Abstract
Stavudine (d4T) and zidovudine (AZT) are thymidine analogs widely used in the treatment of human immunodeficiency virus type 1 (HIV-1)-infected persons. Resistance to d4T is not fully understood, although the selection of AZT resistance mutations in patients treated with d4T suggests that both drugs have similar pathways of resistance. Through the analysis of genotypic changes in nine recombinant viruses cultured with d4T, we identified a new pathway for d4T resistance mediated by K65R, a mutation not selected by AZT. Passaged viruses were derived from treatment-naïve persons or HIV-1HXB2 and had wild-type reverse transcriptase (RT) or T215C/D mutations. K65R was selected in seven viruses and was associated with a high level of enzymatic resistance to d4T-triphosphate (median, 16-fold; range, 5- to 48-fold). The role of K65R in d4T resistance was confirmed in site-directed mutants generated in three different RT backgrounds. Phenotypic assays based on recombinant single-cycle replication or a whole-virus multiple replication cycle were unable to detect d4T resistance in d4T-selected mutants with K65R but detected cross-resistance to other nucleoside RT inhibitors. Four of the six viruses that had 215C/D mutations at baseline acquired the 215Y mutation alone or in association with K65R. Mutants having K65R and T215Y replicated less efficiently than viruses that had T215Y only, suggesting that selection of T215Y in patients treated with d4T may be favored. Our results demonstrate that K65R plays a role in d4T resistance and indicate that resistance pathways for d4T and AZT may not be identical. Biochemical analysis and improved replication assays are both required for a full phenotypic characterization of resistance to d4T. These findings highlight the complexity of the genetic pathways of d4T resistance and its phenotypic expression.
Treatment of human immunodeficiency virus type 1 (HIV-1)-infected persons with reverse transcriptase (RT) and protease inhibitors has significantly reduced the rate of HIV and AIDS-associated morbidity and mortality. However, specific patterns of mutations in the RT and protease have been observed following treatment and have been associated with decreased susceptibility to the antiretroviral drug and loss of clinical benefit (4). For instance, the Met184Val (M184V) mutation observed following treatment with lamivudine (3TC) confers high-level (>100-fold) resistance to 3TC, while the stepwise accumulation of mutations such as M41L, D67N, K70R, L210W, T215Y/F, and K219Q seen in zidovudine (AZT)-treated patients results in increasing resistance to AZT (3, 18, 19, 43).
Stavudine (d4T) is a nucleoside RT inhibitor (NRTI) that was approved for the treatment of HIV-1-infected persons in 1994. Despite the long experience with d4T, both the genotypic and phenotypic correlates of resistance to d4T and their relationship to treatment failure remain poorly understood. Mutations conferring multi-nucleoside analog resistance, including the Q151M mutation and amino acid insertions at position 69 of the RT, are found in d4T-treated patients (32, 38, 46). However, these mutations are only seen in a small proportion of clinical isolates (20, 21).
In recent years, accumulating evidence has shown that AZT-selected mutations may also contribute to d4T resistance. These mutations can be selected in AZT-naïve patients who are treated with d4T (5, 21, 22, 30, 36, 37). The presence of AZT mutations has also been shown to reduce the virologic benefit derived from d4T treatment (42). Thus, these data suggest a selective advantage for these mutations in d4T-treated persons. Because both AZT and d4T are thymidine analogs, these mutations have been referred to as thymidine analog mutations (TAMs). However, despite the association between TAMs and d4T treatment failure, clinical isolates with TAMs show minimal or no resistance to d4T in culture-based assays compared to >50-fold increases in resistance to AZT (37, 40). The discordance between the selection of TAMs in patients and the lack of detectable d4T resistance seen with these mutants in vitro is puzzling and has raised questions regarding the ability of current replication assays to detect phenotypic resistance to d4T (25, 40).
While increased discrimination against nucleoside analogs has been known to be the mechanism of NRTI resistance in enzymes containing mutations such as M184V, L74V, K65R, and Q151M, recent work suggests a distinct mechanism for AZT resistance in viruses with TAMs (23-25). RTs containing TAMs have been shown to have enhanced ability to remove AZT-monophosphate from AZT-terminated primers and transfer it to an acceptor such as a nucleotide triphosphate (ATP) (23-25). TAMs have also been shown to confer elevated unblocking activity for most chain terminators, including d4T, although removal of d4T-monophosphate was found to be strongly inhibited by high concentrations of the next complementary deoxynucleoside triphosphate (dNTP) (25).
Selection of resistance mutations by d4T in vitro can provide important information on the resistance mutations, the kinetics of selection, and the impact on resistance phenotype. However, only two studies have attempted to select mutations by d4T by using a single HIV-1 strain (HXB2) (13, 17). No TAMs were observed in these studies. In contrast, V75A/T was seen in one study, and I50T was selected in the second. The reduction in d4T susceptibility conferred by these mutations was substantial (30-fold) for I50T but modest (7-fold) for V75T (13, 17). The V75T mutation conferred resistance to d4T when present in HIV-1HXB2 but not in HIV-1RF or HIV-1NL4-3, suggesting that the viral genetic background plays a role in the phenotypic expression of d4T resistance mediated by V75T (20). The clinical importance of V75A/T and I50T is unclear, however, since these mutations occur infrequently in patients with d4T treatment failure (13, 17, 20, 21). Therefore, additional studies with patient-derived viruses that have different genetic backgrounds are needed to fully understand the genotypic and phenotypic evolution of d4T resistance in vitro.
In the present study, we investigated in vitro the pathways of acquisition of resistance to d4T in a selected panel of viruses. These viruses included both HIV-1HXB2 and patient-derived viruses that were wild type (WT) or had T215D or C mutations. Viruses with 215D/C are revertants of 215Y and are known to have increased capacity to acquire 215Y in the presence of AZT (9). The increased capacity to acquire 215Y and become AZT resistant is associated with the requirement of only a 1-nucleotide change to evolve from 215D/C to 215Y compared to a 2-nucleotide change required for the WT T215 (9). The increased potential of these viruses to acquire 215Y provides a good opportunity to investigate the role of 215Y in d4T resistance in vitro. We also assessed changes in d4T susceptibility in d4T-selected mutants by using both enzymatic and culture-based assays, and we examined the impact of mutations on replication capacity and fitness.
MATERIALS AND METHODS
Generation of recombinant viruses with cloned RT sequences from patients.
Full-length RT sequences from plasma of treatment-naïve, recently diagnosed HIV-1-infected patients were amplified in duplicate by RT-nested PCR and then cloned using the TA cloning kit (Invitrogen) as described previously (9). A total of 7 recombinant viruses were generated, each from a single cloned RT sequence and an RT-lacking proviral molecular clone (15). The generation and characterization of these viruses have been described elsewhere (9). Of these viruses, two were WT (RD 22wt and RD 23wt), three had the 215C mutation alone or associated with 41L and/or 210W (RD 01215C, RD 02210W/215C, and RD 0341L/210W/215C), and two had the 215D mutation associated with 41L or 210W (RD 04210W/215D and RD 0541L/215D). RT sequences from all these isolates are available in the GenBank database (9).
Generation of site-directed mutants in HXB2 and patient-derived RT backgrounds.
