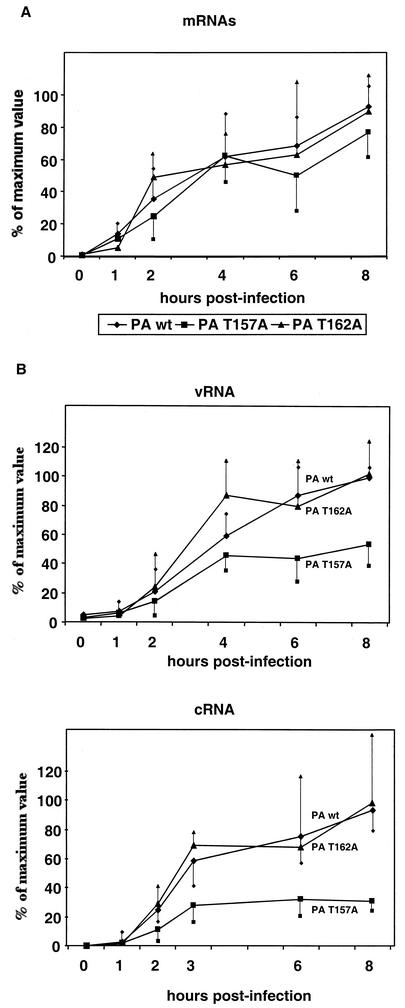
The phenotype of the PA mutants was also studied with regard to viral RNA synthesis. MDCK cells were infected with wild-type or PA mutant viruses, and the accumulation of mRNA, cRNA, and vRNA was determined at various times after infection by real-time RT-PCR. Transcription was studied with NP and M mRNAs as representatives of early and late viral messengers. The results obtained for M mRNA accumulation are presented in Fig. 4A and are similar to those obtained for NP mRNA (data not shown). A small decrease in mRNA accumulation was only observed in mutant PAT157A-infected cells, whereas transcription was indistinguishable in cells infected with the wild-type and PAT162A viruses. These results are compatible with those obtained with recombinant RNPs (32) and with the fact that comparable levels of protein synthesis were observed in cells infected with these rescued viruses (Fig. 3). The analysis of replication activity showed that vRNA was first detectable at 2 h postinfection and its accumulation progressed with the time of infection. No differences were observed between the wild-type and PAT162A viruses, but decreased accumulation was observed at a late time of infection for the PAT157A virus (Fig. 4B, top panel). The accumulation of cRNA started early in infection and peaked at 3 h postinfection (Fig. 4B, bottom panel). The wild-type and PAT162A viruses produced similar amounts of cRNA, whereas the PAT157A mutant produced only around 25% of the cRNA accumulated by the wild-type virus.
FIG. 4.
Kinetics of viral RNA accumulation. MDCK cells were infected at 10 PFU/ml with wild-type and PA mutant viruses. At the times indicated, cells were collected, total RNA was isolated, and the accumulation of the different viral RNA species of segment 7 was determined by real-time RT-PCR amplification, as described in Ma-terials and Methods. (A) mRNA. (B) Top, vRNA. Bottom, cRNA. RNA levels are indicated as the mean ± half of the standard deviation values from duplicates samples from at least seven independent experiments.
Recombinant virus PAT157A is less pathogenic than wild-type PA in the murine model.
Recombinant PAT157A virus presents defects in RNA replication and virus amplification in infected cells. The next step was to study the behavior of this mutant in an animal model. With that aim, mice were infected intranasally with 5 × 105 PFU of influenza virus A/Victoria/3/75 strain or with recombinant viruses containing PAwt or PAT157A or were mock infected. Both series of recombinant viruses described above (see Table 1) were used. Six mice were used for each condition, two of them were sacrificed every 2 or 3 days, and viral titers in the lungs were measured.
The results with series A are shown in Fig. 5A. No loss of body weight was observed in mice infected with Victoria wild-type virus or mock infected. Mice infected with recombinant PAwt virus showed a 20 to 30% body weight loss 5 to 6 days after inoculation, whereas mice infected with recombinant PAT157A virus did not show any significant loss of weight. Figure 5A (right panel) shows the viral titers in the lungs of inoculated mice. Two days after inoculation, mice infected with wild-type Victoria or recombinant PAT157A virus had around 10 to 20 times lower titers than mice infected with recombinant PAwt virus. The lung titers of rescued PAwt-inoculated mice increased 2 to 5 days postinoculation and decreased after 7 days. At days 5 and 7, the differences between mice infected with the Victoria strain and recombinant PAT157A or recombinant PAwt virus were 100 and 300 times lower, respectively. These results indicate that recombinant PAwt virus is more pathogenic than wild-type virus from the Victoria strain, probably as a consequence of the genes from WSN strain, which confer a higher capacity for replication in mice. They also show that recombinant PAT157A replicated more poorly than recombinant PAwt virus.
