Abstract
Coronavirus defective RNAs (D-RNAs) have been used for site-directed mutagenesis of coronavirus genomes and for expression of heterologous genes. D-RNA CD-61 derived from the avian coronavirus infectious bronchitis virus (IBV) was used as an RNA vector for the expression of chicken gamma interferon (chIFN-γ). D-RNAs expressing chIFN-γ were shown to be capable of rescue, replication, and packaging into virions in a helper virus-dependent system following electroporation of in vitro-derived T7 RNA transcripts into IBV-infected cells. Secreted chIFN-γ, under the control of an IBV transcription-associated sequence derived from gene 5 of the Beaudette strain, was expressed from two different positions within CD-61 and shown to be biologically active. In addition, following infection of 10-day-old chicken embryos with IBV containing D-RNAs expressing chIFN-γ, the allantoic fluid was shown to contain biologically active chIFN-γ, demonstrating that IBV D-RNAs can express heterologous genes in vivo.
Infectious bronchitis virus (IBV) is a highly infectious and economically important pathogen of chickens that causes respiratory disease, diminished growth rate, and substantial decline in egg production. Although infectious bronchitis is considered primarily a disease of the respiratory system, strains of IBV have wide and variable tropisms and the clinical manifestations of the disease can be diverse (9). Genetically very similar viruses cause disease in turkeys (6) and pheasants (7). IBV is a group 3 member of the genus Coronavirus of the family Coronaviridae in the order Nidovirales (12), being an enveloped RNA virus with an unsegmented, 5′-end-capped, 3′-end-polyadenylated, single-stranded, positive-sense RNA genome of 27,608 nucleotides (nt) (4). Coronaviruses produce a 3′-coterminal nested set of subgenomic mRNAs (sg mRNAs) that are polycistronic. These are produced by a discontinuous transcription process during synthesis of the negative strand and contain identical 5′ ends due to the addition of a leader sequence derived from the 5′ end of the genomic RNA (gRNA) (35, 36). Preceding the body sequence of each sg mRNA is a consensus sequence, the transcription-associated sequence (TAS) (15), involved in the acquisition of the leader sequence. All coronavirus envelopes contain at least three membrane proteins, the spike glycoprotein, a small membrane protein, and an integral membrane protein. In addition, the coronavirus virion also contains a nucleocapsid protein that interacts with the gRNA.
Coronavirus defective RNAs (D-RNAs), which lack large parts of the genome, are produced following virus passage at a high multiplicity of infection. While all D-RNAs contain cis-acting sequences necessary for replication, only a subset of D-RNAs contain sequences necessary for packaging into virions in the presence of a helper virus. Coronavirus D-RNAs have been used for site-directed mutagenesis of the virus genome (25) and for expression of heterologous genes (1, 2, 13, 17, 20, 22, 39, 42, 43).
Cytokines are regulatory proteins that act as a communication network between cells throughout immunological development. Avian homologues of several mammalian cytokines have been isolated, including chicken gamma interferon (chIFN-γ) (10). Mammalian IFN-γ is a pleiotropic cytokine initially produced by natural killer cells, during immune induction, and then by committed T helper 1 (Th1) cells as a regulator and effector molecule for driving inflammatory responses (for a review, see reference 8). IFN-γ has a major role in activating antiviral immune responses through augmentation of major histocompatibility complex expression on antigen-presenting cells for interaction with T cells, stimulation of antibody (Ab) formation, promotion of Ab isotype class switching, and development of cytotoxic T cells. Several studies have been undertaken to investigate the in vivo potential of IFN-γ as a vaccine adjuvant. Coadministration of bovine IFN-γ with vesicular stomatitis virus G glycoprotein to cattle resulted in an increased formation of protective Ab against vesicular stomatitis virus (41). In a mouse model, fusion of IFN-γ to human immunodeficiency virus gp120 resulted in enhanced primary Ab responses against gp120, enhanced antigen-specific T-cell proliferation, and IFN-γ production (26). The use of recombinant feline IFN-γ as a vaccine adjuvant increased Ab responses against rabies virus and calicivirus antigens (37).
The role of recombinant chIFN-γ as an adjuvant and therapeutic agent has been investigated. Coadministration of chIFN-γ with sheep red blood cells (SRBCs) resulted in an increased secondary Ab response with amounts of SRBCs 10-fold smaller than the dose of SRBCs given alone (23, 24). Administration of recombinant chIFN-γ prior to challenge with avian coccidia resulted in decreased intracellular sporozoite development and oocyst production, with an enhanced level of body weight gain (21). chIFN-γ had an adjuvant effect, reducing parasite replication, when used in a DNA vaccine regimen against Eimeria acervulina (27). Coexpression of Newcastle disease virus antigens with chIFN-γ by using fowlpox virus resulted in an earlier Ab response, with the best protective immune response of the recombinant fowlpox virus vaccines (32).
In this study we describe both the in vitro and in ovo expression of biologically active chIFN-γ from an IBV D-RNA. We demonstrate for the first time the expression of a chicken cytokine from an IBV D-RNA and the in ovo expression of a biologically active heterologous gene from an IBV D-RNA.
MATERIALS AND METHODS
Cells and viruses.
IBV Beaudette was grown in 11-day-old embryonated domestic fowl eggs, harvested from allantoic fluid 24 h postinfection, and used as helper virus for the rescue of IBV D-RNAs (31). IBV was passaged and titrated on primary chick kidney (CK) cells (30). HD11 cells, an avian leukosis virus (MC29)-transformed cell line (3), were cultured in RPMI 1640 medium (Life Technologies) containing 2.5% fetal calf serum, 2.5% chick serum, 10% tryptose phosphate broth, 20 mM l-glutamine, 0.225% NaHCO3, 1 U of penicillin/ml, and 1 μg of streptomycin/ml.
Oligonucleotides.
The oligonucleotides used in this work were obtained from Invitrogen and are listed in Table 1.
TABLE 1.
Oligonucleotides used for generation of TAS-chIFN-γ cassette and hybridization probes
| Oligonucleotide | Sequencea | Positionb | Polarity |
|---|---|---|---|
| IBV5IFN-γSTART | TCC CCC GGG CACGTG TTT TAC TTA ACA AAA ACT TAA CAA ATA CGG ACG ATG ACT TGC CAG ACT | NA | + |
| SmallFN-γ-END | ACC CCC GGG GGT TAG CAA TTG CAT CT | NA | − |
| BG-67 | GGC TGG TTC GAG TGC GAG | Beaudette 265-282 | + |
| BG-2 | TCA GGG GTT GTT TGG CAC T | Beaudette 455-473 | − |
| IFN/3 | ATG ACT TGC CAG ACT TAC AA | chIFN-γ 1-20 | + |
| IFN/7 | CAG GTC CAT GAT ATC TTT CAC | chIFN-γ 340-360 | − |
Underlined sequences correspond to the IBV sequence. Nucleotides marked in bold correspond to the IBV canonical TAS, and those in italics correspond to restriction endonuclease sites used for cloning.
