Abstract
Herpesviruses utilize different origins of replication during lytic versus latent infection. Latent DNA replication depends on host cellular DNA replication machinery, whereas lytic cycle DNA replication requires virally encoded replication proteins. In lytic DNA replication, the lytic origin (ori-Lyt) is bound by a virus-specified origin binding protein (OBP) that recruits the core replication machinery. In this report, we demonstrated that DNA sequences in two noncoding regions of the Kaposi's sarcoma-associated herpesvirus (KSHV) genome, between open reading frames (ORFs) K4.2 and K5 and between K12 and ORF71, are able to serve as origins for lytic cycle-specific DNA replication. The two ori-Lyt domains share an almost identical 1,153-bp sequence and a 600-bp downstream GC-rich repeat sequence, and the 1.7-kb DNA sequences are sufficient to act as a cis signal for replication. We also showed that an AT-palindromic sequence in the ori-Lyt domain is essential for the DNA replication. In addition, a virally encoded bZip protein, namely K8, was found to bind to a DNA sequence within the ori-Lyt by using a DNA binding site selection assay. The binding of K8 to this region was confirmed in cells by using a chromatin immunoprecipitation method. Further analysis revealed that K8 binds to an extended region, and the entire region is 100% conserved between two KSHV ori-Lyt's. K8 protein displays significant similarity to the Zta protein of Epstein-Barr virus (EBV), which is a known OBP of EBV. This notion, together with the ability of K8 to bind to the KSHV ori-Lyt, suggests that K8 may function as an OBP in KSHV.
Kaposi's sarcoma-associated herpesvirus (KSHV), also referred to as human herpesvirus-8 (HHV-8), is a newly identified human herpesvirus which is evolutionarily related to herpesvirus saimiri and Epstein-Barr virus (EBV) (7). KSHV is associated with Kaposi's sarcoma (KS) (1), primary effusion lymphoma (5), and multicentric Castleman's diseases (24). As a gamma-herpesvirus, KSHV has both latent and lytic replication cycles (17, 21). During latency, only a limited number of viral genes is expressed, and no infectious virus is produced. In latently infected cells, multiple copies of the viral genome are maintained as extrachromosomal episomes (plasmids) and are replicated in synchrony with cell division (4). The terminal repeat (TR) sequence in the KSHV genome is necessary and sufficient for episome persistence and likely serves as the origin of latent plasmid replication (ori-P) (3). Virally encoded latency-associated nuclear antigen specifically binds to cis-acting KSHV TR DNA and acts in trans on the ori-P to mediate episome persistence (3). When latency is disrupted, KSHV switches to a lytic life cycle (17, 21). In the lytic phase, the virus expresses most or all of its genes, and viral DNA is amplified by a replication mechanism distinct from that for latent viral DNA replication (12, 26). However, cis-acting and trans-acting elements required for KSHV lytic DNA replication have not been identified.
The lytic DNA replication of EBV has been extensively studied. In the lytic cycle, multiple rounds of DNA replication are initiated from an origin, ori-Lyt (origin of lytic DNA replication) which is distinct from that used for latent plasmid DNA replication (ori-P). Two functional copies of ori-Lyt are present in the EBV genome in most strains (13). Unlike EBV latent DNA replication, which depends on EBV nuclear antigen-1 binding to ori-P, the lytic DNA replication of EBV requires many viral proteins, including POL (polymerase), PPF (polymerase processivity factor), SSB (single-stranded DNA binding protein), PRI (primase), HEL (helicase), PAF (primase-associated factor), and Zta (origin binding protein) (9, 10). The genes for homologs to six EBV core lytic replication proteins were found in the KSHV genome (22, 26). They include ORF9 (POL), ORF59 (PPF), ORF6 (SSB), ORF56 (PRI), ORF44 (HEL), and ORF40/41 (PAF). It was demonstrated that these six KSHV-encoded proteins can substitute for their EBV counterparts and replicate a plasmid containing EBV ori-Lyt in the presence of the EBV origin binding protein (OBP), Zta (26). However, no KSHV OBP has been identified.