FIG. 5.
Pathogenicity of wild-type and mutant recombinant viruses in mice. A total of 5 × 105 PFU of influenza virus A/Victoria/3/75 strain or recombinant (R) viruses corresponding to series A (A) and B (B) were inoculated intranasally into mice. Six mice were used for each condition, and every 2 or 3 days, two of them were sacrificed and viral titers in the lung extracts were measured. Left panels, body weight. Right panels, viral titers.
The results with the viruses of the B series are presented in Fig. 5B. No significant loss of body weight could be observed in any of the inoculated mice. The viral titers in the lungs of mice inoculated either with Victoria wild-type or PAwt rescued viruses were similar and decreased at day 5 postinoculation. In all cases the titers in the lungs of mice inoculated with recombinant PAT157A were around 100 times lower than the titers in the lungs of mice inoculated with the wild type. At day 7 postinoculation, no viruses were found in the lungs of any of the inoculated mice. With this series of viruses, no differences were found between the Victoria wild-type and recombinant virus PAwt. For unknown reasons, different viral titers were detected in mice inoculated with the Victoria wild-type in the A and B series. Probably this discrepancy represents differences between litters. These results indicate that a mutation at position 157 of the PA subunit produces a decrease in viral replication both in infected cells and in an animal model.
DISCUSSION
In this report we analyzed the phenotype of recombinant virus mutants affected at positions located between the two domains that define the PA protein nuclear localization signal (30). Mutations at these sites, particularly at position T157, were previously shown to alter several properties of PA protein, such as its phosphorylation state, its protease activity, and the capacity of recombinant RNPs to synthesize cRNA (32). Recombinant virus containing the PAT162A mutation showed a phenotype undistinguishable from that of the PAwt rescued virus. The decrease in protease activity and RNA replication of the PAT162A mutant in reconstituted RNPs was only moderate. This reduction could not be enough to produce an attenuated phenotype in infected cells. Some defects in viral replication could perhaps be observed in mice due to the higher requirements for viral function in the animal model. Recombinant virus containing the PAT157A mutation showed several alterations both in infected cells and in inoculated mice.
PAT157A mutant virus is affected in nuclear transport of PA.
Immunofluorescence analysis of cells infected with the PAT157A virus indicated that transport of mutant PA to the nucleus is delayed compared to the other RNP components (Fig. 2). This delay was more prominent than the normal behavior of wild-type PA, whose nuclear transport is also delayed compared with that of PB1, PB2, and NP on wild-type influenza virus (1, 31). This observation indicates that nuclear transport is particularly uncoupled from the other polymerase subunits in PAT157A mutant virus-infected cells, in spite of the fact that PA-PB1 interaction is not affected by the T157A mutation, as measured by two-hybrid assays (T. Zurcher, unpublished results).
Nuclear localization signals have been identified in all three polymerase subunits, and individually expressed PB1, PB2, and PA proteins can enter the nucleus (1, 17, 25, 27, 30, 40). These data, together with the results presented here with PAT157A recombinant virus, open the possibility that the polymerase complex might form in the nucleus. The observation that mutant PAT157A protein is underphosphorylated compared to wild-type protein suggests that nuclear transport and/or proper formation of the polymerase complex may be phosphorylation dependent. In this context it should be mentioned that other viral proteins such as NS1, NP, and M1 are also phosphorylated (2, 3, 13, 18, 36) and that nucleocytoplasmic transport of the NP and M1 proteins is affected by phosphorylation (28, 44).
Mutation PAT157A leads to defective production of cRNA in virus-infected cells.
The phenotype of the PAT157A mutant virus with regard to virus-specific RNA accumulation fully confirms previous results obtained with recombinant RNPs (32), indicating that the synthesis of cRNA is affected by the mutation. The delay in PAT157A nuclear transport cannot be responsible for the defects in RNA synthesis because similar mRNA accumulation was observed for the PAT157A mutant and the wild-type virus. Furthermore, increased accumulation of cRNA and vRNA in PAT157A virus was not observed between 3 or 4 h postinfection and 8 h postinfection, although mutant PA was not completely nuclear until late infection times (Fig. 2).
The fact that the mutant virus was able to sustain normal mRNA synthesis (Fig. 4A) rules out that the primary consequence of PAT157A mutation is in the transcription step. Moreover, the accumulation of vRNA was less affected than that of cRNA (Fig. 4B), suggesting that PA is involved in the transcription-replication shift. cRNA synthesis requires (i) the switch from capped RNA-primed initiation to unprimed initiation, (ii) the readthrough of the polyadenylation signal, and (iii) the encapsidation of the nascent RNA chain with newly synthesized NP. The interaction of the polymerase complex with the 5′ end of vRNA is essential in the transcriptase mode of the polymerase, which requires capped RNA binding and endonuclease activity (9, 35, 41). In contrast, the synthesis of cRNA is initiated de novo and would not require these activities.