Recombinant DNA techniques.
Standard procedures were used to produce recombinant DNA (33), or methods were according to the manufacturers' instructions.
Construction of TAS-chIFN-γ gene cassette.
A TAS-chIFN-γ cassette was produced by PCR for insertion into the IBV D-RNA CD-61 cDNA sequence in pIBV-Vec (13). Oligonucleotides IBV5IFN-γSTART and SmaIIFN-γ-END, corresponding to the 5′ end and complementary to the 3′ end, respectively, of the chIFN-γ sequence, were used to generate the TAS-chIFN-γ cassette. Oligonucleotide IBV5IFN-γSTART contained the restriction endonuclease SmaI and PmaCI sites, the Beaudette-derived gene 5 TAS (39), and the first 15 nt of the chIFN-γ sequence. Oligonucleotide SmaIIFN-γ-END consisted of the last 15 nt (complementary) of the chIFN-γ gene, including the termination codon, followed by a SmaI site. The chIFN-γ sequence was amplified by PCR by using Taq/Pwo DNA polymerase (Hybaid) from plasmid pGEM-T-IFN-γ (19) containing a cDNA derived from chIFN-γ mRNA. The TAS-chIFN-γ cassette was digested with XmaI and ligated into XmaI-digested pBluescript II SK(+) (Stratagene), resulting in pBS-IFN-γ+ENDS, in which the TAS-chIFN-γ sequence was verified by sequence analysis. The TAS-chIFN-γ cassette was removed from pBS-IFN-γ+ENDS by using PmaCI and SmaI and inserted into either the PmaCI site or the SnaBI site in the CD-61 sequence of pIBV-Vec.
In vitro rescue of chIFN-γ-containing D-RNAs by IBV Beaudette.
In vitro T7-derived D-RNA transcripts were synthesized from 1 μg of plasmid DNA and electroporated into IBV-infected CK cells (passage 0 [P0]) (39). Virus V1 in 1 ml of cell medium was used to infect CK cells, and viruses V2 to V6 were serially passaged every 24 h for six passages (P1 to P6).
In ovo rescue of chIFN-γ-containing D-RNAs.
Cell medium (100 μl) from P3 CK cells, corresponding to peak D-RNA levels, was used to infect 10-day-old Rhode Island Red specific-pathogen-free embryos. The infected embryos were incubated at 37°C for 16 h and cooled to 4°C overnight. Allantoic fluid was collected from the infected eggs, centrifuged at 1,500 × g for 10 min, and stored at −70°C. Allantoic fluid (100 μl) containing IBV and any potential D-RNA was used to infect CK cells.
Identification of IBV-derived RNAs.
Total cytoplasmic RNA was extracted from infected CK cells by using RNeasy (Qiagen) and electrophoresed in denaturing 1% agarose-2.2 M formaldehyde gels. The RNAs were Northern blotted onto Hybond XL nylon membranes (Amersham), and IBV-derived RNAs were detected with probes covalently labeled with psoralen-biotin (BrightStar; Ambion) (13). The probes were hybridized to the RNAs at 42°C for 16 h, detected with streptavidin-alkaline phosphatase conjugate in the presence of an alkaline phosphatase 1,2-dioxetane chemiluminescent substrate (CDPStar and BrightStar Biodetect; Ambion), and exposed to film at room temperature for 2 h. An IBV-specific 309-nt 3′-untranslated region (UTR) probe, corresponding to the last 309 nt of the 3′ end of the IBV genome, was used to detect all IBV-derived RNA species, including IBV-derived D-RNAs (13). A 209-nt 5′ IBV-specific probe, produced by PCR with oligonucleotides BG-67 and BG-2 (Table 1) corresponding to nt 265 to 474 within the 5′ UTR of the Beaudette genome, was used to detect IBV gRNA and D-RNAs. A 360-nt chIFN-γ-specific probe, produced by PCR with primers IFN/3 and IFN/7 (Table 1), was used to detect RNAs containing the chIFN-γ sequence.
chIFN-γ bioassays and neutralization assays.
The chIFN-γ bioassays and neutralization assays were carried out as described by Lawson et al. (19). Essentially, triplicate samples (200 μl) of serially diluted CK cell medium or allantoic fluid, either from mock-infected embryos or embryos infected with IBV, were added to 4 × 104 HD11 cells in 96-well flat-bottomed plates. Recombinant chIFN-γ, prepared as described by Lawson et al. (19), was serially diluted twofold and used as a positive control. chIFN-γ neutralization assays were carried out by using an anti-chIFN-γ neutralizing monoclonal Ab (MAb), 1E-12 (18), and an anti-bovine granulocyte-macrophage colony-stimulating factor polyclonal Ab, CC305 (kindly provided by Paul Sopp, Institute for Animal Health, Compton, United Kingdom). Both antibodies were diluted 1:1,000 and incubated with CK cell medium and allantoic fluid samples for 2 h at room temperature before being added to HD11 cells. For controls, all samples were also preincubated with the isotype control Ab CC305 and media alone. All HD11 cells were incubated at 41°C in 5% CO2 for 48 h for the chIFN-γ bioassays.
Nitric oxide (NO) produced from HD11 cells, resulting from induction with chIFN-γ, was measured in HD11 cell medium (100 μl) as nitrite (NO2−) by using a modification of the Griess assay described by Lawson et al. (19) and a Spectra Max 250 enzyme-linked immunosorbent assay reader (Molecular Devices, Wokingham, United Kingdom). The amount of NO present in the HD11 cell media was expressed as the concentration of NO2− (millimolar) for the chIFN-γ bioassays or as the optical density at 543 nm for the neutralizing assays.
RESULTS
D-RNAs containing TAS-chIFN-γ.