Five additional recombinant viruses were generated with the WT RT sequences of HXB2, namely, HXB2wt, HXB2215D, HXB2K65R, HXB2K65R/T215Y, and HXB2T215Y. The T215D, K65R, and T215Y mutations were introduced in the pHXB2RIP7-based infectious clone pSUM9 by site-directed mutagenesis as previously described (8). The K65R mutation was also introduced in the WT RT from recombinants RD 22wt and RD 23wt to generate RD 22K65R and RD 23K65R, respectively. The 50% cell culture infectious dose (CCID50) in each virus stock was determined in MT-4 cells by using the method of Reed and Muench (33).
In vitro selection of d4T resistance.
d4T was obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health (Bethesda, Md.). The purity of the preparation of d4T was confirmed by duplicate liquid chromatography and tandem mass spectrometry analysis at three different collision energies. The spectra obtained were identical to those previously published for d4T (27).
For in vitro selection of d4T resistance, we used an approach similar to that described previously for AZT (9). Briefly, 1.5 × 106 MT-4 cells were exposed to 1,500 CCID50 (multiplicity of infection [MOI], 0.001) of each virus for 2 h at 37°C. After two washes with phosphate-buffered saline (PBS), cells were resuspended in 10 ml of complete medium containing 0.7 μM d4T, a concentration close to the 50% inhibitory concentration (IC50) value for HXB2wt determined by the MT-4/MTT assay. Cultures were then incubated at 37°C, and media containing d4T were changed every 3 to 4 days as required. Virus production was monitored by microscopic assessment of syncytium formation. Once virus production was evident at a given concentration of d4T, 500 μl of clarified supernatant was added to 1.5 × 106 fresh cells and cells were cultured with the same or a higher concentration of d4T (generally twofold). Genotypic changes in HIV-1 RT were monitored by sequence analysis of the RT from culture supernatant in selected passages as indicated below.
Phenotypic testing by culture-based assays.
Susceptibility to d4T and other NRTIs was determined by two different culture-based methods: the PhenoSense HIV assay (ViroLogic, South San Francisco, Calif.) (31) and the MT-4/MTT assay (29, 44). The PhenoSense HIV assay was done at ViroLogic. This assay uses resistance test vectors that are constructed by inserting amplified protease and RT sequences into a modified HIV-1 vector derived from the NL4-3 molecular clone. Susceptibility to NRTIs is determined by measuring IC50 values in 293 cells following a single round of virus replication (31). Changes in drug susceptibility were determined by comparing the IC50 values with the IC50 value of a WT reference strain (NL4-3). Results were interpreted according to the assay cutoff values established by the manufacturer (16). Assay cutoff values used were 1.7-fold for ddI, d4T, and ddC, 2.5-fold for AZT and 3TC, 4.5-fold for abacavir, and 1.4-fold for tenofovir.
The MT-4/MTT assay involves multiple rounds of virus replication, and evaluates the ability of an HIV-1 isolate to replicate in the presence of serial dilutions of drug (29, 44). Briefly, MT-4 cells (3 × 104) were exposed to 200 CCID50 of each virus in triplicate both in the absence and in the presence of serial dilutions of d4T. The concentration of d4T required to inhibit 50% of virus-induced cell killing (50% effective concentration [EC50]) was calculated after 5 days of culture, as described previously (29, 44). Phenotypic changes in d4T susceptibility were determined by comparing EC50 values of baseline isolates with those from isolates collected during selection with d4T.
Enzymatic susceptibility to d4T-TP.
Susceptibility to d4T-triphosphate (d4T-TP; Moravek Biochemicals, Brea, Calif.) of virion-associated RTs was measured by using a nonradioactive microtiter plate-based RT assay (Roche Diagnostics GmbH, Mannheim, Germany). Briefly, viruses normalized by their levels of RT activity were exposed for 30 min to serial threefold dilutions of d4T-TP prepared in lysis buffer. RT reaction was done for 2 h at 37°C in an RT buffer containing poly(A) × oligo(dT)15 template/primer, digoxigenin-dUTP, biotin-dUTP, and dTTP. Levels of RT activity were quantified by using an enzyme-linked immunosorbent assay-based chemiluminescence assay according to the manufacturer's instructions. Percentage of RT inhibition was determined by dividing the signal obtained in reactions done with d4T-TP by that seen in reactions done in the absence of drug; IC50 values were calculated as previously described (8). All determinations were done in duplicate, and the results reflect the mean IC50 value obtained in at least two separate experiments. Changes in d4T-TP susceptibility were determined by comparing IC50 values between baseline isolates and isolates collected during selection with d4T.
Replication kinetics in MT-4 cells.
Inocula of 300 CCID50 were used to infect 3 × 105 MT-4 cells (MOI, 0.001). After incubation for 2 h at 37°C, cells were washed twice with PBS and resuspended in complete medium at 7.5 × 104 cells/ml. Two-milliliter cultures were done in duplicate with 24-well tissue culture plates (Costar). Supernatants (200 μl) from each culture were collected at different days, and then an equal volume of culture medium was added. Levels of p24 antigen were quantified in cell-free culture supernatants by using the Coulter HIV-1 p24 antigen assay and were used to monitor replication kinetics.
Analysis of fitness difference between HIV-1T215Y and HIV-1K65R/T215Y.
The fitness difference between HIV-1T215Y and HIV-1K65R/T215Y was analyzed in growth competition experiments done in the presence of d4T. Briefly, HXB2T215Y and HXB2K65R/T215Y were mixed at different proportions and were used to infect 1.5 × 106 MT4-cells at an MOI of 0.001. After a 2-h incubation at 37°C, cells were washed with PBS and resuspended in 10 ml of complete medium containing 12 μM d4T. Cultures were then incubated at 37°C and diluted in complete medium containing d4T every 3 to 4 days. At this time, an aliquot was obtained and was used to estimate the proportions of the two competing variants. Proportions were determined based on the relative peak heights seen by dye-terminator sequencing of the HIV-1 RT from culture supernatants (8). The fitness difference s between the two viruses was calculated according to the formula s = 1/t ln [q(t) p(0)/p(t) q(0)], where p(0) and q(0) are the proportions of the less and the more fit virus at time 0, respectively, and p(t) and q(t) are the proportions of the two viruses at time t (10).
RT-PCR and sequence analysis.
Full-length HIV-1 RT sequences (from nucleotides 2529 to 4128 of HXB2; amino acids 7 to 540) from culture supernatants were obtained from amplified RT-nested PCR products by using an ABI373 automated sequencer. Briefly, HIV-1 RNA was extracted by using the QIAmp viral RNA kit (Qiagen). The RT reaction was done for 1 h at 42°C with primer RT2, as described previously (8). After a first round of PCR amplification with primers RT1 and RT2, 4 μl was subjected to a second round of amplification with primers A35 and NE1 (nucleotides 2529 to 3246) or AV180 and IN3 (nucleotides 3192 to 4128). Primers AV36, AV44, A35, NE(1)35, AV180, and AV181 were used for sequence analysis (39, 44). The DNASIS program (version 2.6; 1988) was used to analyze the data and to determine deduced amino acid sequences.
RESULTS
Selection of the 215Y and 75A/T mutations by d4T.