The region of PB1 that is involved in PA interaction overlaps the PB1 region that is responsible for the binding of cRNA and vRNA molecules (12, 33, 42). Although PB1 alone can bind the 5′-terminal vRNA sequence (10), its binding is more efficient upon PB1-PA complex formation (20), suggesting a role for PA in the ability of the polymerase to interact with the vRNA 5′ terminus. A polymerase complex with alterations in the RNA binding properties could be unsuitable for cRNA synthesis. Additionally, conformational changes in the polymerase complex that would allow the transcription-replication shift could be affected by mutations at position T157. In this regard, the proteolytic activity of the PA subunit and/or its phosphorylation state might be important to allow potential changes in the conformation of the polymerase. Finally, interaction of the PA subunit with hCLE, a human protein homologous to transcription activation factors, has been reported in vitro and in vivo in reconstituted RNPs (16). Although PAT157A interacts with hCLE with the same strength as PAwt, as measured by two-hybrid analysis (16), we cannot exclude that mutations at positions T157 alter the interaction of the polymerase with hCLE or other cellular factors required for viral RNA replication.
Acknowledgments
We are indebted to Pablo Gastaminza for critical comments on the manuscript. The technical assistance of A. Beloso, Y. Fernández, and J. Fernández is gratefully acknowledged.
M. Huarte and A. Falcón were fellows of Comunidad Autónoma de Madrid and Programa Sectorial de Promoción General del Conocimiento, respectively. This work was supported by Programa Sectorial de Promoción General del Conocimiento (grants PB97-1160 and PM98-0116).
REFERENCES
- 1.Akkina, R. K., T. M. Chambers, D. R. Londo, and D. P. Nayak. 1987. Intracellular localization of the viral polymerase proteins in cells infected with influenza virus and cells expressing PB1 protein from cloned cDNA. J. Virol. 61:2217-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almond, J. W., and V. Felsenreich. 1982. Phosphorylation of the nucleoprotein of an avian influenza virus. J. Gen. Virol. 60:295-305. [DOI] [PubMed] [Google Scholar]
- 3.Arrese, M., and A. Portela. 1996. Serine 3 is critical for phosphorylation at the N-terminal end of the nucleoprotein of influenza virus A/Victoria/3/75. J. Virol. 70:3385-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bárcena, J., M. Ochoa, S. de la Luna, J. A. Melero, A. Nieto, J. Ortín, and A. Portela. 1994. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J. Virol. 68:6900-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, S. K., and D. P. Nayak. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaas, D., E. Patzelt, and E. Keuchler. 1982. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 10:4803-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blok, V., C. Cianci, K. W. Tibbles, S. C. Inglis, M. Krystal, and P. Digard. 1996. Inhibition of the influenza virus RNA-dependent RNA polymerase by antisera directed against the carboxy-terminal region of the PB2 subunit. J. Gen. Virol. 77:1025-1033. [DOI] [PubMed] [Google Scholar]
- 8.Fodor, E., M. Crow, L. Mingay, T. Deng, J. Sharps, P. Fechter, and G. Brownlee. 2002. A single amino acid mutation in the the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Gastaminza, P., B. Perales, A. M. Falcón, and J. Ortín. 2003. Mutations in the N-terminal region of influenza virus. PB2 protein affect virus RNA replication but not transcription. J. Virol. 77:5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez, S., and J. Ortín. 1998. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 73:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez, S., and J. Ortín. 1999. Distinct regions of influenza virus PB1 subunit recognize vRNA and cRNA templates. EMBO J. 18:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González, S., T. Zürcher, and J. Ortín. 1996. Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and the PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 24:4456-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregoriades, A., T. Christie, and K. Markarian. 1984. The membrane (M1) protein of influenza virus occurs in two forms and is a phosphoprotein. J. Virol. 49:229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara, K., M. Shiota, H. Kido, Y. Ohtsu, T. Kashiwagi, J. Iwahashi, N. Hamada, K. Mizoue, N. Tsumura, H. Kato, and T. Toyoda. 2001. Influenza virus RNA polymerase PA subunit is a novel serine protease with Ser624 at the active site. Genes Cells 6:87-97. [DOI] [PubMed] [Google Scholar]
- 15.Honda, A., K. Mizumoto, and A. Ishihama. 1999. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 8:475-485. [DOI] [PubMed] [Google Scholar]
- 16.Huarte, M., J. J. Sanz-Ezquerro, F. Roncal, J. Ortín, and A. Nieto. 2001. the PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 75:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, I. M., P. A. Reay, and K. L. Philpott. 1986. Nuclear location of all three influenza polymerase proteins and a nuclear signal in polymerase PB2. EMBO J. 9:2371-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kistner, O., H. Müller, H. Becht, and C. Scholtissek. 1985. Phosphopeptide fingerprints of nucleoproteins of various influenza A virus strains grown in different host cells. J. Gen. Virol. 66:465-472. [DOI] [PubMed] [Google Scholar]
- 19.Krug, R. M. 1989. The influenza viruses. Plenum Press, New York, N.Y.