KSHV-encoded K8 protein shares similarities with the EBV OBP (Zta), including limited sequence homology, analogous genome location, and similar splicing pattern. The open reading frame (ORF) K8 was first identified on a 3.6-kb bicistronic immediate-early transcript, namely ORF50/K8 mRNA (27). It is also encoded in a delayed-early transcript of 1.3 kb (15). Both the 3.6-kb and 1.3-kb mRNAs are highly spliced, which generates three splice variants of K8 as a result of alternative splicing. K8α is the major form and codes for a 237-amino-acid protein with a basic leucine zipper domain near its C terminus and an acidic domain near its N terminus (27). Such a structure was found in many transcriptional regulators that compose a large bZip family. Thus, K8α is a protein of the bZip family. The two other variants, K8β and K8γ, encode proteins sharing the N-terminal portion with K8α but lacking the C-terminal Zip domain.
The goal of this study was to decipher the function of the K8 bZip protein and its role in KSHV lytic cycle replication. The EBV counterpart of K8, the Zta protein, has at least two functions. One is transcriptional activation, in which Zta activates the viral lytic gene expression cascade and initiates the switch of EBV from latency to lytic life cycle (16). Despite extensive studies in several laboratories, no transcriptional activity has been found with K8. The other function of Zta involves lytic viral DNA replication. Zta binds to EBV ori-Lyt, and the binding is essential for viral lytic DNA replication (10). To explore the function of K8, we first asked whether K8 protein exerts its function through its DNA binding activity, like Zta. With the aid of an in vitro DNA binding site selection procedure, we identified a K8 binding hot spot in the KSHV genome. Interestingly, this K8 binding region is located within the domain which has been proposed as a putative ori-Lyt of KSHV (19). We showed that this putative ori-Lyt is indeed able to support lytic cycle-specific DNA replication. Overall, this study led to identification of KSHV ori-Lyt and a potential ori-Lyt binding protein.
MATERIALS AND METHODS
Cell culture.
The BCBL-1 cell line, which carries latently infected KSHV, was established by Ganem and his colleagues (21) and was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. The cells were grown in RPMI 1640 medium (Gibco-BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (Gibco-BRL). 293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. All cultures contained penicillin-streptomycin (50 U/ml) and Fungizone (1.25 μg of amphotericin B/ml and 1.25 μg of sodium desoxycholate/ml).
DNA binding site selection.
Six overlapping KSHV cosmid DNAs (namely, GB11, GA29, GB22, GA1, GA2, and GB1; kindly provided by Ren Sun and George Miller, Yale University) were used to generate a pool of KSHV DNA fragments. These cosmid DNAs were pooled, and two aliquots of the DNA pool were digested completely with AluI and RsaI restriction enzymes. The DNA fragment pool was ligated with double-stranded linker which was prepared by annealing oligodeoxyribonucleotide Ad-1A (5′ GATCCCAGTCACGACGAATTCC 3′) and phosphorylated Ad-1B (5′ pGGAATTCGTCGTGACTGG 3′). The linker-ligated DNA fragments were amplified by PCR using oligonucleotide Ad-1A as a primer.