We first evaluated the frequency of selection of the 215Y, 75A/T, and I50T mutations with d4T in the nine recombinant viruses. Four of the six viruses that had 215C/D at baseline acquired the 215Y mutation after a mean of 57 days (range, 32 to 89 days) in culture with d4T (Table 1). In contrast, none of the three WT viruses selected 215Y after a mean of 104 days (range, 94 to 110 days) in culture, indicating that HIV-1215D and HIV-1215C have increased capacity to acquire 215Y compared to WT viruses. A comparison of the kinetics of emergence of 215Y with d4T with those previously seen with AZT (9) showed that, in these viruses, 215Y was selected more rapidly with AZT (mean, 23 days; range, 18 to 27 days) than with d4T. Table 1 also shows that none of the viruses selected the V75T or I50T mutation during this period of time and that only HXB2T215 acquired a V75A intermediate (17). Recombinant RD 01215C also selected a mutation at codon 75 (V75I) in association with the C215Y and H481Y mutations (Table 1).
TABLE 1.
Kinetics of emergence of RT mutations during sequential passages with d4T
| Recombinant and passage | d4T concn (μM) | Cumulative time in culture (days) | RT mutation(s)b |
|---|---|---|---|
| RD 01215C | |||
| p1 | 0.7 | 8 | n.d. |
| p2 | 1.4 | 20 | |
| p3 | 2.8 | 28 | |
| p4 | 2.8 | 54 | 215C/Y,a V75V/I |
| p5 | 6 | 64 | 215C/Y, V75I |
| p6 | 12 | 93 | 215Y, V75I, H481Y |
| RD 02210W/215C | |||
| p1 | 0.7 | 6 | n.d. |
| p2 | 1.4 | 12 | n.d. |
| p3 | 2.8 | 19 | |
| p4 | 6 | 32 | 215C/Y |
| p5 | 6 | 46 | 215Y |
| p6 | 12 | 67 | 215Y |
| RD 0341L/210W/215C | |||
| p1 | 0.7 | 6 | n.d. |
| p2 | 1.4 | 13 | |
| p3 | 2.8 | 20 | K65R |
| p4 | 6 | 33 | K65R |
| p5 | 6 | 41 | K65R |
| p6 | 12 | 54 | 215C/Y, K65R |
| p7 | 12 | 69 | 215Y/C, K65R |
| p8 | 12 | 79 | 215Y, K65R |
| p9 | 20 | 116 | 215Y, K65R |
| RD 04210W/215D | |||
| p1 | 0.7 | 6 | n.d. |
| p2 | 1.4 | 12 | n.d. |
| p3 | 2.8 | 20 | |
| p4 | 6 | 64 | |
| p5 | 6 | 86 | K65R |
| p6 | 12 | 112 | K65R |
| p7 | 12 | 141 | K65R |
| RD 0541L/215D | |||
| p1 | 0.7 | 7 | n.d. |
| p2 | 1.4 | 14 | n.d. |
| p3 | 2.8 | 22 | |
| p4 | 6 | 53 | K65R |
| p5 | 6 | 67 | K65R |
| p6 | 12 | 89 | 215Y, K65R |
| p7 | 20 | 137 | 215Y, K65R |
| HXB2215D | |||
| p1 | 0.7 | 6 | n.d. |
| p2 | 1.4 | 12 | n.d. |
| p3 | 2.8 | 20 | |
| p4 | 2.8 | 38 | |
| p5 | 6 | 64 | |
| p6 | 6 | 82 | K65R |
| p7 | 12 | 111 | K65R |
| RD 22wt | |||
| p1 | 0.7 | 7 | n.d. |
| p2 | 1.4 | 14 | n.d. |
| p3 | 2.8 | 25 | D67G |
| p4 | 6 | 42 | D67G |
| p5 | 6 | 64 | K65R, M202I |
| p6 | 12 | 81 | K65R, M202I |
| p7 | 12 | 110 | K65R, M202I |
| RD 23wt | |||
| p1 | 0.7 | 7 | n.d. |
| p2 | 1.4 | 15 | |
| p3 | 2.8 | 27 | K65R |
| p4 | 6 | 51 | K65R |
| p5 | 6 | 77 | K65R |
| p6 | 12 | 94 | K65R |
| HXB2T215 | |||
| p1 | 0.7 | 6 | n.d. |
| p2 | 1.4 | 12 | n.d. |
| p3 | 2.8 | 20 | |
| p4 | 6 | 55 | V75V/A |
| p5 | 6 | 66 | V75V/A, K65R |
| p6 | 12 | 87 | V75V/A, K65R |
| p7 | 12 | 109 | V75A, K65R |
Mixed genotype. The first amino acid represents the predominant genotype observed in the mixture.
Amino acid changes identified during selection with d4T. n.d., not done.
Frequent selection of K65R by d4T.
We also examined RT mutations selected by d4T at codons other than 215, 50, and 75. This analysis showed that seven of the nine viruses selected the K65R mutation (Table 1). K65R was seen after a mean of 57 days (range, 20 to 86 days) of culture in viruses containing either WT or 215C/D mutations. Of these viruses, three selected K65R only (recombinants RD 23wt, RD 04210W/215D, and HXB2215D), two selected K65R before 215Y (recombinants RD 0341L/210W/215C and RD 0541L/215D), one selected K65R and M202I (recombinant RD 22wt), and one selected K65R after V75A (HXB2T215). These findings indicate that K65R plays an important role in the evolution of HIV-1 in the presence of d4T in vitro.
Enzymatic resistance to d4T-TP in d4T-selected mutants.
We next determined changes in susceptibility to d4T-TP between baseline viruses and viruses collected during selection with d4T. Table 2 shows IC50 values for d4T-TP and changes in d4T-TP susceptibility for the nine viruses. The median IC50 value for d4T-TP in baseline viruses was 0.0059 μM (range, 0.0015 to 0.032 μM) compared to 0.071 μM (range, 0.007 to 0.23 μM) in d4T-selected isolates, with a median increase in IC50 for d4T-TP of 16-fold (range, 1- to 48-fold). A reduction in d4T-TP susceptibility ranging between 5- and 48-fold was seen in all the seven viruses that had K65R, with the highest level of resistance seen in RD 0341L/210W/215C and RD 0541L/215D. In these two viruses, selection of the K65R mutation at passage 5 was associated with 39- and 24-fold reductions in d4T-TP susceptibility, respectively. Interestingly, the accumulation of the 215Y mutation in these two viruses at passages 8 and 6 further increased the level of resistance to 48- and 32-fold, respectively (Table 2). Isolates RD 03p5-AZT and RD 05p4-AZT, which were previously selected with AZT and lacked the K65R mutation, showed little or no reduction in d4T-TP susceptibility (1- and 1.6-fold changes in IC50 values, respectively). Table 2 also shows that selection of K65R only (recombinants RD 04210W/215D, RD 22wt, RD 23wt, HXB2215D, and HXB2T215) was associated with a 5- to 10-fold resistance to d4T-TP.
TABLE 2.