Expression of heterologous genes from coronavirus D-RNAs requires the genes to be under the control of a TAS for synthesis of an sg mRNA. The chIFN-γ sequence was placed under the control of the IBV Beaudette gene 5 TAS (39) by using PCR and a chIFN-γ mRNA-derived cDNA (19), generating a TAS-chIFN-γ cassette for IBV-controlled expression. The gene 5 TAS was originally chosen for expression of heterologous genes because it has the shortest sequence between the 3′ end of the TAS and the AUG of open reading frame (ORF) 5a and also because the Beaudette sg mRNA 5 is one of the most abundantly expressed sg mRNAs (39). The TAS-chIFN-γ cassette was initially inserted into pBluescript II SK(+) under the control of the T7 promoter. A protein with a size corresponding to that of chIFN-γ was produced in vitro by using the TNT T7 coupled wheat germ extract system (Promega; data not shown), indicating that a product of the expected size could be produced from the TAS-chIFN-γ cassette.
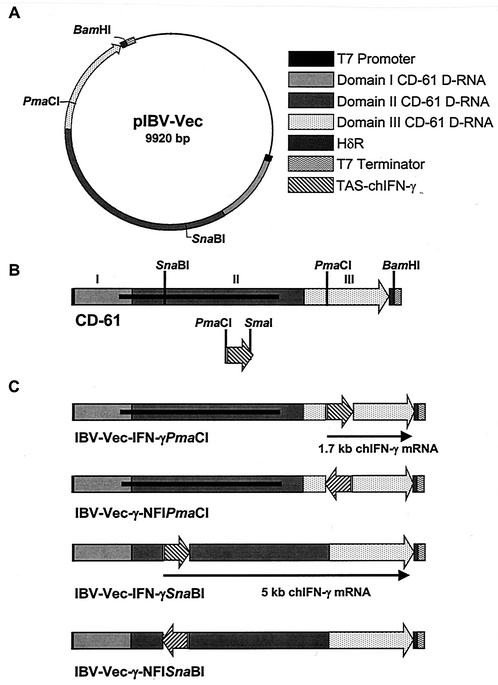
Two restriction endonuclease sites, PmaCI and SnaBI, within the IBV D-RNA CD-61 have been used for the expression of heterologous genes (13, 39). The PmaCI site is within domain III of CD-61 and not within the D-RNA-specific ORF (31), whereas the SnaBI site, within domain II of CD-61, interrupts the D-RNA-specific ORF (Fig. 1B). The TAS-chIFN-γ cassette excised from pBS-IFN-γ+ENDS was inserted into the PmaCI site or the SnaBI site of the CD-61 sequence in pIBV-Vec. Four plasmids, pIBV-Vec-IFN-γPmaCI and pIBV-Vec-IFN-γSnaBI, with the TAS-chIFN-γ gene cassette in the correct orientation, and pIBV-Vec-γ-NFIPmaCI and pIBV-Vec-γ-NFISnaBI, with the TAS-chIFN-γ gene cassette in the opposite (incorrect) orientation, were identified for generating D-RNAs (Fig. 1).
FIG. 1.
Schematic diagrams of the IBV D-RNAs containing the TAS-chIFN-γ sequence. (A) pIBV-Vec showing the PmaCI and SnaBI sites within CD-61 for insertion of heterologous genes. HδR, hepatitis delta virus antigenomic ribozyme. (B) IBV D-RNA CD-61, showing the positions of the restriction sites. The BamHI site was used for determining the orientation of inserts. The TAS-chIFN-γ gene cassette was used for insertion into CD-61. The 998-amino-acid CD-61-specific ORF (30, 31) is indicated as a thick black line. (C) The D-RNAs resulting from insertion of the TAS-chIFN-γ cassette into the two restriction endonuclease sites, in both orientations, within CD-61. The T7 promoter, HδR, and T7 termination sequences of pIBV-Vec (13) and the TAS-chIFN-γ sequences are as indicated in panel A. The chIFN-γ mRNAs of 1.7 and 5.0 kb derived from D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI, respectively, are indicated as black arrows.
In vitro rescue of chIFN-γ-containing D-RNAs.
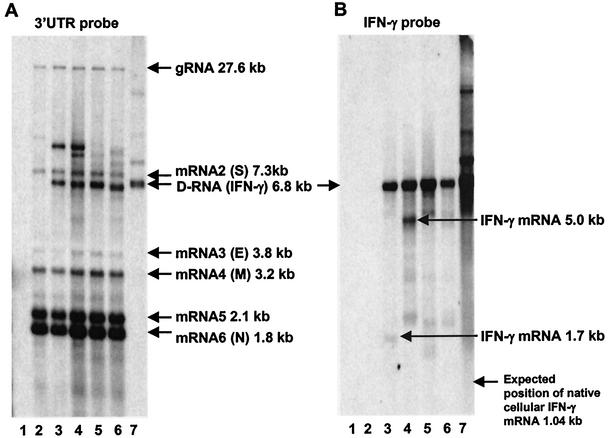
In vitro T7-derived transcripts of D-RNAs IBV-Vec-IFN-γPmaCI, IBV-Vec-IFN-γSnaBI, IBV-Vec-γ-NFIPmaCI, and IBV-Vec-γ-NFISnaBI were electroporated into IBV-infected CK cells, and progeny virus with D-RNAs was serially passaged on CK cells (P0 to P6). Northern blot analyses using the IBV 3′-UTR and chIFN-γ probes on RNA isolated from P3 cells identified an RNA species of 6.8 kb (Fig. 2). This RNA was not present in cells infected with IBV only and corresponded in size to the in vitro T7-derived D-RNA transcript. The chIFN-γ-specific probe did not hybridize to IBV gRNA or sg mRNAs, indicating that the 6.8-kb RNA represented D-RNAs containing the TAS-chIFN-γ sequence. As expected, the chIFN-γ probe detected D-RNAs IBV-Vec-γ-NFIPmaCI and IBV-Vec-γ-NFISnaBI, with the TAS-chIFN-γ sequence in the opposite orientation. The chIFN-γ probe also detected two other RNAs of 1.7 kb (Fig. 2B, lane 3) and 5 kb (Fig. 2B, lane 4) following the rescue of D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI, respectively, in addition to the 6.8-kb D-RNA. These RNAs corresponded to the expected sizes of chIFN-γ mRNAs expressed from the TAS-chIFN-γ sequence in the D-RNAs. The IBV-Vec-IFN-γPmaCI 1.7-kb chIFN-γ mRNA was routinely observed in smaller amounts than the IBV-Vec-IFN-γSnaBI 5-kb chIFN-γ mRNA. The IBV 3′-UTR probe detected RNAs larger than the 6.8-kb D-RNA and the 7.3-kb IBV sg mRNA 2 (Fig. 2A) which were not detected by the chIFN-γ probe (Fig. 2B). These RNAs were shown, by using Northern blot analysis and an IBV 5′-UTR probe (Fig. 3), to be new IBV-derived D-RNAs not containing the TAS-chIFN-γ sequence.