The KSHV DNA fragment pool was incubated with a whole-cell extract prepared from phorbol-12-tetradecanoate-13-acetate (TPA)-induced BCBL-1 cells in a binding buffer (50 μl) containing 20 mM HEPES (pH 7.9), 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.1% NP-40, 10 μg of bovine serum albumin/ml, 62.5 μg of poly(dI-dC)/ml, and proteinase inhibitors. The binding reaction was carried out on ice for 30 min. Then, 1 μl of anti-K8 antibody (a gift from Jae Jung at the New England Regional Primate Research Center) was added to the reaction mixture, followed by incubation for an additional 30 min. The mixture was transferred to a new tube containing protein A-Sepharose (Sigma) and incubated for 1 h. The immunoprecipitated complexes were washed at least four times with 500 μl of cold binding buffer without BSA and poly(dI-dC). K8-bound DNA was eluted at 50°C in elution buffer (50 mM Tris-HCl [pH 7.6], 100 mM NaOAc, 5 mM EDTA, 0.5% sodium dodecyl sulfate [SDS]) for 1 h. Recovered DNA was amplified by PCR with the oligonucleotide Ad-1A as a primer and used for the next cycle of selection.
After each round of selection, the PCR-amplified DNA pool was analyzed by electrophoresis on a 1.5% agarose gel. After seven cycles of selection, the DNA was digested with EcoRI and cloned into pBluescript at the EcoRI site. Twenty clones were randomly picked up for sequencing analysis.
Chromatin immunoprecipitation (ChIP) assay.
BCBL-1 cells were treated with 20 ng of TPA/ml for 48 h. The cells were fixed by addition of 1% formaldehyde to the medium for 10 min and collected by centrifugation. After washing with cold phosphate-buffered saline, the cells (107) were suspended in 1 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0]) supplemented with 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg of pepstatin A/ml, and 1 μg of leupeptin/ml and let sit on ice for 10 min. Then, the cells were sonicated eight times for 10 s each, and the lysates were cleared by centrifugation for 10 min. The cell lysates were diluted 10-fold with dilution buffer (0.01% SDS, 1.1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 167 mM NaCl). Chromatin solution (1 ml) was incubated with 2 μl of anti-K8 antibody or prebleed sera overnight at 4°C. Immune complexes were collected on protein A beads preadsorbed with sonicated single-stranded DNA. Beads were washed sequentially twice each in low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl), high-salt wash buffer (500 mM NaCl), LiCl wash buffer (0.25 mM LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0]), and Tris-EDTA (TE) buffer (pH 8.0). Immune complexes were eluted by incubation in 150 μl of TE buffer-1% SDS. Cross-links were reversed by heating at 65°C for 7 h up to overnight, followed by digestion with proteinase K (25 μg/ml) for 2 h at 50°C. DNA was extracted with phenol-chloroform-isoamyl alcohol and ethanol precipitated. PCR analysis of immunoprecipitated DNA was carried out using oligonucleotides K4.2P1 (5′ TGCGCTAATCCTTTTATGTGCA 3′; nucleotides 23147 to 23168) and K4.2P5 (5′ GATGGGCCAATGGCGGCTCG 3′; nucleotides 23405 to 23386). A PCR with oligonucleotides ORF45-RACE1 (5′ GGCGTCCATGGGATGGGTTAGTCAGGAT 3′; nucleotides 68097 to 68070) and ORF45-RACE2 (5′ ACGTCCGGAGAGTTGGAACTGTCATCGC 3′; nucleotides 67813 to 67840) was performed as a control.
In vitro DNA binding assay.
Various DNA fragments were synthesized using PCR with pOri-A DNA as a template and two oligonucleotides as primers. One of the oligonucleotides was biotinylated at its 5′ end. The resultant biotinylated PCR fragments were coupled to steptavidin-conjugated magnetic beads (Dynal, Oslo, Norway) and then incubated with nuclear extracts prepared from TPA-induced (and uninduced) BCBL-1 cells for 45 min at 25°C. The bound material was washed four times in D150 buffer (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 150 mM KCl, 1 mM dithiothreitol, 1 mM PMSF, 0.05% NP-40) and then progressively eluted with D300 (same as above, except 300 mM KCl), D500 (500 mM KCl), and D1000 (1 M KCl). The affinity-purified materials were assayed by Western blotting analysis with anti-K8 antibody.
Plasmids and DNA transfection.