Changes in d4T susceptibility in recombinant viruses measured by an RT assay and two culture-based assays (MT-4/MTT and PhenoSense)
| Virus and passage | RT mutation(s) | IC50 or EC50 (μM) (fold change) bya:
|
||
|---|---|---|---|---|
| Poly(A) × oligo(dT)15 | MT-4/MTT | PhenoSense | ||
| RD 01 | ||||
| p0 | 215C | 0.002 | n.d. | 0.50 (0.9) |
| p6 | V75I, C215Y, H481Y | 0.033 (16) | n.d. | 0.92 (1.6) |
| RD 02 | ||||
| p0 | 210W, 215C | 0.007 | 0.33 | 0.61 (1.1) |
| p6 | 210W, C215Y | 0.007 (1) | 0.70 (2.1) | 1.02 (1.8) |
| RD 03 | ||||
| p0 | 41L, 210W, 215C | 0.0015 | 0.31 | 0.57 (0.9) |
| p5 | 41L, 210W, 215C, K65R | 0.059 (39) | n.d. | 0.81 (1.3) |
| p8 | 41L, 210W, C215Y, K65R | 0.072 (48) | 0.75 (2.4) | 0.97 (1.6) |
| p5-AZTb | 41L, 210W, C215Y | 0.0014 (1) | n.d. | n.d. |
| RD 04 | ||||
| p0 | 210W, 215D | 0.013 | 0.50 | 0.63 (1.1) |
| p6 | 210W, 215D, K65R | 0.082 (6) | 0.53 (1.1) | 0.90 (1.6) |
| RD 05 | ||||
| p0 | 41L, 215D | 0.006 | 0.23 | 0.46 (0.8) |
| p5 | 41L, 215D, K65R | 0.14 (24) | n.d. | 0.72 (1.2) |
| p6 | 41L, D215Y, K65R | 0.19 (32) | 0.22 (1.0) | 0.99 (1.8) |
| p4-AZTb | 41L, D215Y | 0.009 (1.6) | n.d. | n.d. |
| RD 22wt | ||||
| p0 | 0.0036 | 0.30 | 0.48 (0.8) | |
| p6 | K65R, M202I | 0.036 (10) | 0.28 (0.9) | 0.77 (1.4) |
| RD 23wt | ||||
| p0 | 0.0046 | n.d. | 0.52 (0.9) | |
| p6 | K65R | 0.039 (8) | n.d. | 0.88 (1.6) |
| HXB2215D | ||||
| p0 | 215D | 0.032 | n.d. | 0.47 (0.8) |
| p6 | 215D, K65R | 0.148 (5) | n.d. | 0.95 (1.7) |
| HXB2T215 | ||||
| p0 | 0.011 | 0.52 | 0.48 (0.9) | |
| p7 | V75A, K65R | 0.21 (18) | 0.62 (1.2) | 1.13 (1.7) |
Values in parentheses are changes in IC50 values relative to the IC50 value of the baseline isolate (p0) for the RT and MT4/MTT assays or a reference WT NL4-3 isolate for the PhenoSense assay. In boldface are indicated the isolates that had evidence of reduced susceptibility to d4T by the PhenoSense assay. n.d., not done.
Virus isolated with AZT at the indicated passage.
Of the two isolates that did not select K65R (RD 01 and RD 02), only isolate RD 01 showed evidence of enzymatic resistance to d4T-TP (18-fold) at passage 6. The high-level resistance to d4T-TP seen in this isolate was associated with the selection of the V75I, 215Y, and H481Y mutations (Table 2). Isolate RD 02, which acquired the 215Y mutation only, did not have detectable d4T-TP resistance (Table 2).
Undetectable resistance to d4T in replication-based assays.
Table 2 also shows changes in d4T susceptibility by the PhenoSense and the MT-4/MTT assays. In the PhenoSense assay, the median IC50 value in baseline isolates was 0.50 μM (range, 0.47 to 0.63 μM) compared to 0.92 μM (range, 0.72 to 1.13 μM) in the d4T-selected isolates, with a median change in IC50 values of only 1.6-fold (range, 1.2- to 1.8-fold). Only two isolates (RD 02-p6 and RD 05-p6) had changes in IC50 for d4T (1.8-fold) that were close to the assay cutoff value of 1.7-fold for biological d4T resistance in the PhenoSense assay.
Low or undetectable resistance to d4T was also seen by the MT-4/MTT assay. The median EC50 value calculated in baseline isolates was 0.32 μM (range, 0.23 to 0.52 μM) compared to 0.57 μM (range, 0.22 to 0.75 μM) in d4T-selected isolates (median change in EC50 of 1.13-fold; range, 0.9- to 2.4-fold). Only two isolates (RD 02-p6 and RD 03-p8) showed more than a twofold reduction in d4T susceptibility (2.1- and 2.4-fold, respectively) (Table 2). Overall, these findings show the inability of the PhenoSense and the MT-4/MTT assays to detect d4T resistance in these viruses.
Susceptibility to d4T in site-directed mutants carrying K65R, T215Y, or K65R/T215Y.
The phenotypic effect of K65R and T215Y in d4T susceptibility was also evaluated in site-directed mutants carrying these mutations alone or in combination. Table 3 shows that in the HXB2 background, the K65R mutation conferred a 16-fold reduction in d4T-TP susceptibility and that the addition of T215Y increased the level of resistance to 21-fold. The presence of T215Y only in HXB2T215Y was associated with a threefold reduction in d4T-TP susceptibility. Table 3 also shows that the introduction of K65R in the RTs from the WT isolates RD 22wt and RD 23wt resulted in 17- and 10-fold resistance to d4T-TP, respectively, indicating that the effect of K65R was not limited to the genetic background of HXB2. In contrast, both the PhenoSense and the MT4/MTT were unable to detect d4T resistance in all these mutant viruses (Table 3).
TABLE 3.
Comparison between d4T susceptibility in WT viruses and site-directed mutants carrying K65R, T215Y, or K65R/T215Ya
| Virus | RT mutation(s) | RT assay [poly(A) × oligo(dT)15] result (fold change) | Replication assay result (fold change)
|
|
|---|---|---|---|---|
| MT-4/MTT | PhenoSense | |||
| HXB2wt | 0.011 | 0.52 | 0.48 (0.9) | |
| HXB2T215Y | T215Y | 0.036 (3) | 0.59 (1.1) | 0.86 (1.3) |
| HXB2K65R | K65R | 0.19 (16) | 0.60 (1.2) | 0.90 (1.4) |
| HXB2K65R/T215Y | K65R/T215Y | 0.24 (21) | 0.48 (0.9) | 1.04 (1.6) |
| RD 22wt | 0.0036 | 0.30 | 0.48 (0.8) | |
| RD 22K65R | K65R | 0.06 (17) | n.d. | 0.71 (1.1) |
| RD 23wt | 0.0046 | n.d. | 0.52 (0.9) | |
| RD 23K65R | K65R | 0.044 (10) | n.d. | 1.12 (1.4) |
Mean IC50 or EC50 values (micromolar) are shown. Values in parentheses are changes in IC50 values relative to the IC50 value of the WT isolate for the RT and MT4/MTT assays or a reference WT NL4-3 isolate for the PhenoSense assay. n.d., not done.
Detectability of cross-resistance to other NRTIs in d4T-selected viruses carrying K65R and other mutations.
We also evaluated patterns of cross-resistance to NRTIs in d4T-selected viruses. Table 4 shows changes in IC50 values for abacavir, didanosine (ddI), 3TC, tenofovir, zalcitabine (ddC), and AZT observed with the PhenoSense assay. Selection of the K65R mutation alone or with T215Y or V75A was associated with a median increase in IC50 values of 11-fold for 3TC, 2.5-fold for ddC, 2.4-fold for abacavir, 1.8-fold for ddI, and 1.7-fold for tenofovir. Among the nine d4T-selected isolates, eight had reduced susceptibility to 3TC, seven had reduced susceptibility to ddC, five had reduced susceptibility to tenofovir, and five had reduced susceptibility to ddI (Table 4). These findings indicate that the inability of the PhenoSense assay to detect resistance mediated by K65R was limited to d4T and that selection of this mutation is associated with cross-resistance to other NRTIs. We also evaluated patterns of resistance reversal in these viruses. Table 4 shows that resistance to AZT was undetectable in viruses that had K65R in association with 41L/210W/215Y (RD03-p8), 41L/215Y (RD05-p6), or 215Y (HXB2K65R/215Y), supporting previous findings showing that coexistence of K65R with 41L, 210W, and/or 215Y results in the loss of phenotypic resistance to AZT (2).