FIG. 2.
Northern blot analysis of IBV-specific RNAs following rescue of chIFN-γ-containing D-RNAs. Following electroporation of IBV-infected CK cells with in vitro T7-derived D-RNAs, progeny virus and D-RNAs were serially passaged (P0 to P6) on CK cells and total cytoplasmic RNA was extracted. RNA from P3 cells was electrophoresed in denaturing formaldehyde-agarose gels and subjected to Northern blotting, and IBV-derived RNAs were detected by using the 309-bp IBV 3′-UTR probe (A) and the 360-bp chIFN-γ probe (B). The P3-derived RNA samples analyzed were isolated from uninfected cells (lane 1); IBV-infected cells (lane 2); cells containing D-RNAs IBV-Vec-IFN-γPmaCI, IBV-Vec-IFN-γ-SnaBI, IBV-Vec-γ-NFIPmaCI, and IBV-Vec-γ-NFISnaBI (lanes 3 to 6, respectively); and in vitro T7-derived IBV-Vec-IFN-γPmaCI transcripts (lane 7). Arrows indicate IBV gRNA, sg mRNAs 2 to 6, TAS-chIFN-γ-containing D-RNAs, and D-RNA-derived chIFN-γ mRNAs. The RNAs detected between sg mRNAs 4 and 5 are observed routinely for all strains of IBV, as originally identified by Stern and Kennedy (38), and are of unknown origin. The RNAs migrating slower than IBV sg mRNA 2 were later shown to be new IBV-derived D-RNAs. The expected position of the full-length native cellular mRNA for chIFN-γ (1,040 nt) is indicated on panel B. S, spike glycoprotein; E, small membrane protein; M, integral membrane protein; N, nucleocapsid protein.
FIG. 3.
Northern blot analysis of IBV-specific RNAs following serial passage of D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI. RNAs isolated from P0 to P6 CK cells were analyzed for the presence of D-RNAs IBV-Vec-IFN-γPmaCI (A to C) and IBV-Vec-IFN-γSnaBI (D to F). IBV-derived RNAs were detected by using the 309-bp IBV 3′-UTR probe (A and D), the 209-bp IBV 5′-UTR probe (B and E), and the 360-bp chIFN-γ probe (C and F). The RNA samples analyzed were isolated from IBV-infected cells (lane 1), uninfected cells (lane 2), and P0 to P6 cells containing D-RNAs IBV-Vec-IFN-γPmaCI (A to C, lanes 3 to 9) and IBV-Vec-IFN-γSnaBI (D to F, lanes 3 to 9). (A to C) Lanes 10 correspond to in vitro T7-derived IBV-Vec-IFN-γPmaCI. Arrows indicate IBV gRNA, sg mRNAs, and TAS-chIFN-γ-containing D-RNAs. The RNAs migrating slower than IBV sg mRNA 2, detected in RNA samples isolated from P3 to P6 cells (A, B, D, and E, lanes 6), are new IBV-derived D-RNAs. Analysis of RNA following passage of IBV-Vec-IFN-γSnaBI identified the D-RNA-derived chIFN-γ mRNA (F, lanes 5 to 8). S, spike glycoprotein; E, small membrane protein; M, integral membrane protein; N, nucleocapsid protein.
To investigate the rescue profile of the D-RNAs, Northern blot analyses were carried out on RNA isolated from P0 to P6 CK cells containing D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI by using the IBV 5′-UTR, 3′-UTR, and chIFN-γ probes (Fig. 3). The 6.8-kb D-RNAs were detected by the two IBV probes in RNA isolated from P1 to P6 CK cells (Fig. 3A, B, D, and E), and analyses using the chIFN-γ probe confirmed that they contained the chIFN-γ sequence (Fig. 3C and F). Both D-RNAs, irrespective of whether the chIFN-γ sequence was inserted into the PmaCI or SnaBI site, were initially detected at P1 and increased in amount upon serial passage, with the largest amount in P4 CK cells, after which the amount of D-RNA decreased.
RNAs larger than the 6.8-kb chIFN-γ-containing D-RNA and the IBV sg mRNA 2 were detected from P3 to P6 by using both IBV probes. The use of the IBV 5′ probe indicated that the RNAs corresponded to new IBV-derived D-RNAs (Fig. 3A, B, D, and E). The new D-RNAs did not contain the chIFN-γ sequence (Fig. 3C and F), indicating that they were derived from IBV gRNA, and were observed in increasing amounts in RNA isolated from P3 to P6 cells, with concomitant gradual loss of the 6.8-kb chIFN-γ-containing D-RNAs (Fig. 3).
Analysis of P3-derived RNA following the rescue of D-RNA IBV-Vec-IFN-γSnaBI identified a 5-kb D-RNA-derived chIFN-γ mRNA. This mRNA was detected in RNA from P1 to P6 CK cells containing IBV-Vec-IFN-γSnaBI, with the amounts detected varying in accordance with the amounts of D-RNA present, the largest amount being detected at P4 (Fig. 3F). An RNA corresponding to the IBV-Vec-IFN-γPmaCI 1.7-kb chIFN-γ mRNA was not detected following serial passage of the D-RNA (Fig. 3C). However, from the amounts of the RNA detected, the most likely explanation for this result was that the amount of the 1.7-kb chIFN-γ mRNA was below the detection level of the analysis.
The Northern blot analyses showed that the two chIFN-γ-containing D-RNAs with the TAS-chIFN-γ insert in the correct orientation were rescued in IBV-infected CK cells upon serial passage. The analyses showed that D-RNA-derived chIFN-γ mRNAs were transcribed, irrespective of the position of the TAS-chIFN-γ insert, from the D-RNAs.
Analysis of chIFN-γ expression from D-RNAs.
The Northern blot analyses showed that D-RNAs containing the TAS-chIFN-γ sequences in the correct orientation were rescued upon serial passage, but the analyses could not show whether biologically active chIFN-γ was expressed. The biological assay for IFN-γ is based on the fact that macrophages stimulated with IFN-γ produce NO, along with other reactive nitrogen species, as one of several mechanisms to destroy intracellular pathogens. The chIFN-γ bioassay involves stimulation of HD11 cells, a chicken macrophage cell line (3), with chIFN-γ for the induction of NO, which accumulates as stable and quantifiable NO2− in the HD11 culture medium.