Plasmid pOri-A was constructed by cloning an EcoRI-PstI fragment (nucleotides 22409 to 26491) of KSHV DNA in pBluescript at the EcoRI/PstI site. pOri-B was constructed by cloning a KpnI fragment (nucleotides 117594 to 120163) in pBluescript at the KpnI site. A series of deletion mutants of pOri-A were generated using appropriate restriction endonucleases and standard cloning methods.
The internal deletion mutants of pOri-A (pOri-ID2, pOri-ID4, and pOri-ID9) were generated using a PCR-based mutagenesis system, namely ExSite (Stratagene). In brief, a pair of phosphorylated oligonucleotides toward opposite directions was used in a high-fidelity PCR with pOri-A plasmid as a template. After PCR, the template DNA (wild-type pOri-A) was removed by a complete digestion with DpnI, which does not degrade PCR-synthesized DNA. The PCR products were self-ligated and used to transform Escherichia coli competent cells.
pCR3.1-ORF50 was constructed by cloning the cDNA sequence of the ORF50 coding region into pCR3.1 vector (Invitrogen). The cDNA was generated by reverse transcription-coupled PCR using mRNA from butyrate-induced BC-1 cells and two specific primers, ORF50N (5′ ATGGCGCAAGATGACAAGGGTAAGA 3′) and ORF50C (5′ TCAGTCACGGAAGTAATTACGCCAT 3′).
To transfect cells, 5 μg of ori-Lyt-carrying plasmid and 5 μg of pcR3.1-ORF50 (or pCR3.1 vector) were mixed with 107 BCBL-1 cells in OPTI-MEM medium (Gibco-BRL) and electroporated (200 V, 960 μF) with a Genepulser II (Bio-Rad, Hercules, Calif.). Electroporated cells were then transferred to RPMI 1640 medium supplemented with 10% serum and grown for 72 h.
DNA replication assay.
Total DNAs were isolated from transfected cells using the DNeasy DNA isolation kit (Qiagen). In some experiments, extrachromosomal DNAs were prepared from cells using the Hirt DNA extraction method as follows. Cells were lysed in 700 μl of lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, and 0.6% SDS). Chromosomal DNA was precipitated at 4°C overnight by adding 5 M NaCl to a final concentration of 0.85 M. Cell lysates were centrifuged at 4°C at 14,000 rpm in an Eppendorf microcentrifuge for 30 min. The supernatant containing extrachromosomal DNA was subjected to phenol-chloroform extraction followed by ethanol precipitation. The DNA was treated with RNase A at 25°C for 30 min and then with proteinase K at 50°C for 30 min. Five micrograms of DNA was digested with KpnI/SacI or KpnI/SacI/DpnI (New England Biolabs). The DNAs were separated by electrophoresis on 1% agarose gels and transferred onto Nytran membranes (Schleicher & Schuell, Keene, N.H.). The Southern blots were hybridized with 32P-labeled pBluescript plasmid in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2× Denhardt's solution, 1% SDS, and 50 μg of denatured salmon sperm DNA/ml at 68°C.
RESULTS
The K8 bZip protein binds to a region in the KSHV genome.