TABLE 4.
Cross resistance to other NRTIs and resistance reversal in viruses selected with d4T in vitro
| Recombinant and passage | RT genotype | IC50, μM (fold change) fora:
|
|||||
|---|---|---|---|---|---|---|---|
| ABC | ddI | 3TC | Tenofovir | ddC | AZT | ||
| RD 01 | |||||||
| p0 | 215C | 1.44 (0.8) | 4.59 (0.8) | 1.88 (0.9) | 0.87 (0.9) | 0.66 (0.8) | 0.035 (1.3) |
| p6 | V75I, C215Y, H481Y | 2.91 (1.7) | 7.8 (1.3) | 6.42 (3.1) | 0.82 (0.8) | 1.44 (1.7) | 0.050 (1.9) |
| RD 02 | |||||||
| p0 | 210W, 215C | 1.93 (1.1) | 4.85 (0.8) | 3.01 (1.5) | 0.82 (0.8) | 0.99 (1.2) | 0.03 (1.1) |
| p6 | 210W, C215Y | 3.03 (1.8) | 6.52 (1.1) | 4.06 (2.0) | 1.16 (1.2) | 1.06 (1.2) | 0.29 (11) |
| RD 03 | |||||||
| p0 | 41L, 210W, 215C | 2 (0.9) | 4.73 (0.8) | 2.39 (0.6) | 1.02 (1.0) | 0.78 (0.6) | 0.037 (1.9) |
| p5 | 41L, 210W, 215C, K65R | 4.27 (1.9) | 9.1 (1.6) | 29.6 (7.7) | 2.07 (2) | 2.36 (1.7) | 0.023 (1.2) |
| p8 | 41L, 210W, C215Y, K65R | 5.37 (2.4) | 9.47 (1.7) | 43.6 (11) | 2.18 (2.2) | 2.83 (2.1) | 0.033 (1.7) |
| RD 04 | |||||||
| p0 | 210W, 215D | 1.51 (0.9) | 5.98 (1.0) | 3.6 (1.7) | 0.64 (0.7) | 0.93 (1.1) | 0.016 (0.6) |
| p6 | 210W, 215D, K65R | 3.6 (2.1) | 10.9 (1.8) | 38.8 (19) | 1.58 (1.6) | 2.45 (2.9) | 0.011 (0.4) |
| RD 05 | |||||||
| p0 | 41L, 215D | 1.83 (0.8) | 5.45 (1.0) | 5.95 (1.6) | 0.68 (0.7) | 1.22 (0.9) | 0.02 (1.0) |
| p5 | 41L, 215D, K65R | 3.5 (1.6) | 9.35 (1.7) | 27.5 (7.2) | 1.21 (1.2) | 2.9 (2.1) | 0.016 (0.8) |
| p6 | 41L, D215Y, K65R | 4.14 (2.4) | 10.5 (1.8) | 36.8 (18) | 1.68 (1.7) | 2.11 (2.5) | 0.02 (0.7) |
| HXB2215D | |||||||
| p0 | 215D | 1.27 (0.7) | 5.1 (0.9) | 3.9 (1.9) | 0.35 (0.4) | 0.99 (1.2) | 0.005 (0.2) |
| p6 | 215D, K65R | 4.09 (2.4) | 12.5 (2.1) | 31.63 (15) | 1.29 (1.3) | 2.85 (3.3) | 0.01 (0.4) |
| RD 22wt | |||||||
| p0 | 1.12 (0.6) | 4.61 (0.8) | 1.79 (0.9) | 0.56 (0.6) | 0.69 (0.8) | 0.014 (0.5) | |
| p6 | K65R, M202I | 4.18 (2.4) | 12.5 (2.1) | 26.5 (13) | 2.11 (2.2) | 2.88 (3.4) | 0.02 (0.7) |
| p0K65Rb | K65R | 3.39 (1.5) | 10.3 (1.5) | 19.2 (7.2) | 1.7 (1.6) | 2.52 (2.6) | 0.01 (0.4) |
| RD 23wt | |||||||
| p0 | 1.78 (1.0) | 4.52 (0.8) | 2.13 (1.0) | 0.84 (0.9) | 0.84 (1.0) | 0.02 (0.7) | |
| p6 | K65R | 4.87 (2.8) | 8.8 (1.5) | 22.4 (11) | 1.76 (1.8) | 2.09 (2.4) | 0.014 (0.5) |
| p0K65Rb | K65R | 4.50 (1.9) | 12.5 (1.6) | 28.9 (11) | 2.18 (1.9) | 2.7 (2.9) | 0.012 (0.4) |
| HXB2wt | |||||||
| p0 | 1.36 (0.8) | 4.56 (0.8) | 2.39 (1.2) | 0.48 (0.9) | 0.89 (1.0) | 0.007 (0.3) | |
| p7 | V75A, K65R | 3.43 (2.0) | 13.6 (2.3) | 21.1 (11) | 1.19 (1.2) | 2.35 (3.5) | 0.009 (0.4) |
| HXB265Rb | K65R | 3.45 (2.0) | 11.5 (2.0) | 16.9 (8.8) | 1.12 (1.2) | 2.21 (3.3) | 0.007 (0.3) |
| HXB265R/215Yb | K65R, T215Y | 3.35 (2.0) | 11.2 (1.9) | 29.3 (15) | 1.07 (1.1) | 2.51 (3.7) | 0.008 (0.3) |
| HXB2215Yb | T215Y | 1.74 (1.0) | 6.72 (1.2) | 4.46 (2.3) | 0.52 (0.5) | 1 (1.5) | 0.015 (0.6) |
Values in parentheses are changes in drug susceptibility compared to the IC50 value of the reference WT NL4-3 isolate used in the PhenoSense assay. In boldface are indicated the isolates that had evidence of reduced susceptibility using the assay cutoff values of the PhenoSense assay. ABC, abacavir.
Mutations were introduced by site-directed mutagenesis.
Impact of K65R and 215Y on replication capacity in the presence and absence of d4T.