Cell medium from the P0 to P6 CK cells, previously shown to contain the D-RNAs containing chIFN-γ, was assayed for chIFN-γ activity. Medium from cells containing D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI induced significantly larger amounts of NO, following stimulation of HD11 cells, than any of the controls (Fig. 4A). In contrast, medium from cells containing D-RNAs IBV-Vec-γ-NFIPmaCI and IBV-Vec-γ-NFISnaBI did not induce larger amounts of NO than the controls. Controls involved medium from uninfected CK and IBV-infected CK cells. The amounts of NO induced by using medium from control samples and from cells containing D-RNAs with the TAS-chIFN-γ insert in the incorrect orientation were similar and likely to result from the presence of IFN-α or IFN-β. The IFN-γ bioassay also detects IFN-α and IFN-β, previously shown to be produced by IBV-infected cells (16, 28, 29).
FIG. 4.
Analysis of IFN activity in cell medium following in vitro passage of chIFN-γ-containing D-RNAs. (A) NO detected from HD11 cells, following stimulation with cell medium samples from uninfected cells (sample 1); IBV-infected cells (six passages) (sample 2); and P0 to P6 cells containing D-RNAs IBV-Vec-IFN-γPmaCI, IBV-Vec-IFN-γSnaBI, IBV-Vec-γ-NFIPmaCI, and IBV-Vec-γ-NFISnaBI (samples 3 to 6, respectively). (B) chIFN-γ-neutralizing bioassays using anti-chIFN-γ MAb 1E-12. Cell medium samples were preincubated for 2 h with medium alone (black bars), MAb 1E-12 (dark grey bars), or the isotype control Ab CC305 (light grey bars). Antibodies were diluted 1:1,000 for the assay. Ab-treated samples, analyzed for their ability to induce NO from HD11 cells, were from IBV-infected cells (sample 1), cells containing D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI (used at 1:8 dilutions) (samples 2 and 3, respectively), cells containing D-RNAs IBV-Vec-γ-NFIPmaCI and IBV-Vec-γ-NFISnaBI (samples 4 and 5, respectively), medium alone (sample 6), and recombinant chIFN-γ at 1:125 and 1:250 dilutions (samples 7 and 8, respectively). Nitrite resulting from the induced NO in the HD11 cell medium samples was quantified by using a variation of the Griess assay, measuring absorbance at 543 nm. The histograms represent the midpoints taken from the linear portions of titration curves calculated from the means of results for triplicates of each sample (A) and the means of results for triplicate samples (± standard errors) (B). OD543, optical density at 543 nm.
To confirm that the NO detected by the IFN-γ bioassay was due to the induction of HD11 cells by IFN-γ, and not by IFN-α and IFN-β, which also stimulate the production of NO, a neutralizing bioassay was carried out. Samples of cell medium were preincubated with the chIFN-γ-neutralizing MAb 1E-12 and analyzed with the IFN-γ bioassay. Samples were also preincubated with medium and Ab CC305, an isotype control Ab against bovine granulocyte-macrophage colony-stimulating factor. For comparative purposes, recombinant chIFN-γ was included as a positive control sample. NO induction, using medium from cells containing D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI, with the TAS-chIFN-γ sequence in the correct orientation, was reduced following preincubation of the samples with the neutralizing MAb 1E-12 (Fig. 4B). The amount of neutralizing MAb 1E-12 used was sufficient to neutralize all the chIFN-γ in the solution. To confirm this, the neutralizing activity of the MAb had been titrated before use in this experiment. Levels of remaining activity were assumed to be due to IFN-α and IFN-β. These levels were consistent with those seen in a previous study using this neutralizing MAb (19). A similar reduction in NO induction was observed when the recombinant chIFN-γ was preincubated with the MAb 1E-12. In contrast, the NO induced by using medium from cells containing D-RNAs IBV-Vec-γ-NFIPmaCI and IBV-Vec-γ-NFISnaBI, with the TAS-chIFN-γ sequence in the incorrect orientation, was not affected by the addition of the neutralizing MAb 1E-12. This indicated that the NO detected resulted from induction with chicken IFN-α or IFN-β rather than chIFN-γ in the cell medium (Fig. 4B). These results showed that the NO induction observed by using samples taken from cells containing D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI resulted from the presence of chIFN-γ in the cell medium and confirmed that the D-RNAs expressed biologically active chIFN-γ.
Induction of NO, using medium from cells containing D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI, was observed from P0 and increased to P4, followed by a decrease in NO at P5 to P6. This indicated that the amounts of chIFN-γ expressed increased upon serial passage of the D-RNAs, with a peak of activity in P4 CK cells followed by a decrease in activity (Fig. 4A). This result correlated with the Northern blot analyses in which the amounts of the D-RNAs and one of the D-RNA-derived chIFN-γ mRNAs followed the same pattern over serial passage. Results of the bioassay also showed that there were no significant differences in the amounts of NO induced whether the samples came from cells containing TAS-chIFN-γ inserted into the PmaCI or from cells containing TAS-chIFN-γ inserted into the SnaBI site of CD-61 (Fig. 4A), indicating that both D-RNAs expressed similar amounts of chIFN-γ.
In ovo rescue of chIFN-γ-containing D-RNAs.
Previous work to determine whether IBV D-RNAs could be passaged in vivo by using embryonated eggs (in ovo) had required subsequent passage of a D-RNA expressing chloramphenicol acetyltransferase (CAT) on indicator CK cells. Subsequent analysis of cell lysates demonstrated the presence of CAT (5, 11). A limitation of these experiments was that replication may not have occurred in ovo and that rescue on the CK cells could have resulted from virus and D-RNA present in the allantoic fluid from the initial inoculum, although rescue following dilution in the allantoic fluid would be indicative of replication in ovo. The secretion of chIFN-γ into cell medium following expression from the D-RNAs described in this work provided a means of determining whether IBV D-RNAs can be replicated in ovo.
Following serial passage of D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI, the largest amounts of D-RNA detected with concomitant expression of chIFN-γ occurred in P4 cells (Fig. 3 and 4). Consequently, we decided to use progeny virus (V4) containing the D-RNAs from P3 cells to determine whether the D-RNAs could be rescued in ovo. Virus (V4) from P3 cells, representing virus that was one passage away from peak chIFN-γ activity, was used to infect 10-day-old specific-pathogen-free embryonated eggs. At 16 h postinoculation, allantoic fluid was clarified and used to infect CK cells to confirm that the D-RNAs could be passaged from embryonated eggs. The fluid was also used for chIFN-γ bioassays to analyze for the presence of any chIFN-γ resulting from the replication of the D-RNAs in ovo.