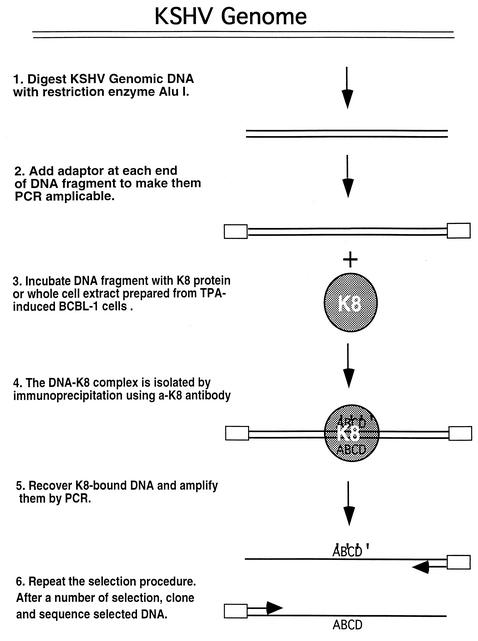
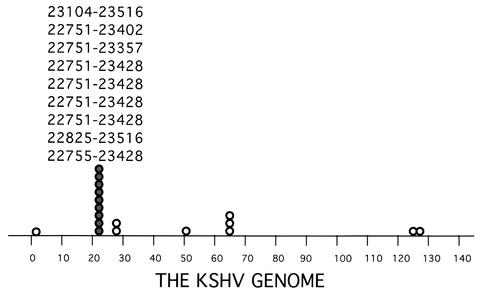
The EBV counterpart of K8, the Zta protein, activates viral genes through binding to a group of DNA sequences (Zta-responsive elements) that are found in many EBV early promoters. In addition, the Zta also directly binds to the EBV lytic replication origin (ori-Lyt), and the binding is essential for viral lytic replication. Does the K8 protein preserve one or both of the functions that Zta possesses, i.e., transcriptional activator and ori-Lyt binding protein? To explore the function of K8, we first asked whether the K8 protein exerts its function through its DNA binding activity. To determine whether K8 binds to a specific DNA sequence(s) and, if so, where K8 binds in the KSHV genome, we performed a DNA binding site selection. The strategy of the selection is illustrated in Fig. 1. KSHV genomic DNA was chopped into small fragments by complete digestions with restriction enzymes AluI or RsaI, followed by ligating a linker to each end of the DNAs to make them PCR amplifiable. The DNA fragments were incubated with whole-cell extract prepared from TPA-induced BCBL-1 cells, a primary effusion lymphoma cell line which is latently infected by KSHV. The considerations for using the cell extract instead of using K8 protein produced and purified from an E. coli expression system are as follows. First, the use of the cell extract in the selection allows identification of DNA sequences that are bound by K8 directly and indirectly. Second, we are not certain whether bacterially synthesized K8 is biologically active. The mixtures were subjected to immunoprecipitation with anti-K8 antibody or preimmune serum. The K8-bound DNAs were eluted, PCR amplified, and used as input for subsequent rounds of selection. The PCR-amplified DNA recovered after each round of selection was subjected to agarose gel electrophoresis. After seven rounds of selection, the resulting DNA displayed a clear pattern in an agarose gel in comparison to a smear staining of the original DNA pool. The pattern was different from that of the control with preimmune serum. Then, the DNAs were cloned, and 20 clones were randomly picked up for sequencing analysis. The locations of the sequences of these clones in the KSHV genome are illustrated in Fig. 2. Interestingly, 9 out of 20 clones were found to derive from the same region of the KSHV genome and shared a common DNA sequence between nucleotides 23104 and 23402 of the KSHV genome (Fig. 2).
FIG. 1.
Scheme for in vitro DNA binding site selection and amplification.
FIG. 2.
The position and frequency of DNA fragments derived from an in vitro DNA binding site selection corresponding to the KSHV genome. The positions of the DNAs from nine clones are indicated (nucleotide numbers are according to numbering of Russo et al. [22]). The data revealed that the KSHV K8 bZip protein binds to a hot spot in the KSHV genome.