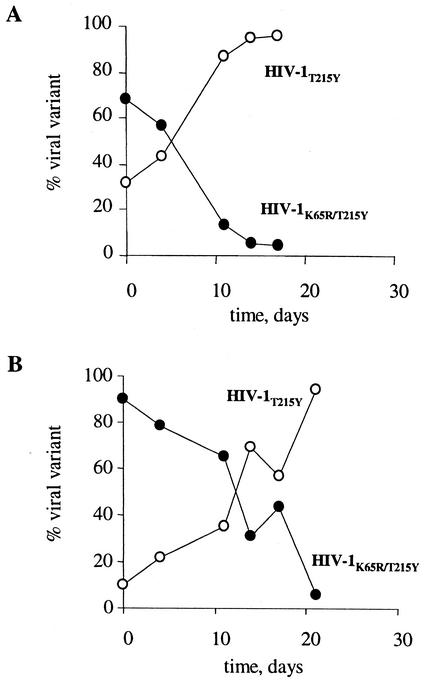
To evaluate the impact of both K65R and T215Y on replication capacity, we determined the fitness difference between HIV-1K65R/T215Y and HIV-1215Y in the presence of 12 μM d4T. Figure 1 shows the relative proportion of HIV-1T215Y and HIV-1T215Y/K65R over time in two experiments in which HIV-1T215Y comprised 10 or 30% of the initial virus mixture. In both cases, HIV-1T215Y outgrew HIV-1T215Y/K65R, indicating that coexistence of T215Y and K65R confers a fitness cost in the presence of d4T compared to viruses that have only T215Y. The fitness difference s was calculated in the two mixing experiments and was found to be 0.17 and 0.24, respectively, indicating that HIV-1T215Y is on average 20% more fit than HIV-1T215Y/K65R in the presence of a high concentration of d4T (Fig. 1).
FIG. 1.
Fitness difference between HIV-1T215Y and HIV-1K65R/T215Y in the presence of 12 μM d4T. The relative proportion of HIV-1T215Y and HIV-1K65R/T215Y mixed at two different initial ratios (30%/70% [A] or 10%/90% [B]) is shown over time.
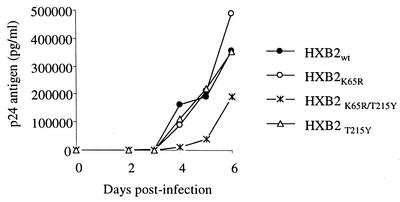
The impact of K65R and 215Y on replication capacity was also evaluated in the absence of d4T. Figure 2 compares the kinetics of p24 antigen production among WT viruses and viruses that have K65R, a combination of K65R and T215Y, or T215Y only. The results indicated that in these assay conditions, HXB2K65R, HXB2wt, and HXB2T215Y had similar replication capacities and were indistinguishable from each other. In contrast, HXB2K65R/T215Y had delayed p24 antigen production, indicating decreased replication capability. These findings further confirm that coexistence of K65R and T215Y has a negative impact on viral fitness compared to WT viruses or viruses that have either K65R or T215Y.
FIG. 2.
Replication kinetics of HIV-1K65R, HIV-1T215Y, and HIV-1K65R/T215Y and comparison with the WT HIV-1T215. Mean p24 values from duplicate cultures are shown.
DISCUSSION
We investigated the genotypic and phenotypic determinants of d4T resistance by monitoring RT mutations and changes in d4T susceptibility in nine different HIV-1 isolates cultured with d4T. We show that K65R was the most frequently selected mutation and was seen in seven isolates. We also demonstrate biochemical resistance to d4T-TP in all seven viruses that acquired the K65R mutation, a finding consistent with the selection of this mutation at high d4T concentrations. We confirm the role of K65R in the observed d4T-TP resistance by showing that introduction of K65R in three different RT backgrounds confers resistance to d4T-TP. These findings provide direct evidence indicating that K65R represents a new pathway for d4T resistance. The selection of K65R in viruses with genetically distinct RT backgrounds and the associated resistance to d4T-TP further suggest that this pathway is not restricted to specific RT backgrounds. The resistance to d4T observed in viruses with K65R implies that the clinical utility of d4T may be reduced in patients that have viruses carrying K65R and who initiate d4T treatment.
Our data also document the inability of replication-based assays to detect d4T resistance. We found that d4T-selected mutants that can replicate well in the presence of high concentrations of d4T and are biochemically resistant to d4T-TP were indistinguishable from WT viruses in these assays. We also found that the inability to detect phenotypic resistance mediated by both K65R and 215Y may be restricted to d4T, since these mutants were found to be cross-resistant to other NRTIs. The failure to detect d4T resistance was seen despite differences in the testing strategies between the two assays in the number of replication cycles or the use of recombinant or whole virions, suggesting that other factors play a role in this phenomenon.
Our study documents the first instance in which replication assays do not correlate with biochemical assays and thus underscores the importance of both biochemical and replication-based testing to obtain a full phenotypic characterization of resistance mutations. A better understanding of the basis of why replication assays do not detect resistance to d4T-TP may help in the development of modified assays capable of detecting reduced d4T susceptibility. Meyer et al. have proposed that because d4T-MP excision is sensitive to inhibition by high concentrations of the next complementary dNTP, the high intracellular dNTP levels present in cell lines used in replication assays may play a role in the inability of these assays to detect d4T resistance in TAM-containing mutants (25). However, it is not known whether the intracellular concentrations of dNTPs are important for detecting d4T resistance in mutants with K65R.
The fact that our biochemical data were all derived from an RT assay that has not been modified to measure excision of d4T suggests that the underlying mechanism of the d4T resistance detected in the mutants containing K65R may be more through increased discrimination against d4T-TP rather than enhanced excision. These data expand previous findings showing that enzymes with K65R have increased discrimination against 3TC, ddC, and ddI (12). It is also possible that in some mutants that have both 215Y and K65R, enhanced excision of d4T-TP may also be involved in the d4T resistance. However, since our testing was not designed to measure d4T unblocking activity, our data cannot address this possibility.
The K65R mutation has been previously associated with emergence of resistance to tenofovir, ddC, and abacavir in vitro and confers cross-resistance to ddI and 3TC (7, 14, 43, 45). Despite this association, the prevalence of K65R in patients treated with these drugs is low compared to the prevalence of other mutations, suggesting that the emergence of K65R is less favored, probably because of the negative impact of this mutation on replication capacity (5, 20-22, 26, 28). We demonstrate that in the presence of d4T, viruses that have both K65R and T215Y are on average 20% less fit than viruses that have only T215Y. Reductions in fitness of 0.4 to 2.3% have been associated with a shift in virus populations carrying resistance mutations in vivo (11). The reduced fitness in this virus might be related to changes in processivity and fidelity of RT conferred by K65R (1, 41). A lower fitness of HIV-1K65R/T215Y compared to HIV-1T215Y might favor reversion of K65R, and might explain the frequent observation of T215Y but not K65R in patients treated with d4T (5, 20, 21, 30). However, our data raise questions on whether an increased prevalence of K65R will be seen in the future in patients failing regimens containing d4T and other K65R-selecting drugs like abacavir or tenofovir. A surprisingly high incidence of K65R has been seen in patients receiving combination therapy with abacavir, didanosine, and d4T, suggesting that K65R can be consistently selected in vivo if the selective pressure is adequate (35).
Our results showing the selection of the 215Y mutation by d4T in four of six viruses containing 215D/C provide direct evidence for a primary role of this mutation in d4T resistance. We also demonstrate that viruses carrying T215Y can replicate in a concentration of d4T that is 26-fold higher than the IC50 value of the WT isolate. These findings support the association between T215Y and d4T found in d4T-treated patients. The longer time required to select 215Y with d4T than with AZT suggests a higher selective advantage conferred by 215Y for AZT than for d4T (9). The lower selective advantage of HIV-1T215Y with d4T might explain why 215Y is selected less frequently in patients who fail d4T than in patients who fail AZT treatment (5, 21, 30, 36, 37).
Our data showing that HIV-1215C and HIV-1215D can acquire 215Y with d4T more easily than WT viruses expand our previous observations showing the increased ability of these viruses to select the 215Y mutation in the presence of AZT (9). These data may have clinical implications, since recent findings have shown that patients infected with these mutants are at increased risk of virologic failure compared to persons infected with WT viruses (6, 34). Therefore, the use of thymidine analogs in patients infected with 215C/D/S mutants may not be prudent. Additional assessment of clinical responses in patients infected with these viruses and treated with antiretroviral therapy is needed.