IFN-γ bioassays showed that allantoic fluid and CK cell medium samples, following passage of D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI, contained amounts of chIFN-γ that were up to 64-fold and 10-fold larger, respectively, as measured by NO levels, than those of the controls (Fig. 5A), indicating that the D-RNAs had replicated in ovo. The amounts of NO induced following infection with IBV only and passage of D-RNAs containing the TAS-chIFN-γ gene construct in the antisense orientation were all similar, with NO induction presumably due to the presence of IFN-α or IFN-β. The amounts of chIFN-γ, as measured by NO levels, in allantoic fluid following in ovo passage of D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI were larger than those found in CK cell medium following subsequent in vitro passage of the D-RNAs. This result was anticipated, as the in ovo passage was equivalent to P4 and CK passage was equivalent to P5 when equated with in vitro passage of the D-RNAs.
FIG. 5.
Analysis of IFN activity in allantoic fluid following in ovo passage of chIFN-γ-containing D-RNAs. The chIFN-γ-containing D-RNAs were initially passaged in embryonated eggs, and progeny virus containing the D-RNAs in the allantoic fluid was passaged on indicator CK cells. (A) NO detected from HD11 cells following stimulation with allantoic fluid (dark grey bars) or CK cell medium (light grey bars) from passage of D-RNAs IBV-Vec-IFN-γPmaCI, IBV-Vec-IFN-γSnaBI, IBV-Vec-γ-NFIPmaCI, and IBV-Vec-γ-NFISnaBI (samples 1 to 4, respectively); IBV-infected cells (sample 5); and uninfected cells (sample 6). The histogram represents the midpoints taken from the linear portions of titration curves calculated from the means (± standard errors) of results for triplicates of each sample. (B) chIFN-γ-neutralizing bioassays in which samples, preincubated for 2 h with medium alone (black bars), MAb 1E-12 (dark grey bars), or CC305 (light grey bars), were analyzed for their ability to induce NO from HD11 cells consisted of allantoic fluid (samples 1 to 6); CK cell media (samples 9 to 14); recombinant chIFN-γ diluted 1:2,000 (sample 7); and media alone (sample 8). Samples 1 to 4 and 9 to 12 were from allantoic fluid and CK cell medium, respectively, following passage of D-RNAs IBV-Vec-IFN-γPmaCI (diluted 1:8), IBV-Vec-IFN-γSnaBI (diluted 1:8), IBV-Vec-γ-NFIPmaCI, and IBV-Vec-γ-NFISnaBI, respectively. Samples 5 to 6 and 13 to 14 were from allantoic fluid and CK cell medium, respectively, from cells infected with IBV (5 and 13) or uninfected cells (6 and 14). Antibodies were diluted 1:1,000 for the assay. The histogram represents the means (± standard errors) of results for triplicate samples. OD543, optical density at 543 nm.
A neutralizing IFN-γ bioassay was again performed to confirm that the NO induced resulted from IFN-γ and not from IFN-α and IFN-β. Allantoic fluid and CK cell medium samples were preincubated with MAb 1E-12, medium, and the isotype control Ab CC305 for 2 h prior to the IFN-γ bioassay (Fig. 5B). The amounts of NO induced by using samples from embryonated eggs (Fig. 5B, samples 1 and 2) and CK cells (Fig. 5B, samples 9 and 10) infected with IBV containing D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI were similar for both D-RNAs both in ovo and in vitro and were similarly reduced following preincubation with MAb 1E-12. A similar reduction in NO induction was observed following preincubation of recombinant chIFN-γ with MAb 1E-12 (Fig. 5B, sample 7), indicating that the IFN activities detected in the allantoic fluid and CK cell medium samples resulted from IFN-γ. In contrast, the amounts of NO induced by using samples from embryonated eggs and CK cells infected with IBV containing D-RNAs IBV-Vec-γ-NFIPmaCI and IBV-Vec-γ-NFISnaBI and from IBV infection alone were not reduced following preincubation with MAb 1E-12 (Fig. 5B). This indicated that the IFN activities observed following in ovo passage of these D-RNAs and IBV alone resulted from the induction of IFN-α and IFN-β and not from IFN-γ.
These results confirmed that D-RNAs IBV-Vec-IFN-γPmaCI and IBV-Vec-IFN-γSnaBI expressed biologically active chIFN-γ following helper virus-dependent replication in ovo.
DISCUSSION
The construction of a series of IBV-based D-RNAs containing the chIFN-γ sequence under the control of an IBV TAS has been described. We have shown that IBV D-RNAs containing the chIFN-γ gene were incorporated into virus particles and expressed biologically active chIFN-γ in vitro and in vivo.
Serial passage of the chIFN-γ-containing D-RNAs followed the same pattern of both D-RNA replication and heterologous protein expression that we have previously observed for the expression of the heterologous reporter genes encoding CAT (39) and luciferase (13) from IBV-based D-RNAs. Both detection of the D-RNA and expression of chIFN-γ increased from P0 to P4 and decreased thereafter. No differences in the amounts of D-RNA detected or in chIFN-γ activities were observed whether the TAS-chIFN-γ cassette was inserted in domain I (IBV-Vec-IFN-γSnaBI), interrupting the D-RNA-specific ORF, or domain III (IBV-Vec-IFN-γPmaCI), not interrupting the D-RNA-specific ORF, of D-RNA CD-61.
A D-RNA-derived chIFN-γ mRNA synthesized from IBV-Vec-IFN-γSnaBI was detectable upon serial passage of the D-RNA (Fig. 2B and 3F). Although a D-RNA-derived chIFN-γ mRNA synthesized from IBV-Vec-IFN-γPmaCI was initially detected (Fig. 2B), it was synthesized in smaller amounts, in relation to the D-RNA, than the mRNA from IBV-Vec-IFN-γSnaBI. The observed levels of subgenomic RNAs (sgRNAs) transcribed, under the control of the gene 5 TAS, from CD-61-derived D-RNAs were far less than the amounts observed for the transcription of sg mRNA 5 from genomic RNA. Similar observations have been made following transcription of sgRNAs from other coronavirus-derived D-RNAs. The altered transcription levels may be connected to the observation that D-RNAs may replicate in a manner analogous to the transcription of sg mRNAs from genomic RNA (40), thereby affecting the levels of transcription of D-RNA-derived sgRNAs, or the levels may result from the altered context of the TAS within the D-RNA. The sequences flanking the TASs of heterologous genes in transmissible gastroenteritis virus-derived D-RNAs have been shown to alter the transcription of D-RNA-derived sgRNAs (1). The amounts of chIFN-γ secreted from cells containing the D-RNAs were similar, as observed from the induction of NO from HD11 cells, indicating that the D-RNAs expressed similar amounts of active chIFN-γ. Similar results were observed for the expression of β-glucuronidase (GUS) from a transmissible gastroenteritis virus-based D-RNA, in which the D-RNA-derived GUS mRNA was transcribed in smaller amounts in P2 cells than in P5 cells, although the amounts of GUS protein expressed were the same (2).