- 20.Lee, M. T., K. Bishop, L. Medcalf, D. Elton, P. Digard, and L. Tiley. 2002. Definition of the minimal viral components required for the initiation of umprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, M.-L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M. L., B. C. Ramirez, and R. M. Krug. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17:5844-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licheng, S., D. F. Summers, Q. Peng, and J. Galarza. 1995. Influenza A virus polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology 208:38-47. [DOI] [PubMed] [Google Scholar]
- 24.Mahy, B. W. J. 1983. Mutants of influenza virus, p. 192-253. In P. Palese and D. W. Kingsbury (ed.), Genetics of influenza viruses. Springer-Verlag, Vienna, Austria.
- 25.Mukaigawa, J., and D. P. Nayak. 1991. Two signals mediate nuclear localization of influenza virus (A/WSN/33) polymerase basic protein 2. J. Virol. 65:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naffakh, N., P. Massin, and S. van der Werf. 2001. The transcription/replication activity of the polymerase of influenza A viruses is not correlated with the level of proteolysis induced by the the PA subunit. Virology 285:244-252. [DOI] [PubMed] [Google Scholar]
- 27.Nath, S. T., and D. P. Nayak. 1990. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1N1). Mol. Cell. Biol. 10:4139-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann, G., M. R. Castrucci, and Y. Kawaoka. 1997. Import and export of influenza virus nucleoprotein. J. Virol. 71:9690-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hugues, D. R. Perea, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieto, A., S. de la Luna, J. Bárcena, A. Portela, and J. Ortín. 1994. Complex structure of the nuclear translocation signal of the influenza virus polymerase the PA subunit. J. Gen. Virol. 75:29-36. [DOI] [PubMed] [Google Scholar]
- 31.Nieto, A., S. de la Luna, J. Bárcena, A. Portela, J. Valcárcel, J. A. Melero, and J. Ortín. 1992. Nuclear transport of influenza virus polymerase PA protein. Virus Res. 24:65-75. [DOI] [PubMed] [Google Scholar]
- 32.Perales, B., J. J. Sanz-Ezquerro, P. Gastaminza, J. Ortega, J. Fernandez-Santarén, J. Ortín, and A. Nieto. 2000. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol. 74:1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez, D. R., and R. O. Donis. 1995. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J. Virol. 69:6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1990. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritlove, D. C., L. L. Poon, L. J. Devenish, M. B. Leahy, and G. Brownlee. 1999. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J. Virol. 73:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Privalsky, M. L., and E. E. Penhoet. 1977. Phosphorylated protein component present in influenza virions. J. Virol. 24:401-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanz-Ezquerro, J. J., S. de la Luna, J. Ortín, and A. Nieto. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J. Virol. 69:2420-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanz-Ezquerro, J. J., J. Férnandez-Santarén, T. Sierra, T. Aragón, J. Ortega, J. Ortín, G. L. Smith, and A. Nieto. 1998. The PA influenza virus polymerase subunit is a phosphorylated protein. J. Gen. Virol. 79:471-478. [DOI] [PubMed] [Google Scholar]
- 39.Sanz-Ezquerro, J. J., T. Zürcher, S. de la Luna, J. Ortín, and A. Nieto. 1996. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J. Virol. 70:1905-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, G. L., J. Z. Levin, P. Palese, and B. Moss. 1987. Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology 160:336-345. [DOI] [PubMed] [Google Scholar]
- 41.Tiley, L. S., M. Hagen, J. T. Matthews, and M. Krystal. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68:5108-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyoda, T., D. M. Adyshev, M. Kobayashi, A. Iwata, and A. Ishihama. 1996. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J. Gen. Virol. 77:2149-2157. [DOI] [PubMed] [Google Scholar]
- 43.Ulmanen, I., B. A. Broni, and R. M. Krug. 1981. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc. Natl. Acad. Sci. USA 78:7355-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whittaker, G., I. Kemler, and A. Helenius. 1995. Hyperphosphorylation of mutant influenza virus matrix protein, M1, causes its retention in the nucleus. J. Virol. 69:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]