In conclusion, we provide evidence of a distinct pathway of d4T resistance that involves selection of the K65R mutation. We also demonstrate in vitro the association between the 215Y mutation and d4T resistance, supporting a role of TAMs in d4T treatment failure. The inability of culture assays to detect d4T resistance mediated by K65R emphasizes the need for a better understanding of the determinants of d4T resistance in culture. This finding has important implications for phenotypic resistance testing for d4T.
Acknowledgments
We thank Irum Zaidi, Toni Woods, Sara Mirza, and the Sentinel Surveillance for Variant and Drug-Resistant Strains of HIV-1 Study team for their contribution to this work. We also thank John Barr at the CDC for the analysis of the purity of d4T.
Hamish MacInnes was supported by a fellowship from the Oak Ridge Institute for Science and Education, and Patrick Reid was supported by an Emerging Infectious Diseases Laboratory Fellowship.
REFERENCES
- 1.Arion, D., G. Borkow, Z. Gu, M. A. Wainberg, and M. A. Parniak. 1996. The K65R mutation confers increased DNA polymerase processivity to HIV-1 reverse transcriptase. J. Biol. Chem. 271:19860-19864. [DOI] [PubMed] [Google Scholar]
- 2.Bazmi, H. Z., J. L. Hammond, S. C. Cavalcanti, C. K. Chu, R. F. Schinazi, and J. W. Mellors. 2000. In vitro selection of mutations in the human immunodeficiency virus type 1 reverse transcriptase that decrease susceptibility to (−)-β-d-dioxolane-guanosine and suppress resistance to 3′-azido-3′-deoxythymidine. Antimicrob. Agents Chemother. 44:1783-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher, C. A. B., N. Cammack, P. Scipper, R. Schuurman, P. Rouse, M. A. Wainberg, and J. M. Cameron. 1993. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Aquila, R. T., J. M. Schapiro, F. Brun-Vezinet, B. Clotet, B. Conway, L. M. Demeter, R. M. Grant, V. A. Johnson, D. R. Kuritzkes, C. Loveday, R. W. Shafer, and D. D. Richman. 2002. Drug resistance mutations in HIV-1. Top. HIV Med. 10:11-15. [PubMed] [Google Scholar]
- 5.de Mendoza, C., V. Soriano, C. Briones, O. Gallego, P. Barreiro, A. Alvarez, and J. González-Lahoz. 2000. Emergence of zidovudine resistance in HIV-infected patients receiving stavudine. J. Acquir. Immune Defic. Syndr. 23:279-281. [DOI] [PubMed] [Google Scholar]
- 6.de Ronde, A., M. van Dooren, E. de Rooij, B. van Gemen, J. Lange, and J. Goudsmit. 2000. Infection by zidovudine-resistant HIV-1 compromises the virological response to stavudine in a drug-naive patient. AIDS 14:2632-2633. [DOI] [PubMed] [Google Scholar]
- 7.Foli, A., K. M. Sogocio, B. Anderson, M. Kavlick, M. W. Saville, M. A. Wainberg, Z. Gu, J. M. Cherrington, H. Mitsuya, and R. Yarchoan. 1996. In vitro selection and molecular characterization of human immunodeficiency virus type 1 with reduced sensitivity to 9-[2-(phosphonomethoxy)ethyl]adenine (PMEA). Antivir. Res. 32:91-98. [DOI] [PubMed] [Google Scholar]
- 8.García-Lerma, J. G., P. J. Gerrish, A. C. Wright, S. H. Qari, and W. Heneine. 2000. Evidence of a role for the Q151L mutation and the viral background in development of multiple dideoxynucleoside-resistant human immunodeficiency virus type 1. J. Virol. 74:9339-9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Lerma, J. G., S. Nidtha, K. Blumoff, H. Weinstock, and W. Heneine. 2001. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. Proc. Natl. Acad. Sci. USA 98:13907-13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goudsmit, J., A. de Ronde, E. de Rooij, and R. de Boer. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goudsmit, J., A. de Ronde, D. D. Ho, and A. S. Perelson. 1996. Human immunodeficiency virus fitness in vivo: calculations based on single zidovudine resistance mutation at codon 215 of reverse transcriptase J. Virol. 70:5662-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu, Z., R. S. Fletcher, E. J. Arts, M. A. Wainberg, and M. A. Parniak. 1994. The K65R mutant reverse transcriptase of HIV-1 cross-resistant to 2′,3′-dideoxycytidine, 2′,3′-dideoxy-3′-thiacytidine, and 2′,3′-dideoxyinosine shows reduced sensitivity to specific dideoxynucleoside triphosphate inhibitors in vitro J. Biol. Chem. 269:28118-28122. [PubMed] [Google Scholar]
- 13.Gu, Z., Q. Gao, H. Fang, M. A. Parniak, B. G. Brenner, and M. A. Wainberg. 1994. Identification of novel mutations that confer drug resistance in the human immunodeficiency virus polymerase gene. Leukemia 8(Suppl. 1):S166-S169. [PubMed] [Google Scholar]
- 14.Gu, Z., Q. Gao, H. Fang, H. Salomon, M. A. Parniak, E. Goldberg, J. Cameron, and M. A. Wainberg. 1994. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine Antimicrob. Agents Chemother. 38:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertogs, K., M. de Bethune, V. Miller, T. Ivens, P. Schel, A. van Cauwenberge, C. Van den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, V. A., C. J. Petropoulos, C. R. Woods, J. D. Hazelwood, N. T. Parkin, C. D. Hamilton, and S. A. Fiscus. 2001. Vertical transmission of multidrug-resistant human immunodeficiency virus type 1 (HIV-1) and continued evolution of drug resistance in an HIV-1-infected infant. J. Infect. Dis. 183:1688-1693. [DOI] [PubMed] [Google Scholar]
- 17.Lacey, S. F., and B. A. Larder. 1994. Novel mutation (V75T) in human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine in cell culture. Antimicrob. Agents Chemother. 38:1428-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larder, B. A., K. E. Coates, and S. D. Kemp. 1991. Zidovudine-resistant human immunodeficiency virus selected by passage in cell culture J. Virol. 65:5232-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1157. [DOI] [PubMed] [Google Scholar]
- 20.Lin, P. F., C. J. González, B. Griffith, G. Friedland, V. Calvez, F. Ferchal, R. F. Schinazi, D. H. Shepp, A. B. Ashraf, M. A. Wainberg, V. Soriano, J. W. Mellors, and R. J. Colonno. 1999. Stavudine resistance: an update on susceptibility following prolonged therapy. Antivir. Ther. 4:21-28. [PubMed] [Google Scholar]
- 21.Lin, P. F., H. Samanta, R. E. Rose, A. K. Patick, J. Trimble, C. M. Bechtold, D. R. Revie, N. C. Khan, M. E. Federici, H. Li, A. Lee, R. E. Anderson, and R. J. Colonno. 1994. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy J. Infect. Dis. 170:1157-1164. [DOI] [PubMed] [Google Scholar]