Although its main role is in driving a Th1 cell-mediated response against intracellular pathogens, including viruses, IFN-γ can have a direct antiviral effect. Like IFN-α and IFN-β, but to a much lesser extent, it can induce oligoadenylate synthetase and RNA-dependent protein kinase PKR, both important components of IFN-induced antiviral responses (8, 14, 34). It was therefore possible that expression of chIFN-γ from the D-RNAs might interfere with replication of the helper virus. A murine IFN-γ-containing D-RNA of the murine coronavirus mouse hepatitis virus (MHV) expressed murine IFN-γ that resulted in a slight reduction in virus titer (0.5 log10) at low multiplicities of infection, suggesting that the murine IFN-γ had a weak antiviral activity (42). In contrast, we observed no significant differences in virus titers resulting from expression of the chIFN-γ from any of the IBV D-RNAs (data not shown), suggesting that the chIFN-γ did not have an appreciable antiviral effect on the helper IBV.
The murine IFN-γ-containing MHV D-RNA was not detected beyond P4 (42), whereas the IBV-based chIFN-γ-containing D-RNAs were rescued for at least six passages. This observation reflects our previous studies on the expression of heterologous genes from IBV-CD-61-based D-RNAs. Expression of heterologous genes or coronavirus-derived genes from other coronavirus D-RNAs also resulted in loss of the D-RNAs. For example, expression of CAT and hemagglutinin esterase from MHV D-RNAs was not detected beyond P2 (20) and P3 (43), respectively. The most likely explanation for the instability of coronavirus D-RNAs expressing heterologous genes is intolerance of the heterologous sequence within the D-RNA. D-RNAs evolve by the removal of unnecessary sequences. Therefore, the presence of a heterologous sequence may impart some innate instability into the RNA or affect replication and/or packaging of the D-RNA; i.e., D-RNAs containing heterologous or nonrequired sequences are less fitted for replication than D-RNAs with minimal required sequences. We detected additional RNA species, which were larger than the chIFN-γ-containing D-RNAs and IBV sg mRNA 2, from P3 which increased in amount upon serial passage. The new RNAs were not detected by the chIFN-γ-specific probe but by the IBV 5′ probe, showing that they contained sequences derived from the 5′ end of the IBV genome and were therefore new D-RNAs. As determined from their sizes, the new D-RNAs were not from the loss of the chIFN-γ sequence. An explanation for the loss of the chIFN-γ-containing D-RNAs beyond P4 was the generation of more stable D-RNAs from P3 that eventually resulted, due to competition, in the loss of the chIFN-γ-containing D-RNAs.
Overall, we have demonstrated that in vitro passage of the chIFN-γ-containing D-RNAs resulted in expression of biologically active chIFN-γ secreted into cell culture medium. This observation allowed us to demonstrate that IBV D-RNAs can be replicated in ovo from the presence of biologically active chIFN-γ secreted into the allantoic fluid and subsequent passage on indicator CK cells.
Acknowledgments
Karen Hackney held a Research Studentship from the British Egg Marketing Board Research and Education Trust. This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) and the Department of Environment, Food and Rural Affairs (DEFRA) project code OD0712.
REFERENCES
- 1.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, S., I. Sola, J. P. Teifke, I. Reimann, A. Izeta, M. Balasch, J. Plana-Duran, R. J. Moormann, and L. Enjuanes. 2002. In vitro and in vivo expression of foreign genes by transmissible gastroenteritis coronavirus-derived minigenomes. J. Gen. Virol. 83:567-579. [DOI] [PubMed] [Google Scholar]
- 3.Beug, H., A. von Kirchbach, G. Doderlein, J. F. Conscience, and T. Graf. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375-390. [DOI] [PubMed] [Google Scholar]
- 4.Boursnell, M. E., T. D. Brown, I. J. Foulds, P. F. Green, F. M. Tomley, and M. M. Binns. 1987. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 68:57-77. [DOI] [PubMed] [Google Scholar]
- 5.Britton, P., K. Stirrups, K. Dalton, K. Shaw, S. Evans, B. Neuman, B. Dove, R. Casais, and D. Cavanagh. 2001. Use of an infectious bronchitis virus D-RNA as an RNA vector. Adv. Exp. Med. Biol. 494:507-512. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh, D., K. Mawditt, M. Sharma, S. E. Drury, H. L. Ainsworth, P. Britton, and R. E. Gough. 2001. Detection of a coronavirus from turkey poults in Europe genetically related to infectious bronchitis virus of chickens. Avian Pathol. 30:365-378. [DOI] [PubMed] [Google Scholar]
- 7.Cavanagh, D., K. Mawditt, D. D. B. Welchman, P. Britton, and R. E. Gough. 2002. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 31:81-93. [DOI] [PubMed] [Google Scholar]
- 8.De Maeyer, E., and J. De Maeyer-Guinard. 1998. Interferons, p. 491-516. In A. Thomson (ed.), The cytokine handbook, 3rd ed. Academic Press, San Diego, Calif.
- 9.Dhinakar-Raj, G., and R. C. Jones. 1997. Infectious bronchitis virus: immunopathogenesis of infection in the chicken. Avian Pathol. 26:677-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Digby, M. R., and J. W. Lowenthal. 1995. Cloning and expression of the chicken interferon-γ gene. J. Interferon Cytokine Res. 15:939-945. [DOI] [PubMed] [Google Scholar]
- 11.Dove, B. 2002. Ph.D. thesis. University of Reading, Reading, United Kingdom.
- 12.Enjuanes, L., W. J. Spaan, E. J. Snijder, and D. Cavanagh. 2000. Nidovirales, p. 827-834. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, New York, N.Y.
- 13.Evans, S., D. Cavanagh, and P. Britton. 2000. Utilizing fowlpox virus recombinants to generate defective RNAs of the coronavirus infectious bronchitis virus. J. Gen. Virol. 81:2855-2865. [DOI] [PubMed] [Google Scholar]
- 14.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 15.Hiscox, J. A., K. L. Mawditt, D. Cavanagh, and P. Britton. 1995. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J. Virol. 69:6219-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes, H. C., and J. H. Darbyshire. 1978. Induction of chicken interferon by avian infectious bronchitis virus. Res. Vet. Sci. 25:178-181. [PubMed] [Google Scholar]
- 17.Izeta, A., C. Smerdou, S. Alonso, Z. Penzes, A. Mendez, J. Plana-Duran, and L. Enjuanes. 1999. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 73:1535-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambrecht, B., M. Gonze, G. Meulemans, and T. P. van den Berg. 2000. Production of antibodies against chicken interferon-γ: demonstration of neutralizing activity and development of a quantitative ELISA. Vet. Immunol. Immunopathol. 74:137-144. [DOI] [PubMed] [Google Scholar]
- 19.Lawson, S., L. Rothwell, B. Lambrecht, K. Howes, K. Venugopal, and P. Kaiser. 2001. Turkey and chicken interferon-γ, which share high sequence identity, are biologically cross-reactive. Dev. Comp. Immunol. 25:69-82. [DOI] [PubMed] [Google Scholar]
- 20.Liao, C. L., X. Zhang, and M. M. Lai. 1995. Coronavirus defective-interfering RNA as an expression vector: the generation of a pseudorecombinant mouse hepatitis virus expressing hemagglutinin-esterase. Virology 208:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillehoj, H. S., and K. D. Choi. 1998. Recombinant chicken interferon-γ mediated inhibition of Eimeria tenella development in vitro and reduction of oocyst production and body weight loss following Eimeria acervulina challenge infection. Avian Dis. 42:307-314. [PubMed] [Google Scholar]
- 22.Lin, Y. J., and M. M. Lai. 1993. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J. Virol. 67:6110-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowenthal, J. W., T. E. O'Neil, M. Broadway, A. D. Strom, M. R. Digby, M. Andrew, and J. J. York. 1998. Coadministration of IFN-γ enhances antibody responses in chickens. J. Interferon Cytokine Res. 18:617-622. [DOI] [PubMed] [Google Scholar]
- 24.Lowenthal, J. W., J. J. York, T. E. O'Neil, R. A. Steven, D. G. Strom, and M. R. Digby. 1998. Potential use of cytokine therapy in poultry. Vet. Immunol. Immunopathol. 63:191-198. [DOI] [PubMed] [Google Scholar]
- 25.Masters, P. S. 1999. Reverse genetics of the largest RNA viruses. Adv. Virus Res. 53:245-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick, A. L., M. S. Thomas, and A. W. Heath. 2001. Immunization with an interferon-γ-gp120 fusion protein induces enhanced immune responses to human immunodeficiency virus gp120. J. Infect. Dis. 184:1423-1430. [DOI] [PubMed] [Google Scholar]
- 27.Min, W., H. S. Lillehoj, J. Burnside, K. C. Weining, P. Staeheli, and J. J. Zhu. 2001. Adjuvant effects of IL-1β, IL-2, IL-8, IL-15, IFN-α, IFN-γ, TGF-β4 and lymphotactin on DNA vaccination against Eimeria acervulina. Vaccine 20:267-274. [DOI] [PubMed] [Google Scholar]
- 28.Otsuki, K., J. Maeda, H. Yamamoto, and M. Tsubokura. 1979. Studies on avian infectious bronchitis virus (IBV). III. Interferon induction by and sensitivity to interferon of IBV. Arch. Virol. 60:249-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otsuki, K., T. Nakamura, Y. Kawaoka, and M. Tsubokura. 1988. Interferon induction by several strains of avian infectious bronchitis virus, a coronavirus, in chickens. Acta Virol. 32:55-59. [PubMed] [Google Scholar]
- 30.Penzes, Z., K. Tibbles, K. Shaw, P. Britton, T. D. Brown, and D. Cavanagh. 1994. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology 203:286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penzes, Z., C. Wroe, T. D. Brown, P. Britton, and D. Cavanagh. 1996. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J. Virol. 70:8660-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rautenschlein, S., J. M. Sharma, B. J. Winslow, J. McMillen, D. Junker, and M. Cochran. 1999. Embryo vaccination of turkeys against Newcastle disease infection with recombinant fowlpox virus constructs containing interferons as adjuvants. Vaccine 18:426-433. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 37.Schijns, V. E., N. C. Scholtes, H. I. Zuilekom, L. E. Sanders, L. Nicolson, and D. J. Argyle. 2002. Facilitation of antibody forming responses to viral vaccine antigens in young cats by recombinant baculovirus-expressed feline IFN-γ. Vaccine 20:1718-1724. [DOI] [PubMed] [Google Scholar]
- 38.Stern, D. F., and S. I. T. Kennedy. 1980. Coronavirus multiplication strategy. I. Identification and characterisation of virus-specific RNA. J. Virol. 34:665-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stirrups, K., K. Shaw, S. Evans, K. Dalton, R. Casais, D. Cavanagh, and P. Britton. 2000. Expression of reporter genes from the defective RNA CD-61 of the coronavirus infectious bronchitis virus. J. Gen. Virol. 81:1687-1698. [DOI] [PubMed] [Google Scholar]
- 40.Stirrups, K., K. Shaw, S. Evans, K. Dalton, D. Cavanagh, and P. Britton. 2000. Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus. J. Gen. Virol. 81:791-801. [DOI] [PubMed] [Google Scholar]
- 41.Yilma, T., S. Owens, E. H. Fennie, and K. P. Anderson. 1989. Enhancement of primary and secondary immune responses by interferon-γ. Adv. Exp. Med. Biol. 251:145-152. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, X., D. R. Hinton, D. J. Cua, S. A. Stohlman, and M. M. Lai. 1997. Expression of interferon-γ by a coronavirus defective-interfering RNA vector and its effect on viral replication, spread, and pathogenicity. Virology 233:327-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, X., D. R. Hinton, S. Park, B. Parra, C. L. Liao, M. M. Lai, and S. A. Stohlman. 1998. Expression of hemagglutinin/esterase by a mouse hepatitis virus coronavirus defective-interfering RNA alters viral pathogenesis. Virology 242:170-183. [DOI] [PMC free article] [PubMed] [Google Scholar]