- 22.Maxeiner, H. G., W. Keulen, R. Schuurman, M. Bijen, L. de Graaf, A. van Wijk, N. Back, M. W. Kline, C. A. Boucher, and M. Nijhuis. 2002. Selection of zidovudine resistance mutations and escape of human immunodeficiency virus type 1 from antiretroviral pressure in stavudine-treated pediatric patients J. Infect. Dis. 185:1070-1076. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 24.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism Proc. Natl. Acad. Sci. USA. 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, P. R., S. E. Matsuura, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, M. D., N. A. Margot, P. D. Lamy, M. D. Fuller, K. E. Anton, A. S. Mulato, and J. M. Cherrington. 2001. Adefovir and tenofovir susceptibilities of HIV-1 after 24 to 48 weeks of adefovir dipivoxil therapy: genotypic and phenotypic analyses of study GS-96-408. J. Acquir. Immune Defic. Syndr. 27:450-458. [DOI] [PubMed] [Google Scholar]
- 27.Moore, J. D., G. Valette, A. Darque, X.-J. Zhou, and J.-P. Sommadossi. 2000. Simultaneous quantitation of the 5′-triphosphate metabolites of zidovudine, lamivudine, and stavudine in peripheral mononuclear blood cells of HIV infected patients by high-performance liquid chromatography tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 11:1134-1143. [DOI] [PubMed] [Google Scholar]
- 28.Moyle, G. J., and B. G. Gazzard. 2001. Differing reverse transcriptase mutation patterns in individuals experiencing viral rebound on first-line regimens with stavudine/didanosine and stavudine/lamivudine. AIDS 15:799-800. [DOI] [PubMed] [Google Scholar]
- 29.Pauwels, R., J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter, and E. de Clercq. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20:309-321. [DOI] [PubMed] [Google Scholar]
- 30.Pellegrin, I., J. Izopet, J. Reynes, M. Denayrolles, B. Montes, J. L. Pellegrin, P. Massip, J. Puel, H. Fleury, M. Segondy, et al. 1999. Emergence of zidovudine and multidrug-resistance mutations in the HIV-1 reverse transcriptase gene in therapy-naive patients receiving stavudine plus didanosine combination therapy. AIDS 13:1705-1709. [DOI] [PubMed] [Google Scholar]
- 31.Petropoulos, C. J., N. T. Parkin, K. L. Limoni, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Picard, V., E. Angelini, A. Maillard, E. Race, F. Clavel, G. Chene, F. Ferchal, and J. M. Molina. 2001. Comparison of genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 isolates from patients treated with stavudine and didanosine or zidovudine and lamivudine. J. Infect. Dis. 184:781-784. [DOI] [PubMed] [Google Scholar]
- 33.Reed, L. J., and H. Muench. 1938. A simple method for estimating fifty percent end points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 34.Riva, C., M. Violin, A. Cozzi-Lepri, R. Velleca, A. Bertoli, P. Vigano, L. Minoli, A. Orani, G. Rizzardini, T. Zauli, C. F. Perno, A. d'Arminio-Monforte, and C. Balotta for the I.Co.N.A. Study Group. 2002. Transmitted virus with substitutions at position 215 and risk of virological failure in antiretroviral-naive patients starting highly active antiretroviral therapy. Antiviral Ther. 7:S136. [Google Scholar]
- 35.Roge, B. T., T. L. Katzenstein, N. Obel, H. Nielsen, O. Kirk, C. Pedersen, L. Mathisen, J. Lundgren, and J Gerstoft. K65R with and without S68—a new resistance profile in vivo detected in most patients failing abacavir, didanosine, and stavudine. Antivir. Ther., in press. [PubMed]
- 36.Ross, L., K. Henry, D. Paar, P. Salvato, M. Shaefer, R. Fisher, Q. Liao, and M. St Clair. 2001. Thymidine-analog and multi-nucleoside resistance mutations are observed in both zidovudine-naive and zidovudine-experienced subjects with viremia after treatment with stavudine-containing regimens. J. Hum. Virol. 4:217-222. [PubMed] [Google Scholar]
- 37.Ross, L., A. Scarsella, S. Raffanti, K. Henry, S. Becker, R. Fisher, Q. Liao, A. Hirani, N. Graham, M. St Clair, and J. Hernandez for the NZT40012 Study Team. 2001. Thymidine analog and multinucleoside resistance mutations are associated with decreased phenotypic susceptibility to stavudine in HIV type 1 isolated from zidovudine-naive patients experiencing viremia on stavudine-containing regimens. AIDS Res. Hum. Retrovir. 17:1107-1115. [DOI] [PubMed] [Google Scholar]
- 38.Schmit, J. C., K. Van Laethem, L. Ruiz, P. Hermans, S. Sprecher, A. Sonnerborg, M. Leal, T. Harrer, B. Clotet, V. Arendt, E. Lissen, M. Witvrouw, J. Desmyter, E. de Clercq, and A. M. Vandamme. 1998. Multiple dideoxynucleoside analogue-resistant (MddNR) HIV-1 strains isolated from patients from different European countries. AIDS 12:2005-2015. [DOI] [PubMed] [Google Scholar]
- 39.Schmit, J. C., J. Cogniaux, P. Hermans, C. Van Vaeck, S. Sprecher, B. Van Remoortel, M. Witvrouw, J. Balzarini, J. Desmyter, E. de Clercq, and A. M. Vandamme. 1996. Multiple drug resistance to nucleoside analogs and nonnucleoside reverse transcriptase inhibitors in an efficiently replicating human immunodeficiency virus type 1 patient strain. J. Infect. Dis. 174:962-968. [DOI] [PubMed] [Google Scholar]
- 40.Scott, W. A. 2001. The enzymatic basis for thymidine analogue resistance in HIV-1. AIDS Rev. 3:194-200. [Google Scholar]
- 41.Shah, F. S., K. A. Curr, M. E. Hamburgh, M. Parniak, H. Mitsuya, J. G. Arnez, and V. R. Prasad. 2000. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 275:27037-27044. [DOI] [PubMed] [Google Scholar]
- 42.Shulman, N. S., R. A. Machekano, R. W. Shafer, M. A. Winters, A. R. Zolopa, S. H. Liou, M. Hughes, D. A. Katzenstein, and the AIDS Clinical Trials Group 302 Study Team. 2001. Genotypic correlates of a virologic response to stavudine after zidovudine monotherapy. J. Acquir. Immune Defic. Syndr. 27:377-380. [DOI] [PubMed] [Google Scholar]
- 43.Tisdale, N. S., D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of the reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandamme, A. M., M. Witvrouw, C. Pannecouque, J. Balzarini, K. Van Laethem, J. C. Schmit, J. Desmyter, and E. de Clercq. 1999. Evaluating clinical isolates for their phenotypic and genotypic resistance against anti-HIV drugs, p. 223-258. In J. Kinchington and R. F. Schinazi (ed.), Methods in molecular medicine: antiviral chemotherapy protocols, vol. 24. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 45.Wainberg, M. A., M. D. Miller, Y. Quan, H. Salomon, A. S. Mulato, P. D. Lamy, N. A. Margot, K. E. Anton, and J. M. Cherrington. 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir. Ther. 4:87-94. [DOI] [PubMed] [Google Scholar]
- 46.Winters, M. A., K. L. Coolley, Y. A. Girard, D. J. Levee, H. Hamdan, R. W. Shafer, D. A. Katzenstein, and T. C. Merigan. 1998. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J. Clin. Investig. 102:1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]