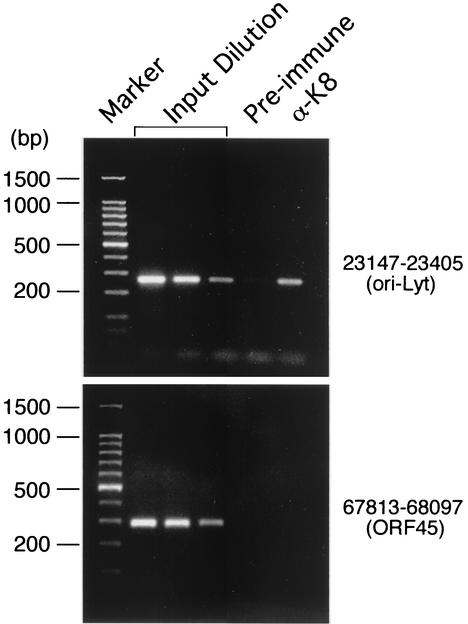
To confirm that the binding of K8 protein to this hot spot in the KSHV genome indeed occurs in vivo, we employed a formaldehyde cross-linking-ChIP technique. In brief, K8-DNA cross-linking was induced by addition of formaldehyde to living TPA-induced BCBL-1 cells. Chromatin from these cells was fragmented by sonication, and the resulting material was immunoprecipitated with anti-K8 antibody. K8-bound DNAs were quantified by PCR by using a pair of primers that were designed to amplify the KSHV sequence between nucleotides 23147 and 23405. The result showed that the potential K8 binding DNA sequence can be coprecipitated by anti-K8 antibody, but not by preimmune serum (Fig. 3, upper panel). Another pair of primers that amplify the viral DNA between nucleotides 67813 and 68097 (within the ORF45 coding region) were used as a control. The ORF45 sequence was not detected in the precipitate brought down by anti-K8 antibody (Fig. 3, lower panel). Thus, K8 protein does bind to this DNA sequence in vivo.
FIG. 3.
Analysis of the association of K8 protein with the ori-Lyt region, using a ChIP assay. The immunoprecipitates by anti-K8 antibody or preimmune serum were analyzed by a PCR which was designed to amplify KSHV DNA between nucleotides 23147 and 23405 (upper panel) and between nucleotides 67813 and 68097 (lower panel).
Characterization of binding of K8 protein to DNA in vitro.
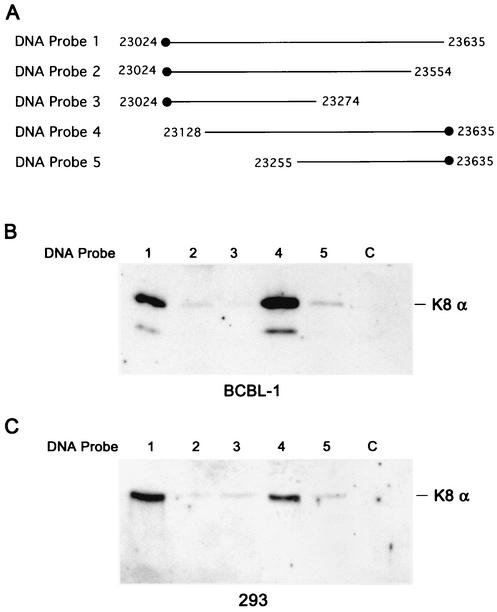
To characterize the DNA binding properties of K8, we first attempted to employ an electrophoretic mobility shift assay (EMSA) with an E. coli-synthesized glutathione S-transferase-K8 fusion protein. Seven double-stranded DNA fragments ranging from 70 to 150 bp around the K8 binding hot spot region (between nucleotides 22925 and 23635) were prepared and used in EMSAs. No specific binding was detected. We thought that there were two possible explanations for this result. (i) The bacterially synthesized K8 is perhaps not able to bind to DNA because it may not be biologically functional. (ii) The stable binding of K8 protein to DNA may require multiple sites that are scattered throughout an extended DNA sequence, and none of the probes that we used in the EMSA (70 to 150 bp in length) contains sufficient DNA sequence for stable K8 binding. Therefore, we decided to use nuclear extract prepared from TPA-induced BCBL-1 cells and relatively long DNA fragments to study the nature of DNA binding by K8. Five DNA fragments, as illustrated in Fig. 4A, were synthesized by PCR, and each DNA was biotinylated at an end. These DNAs were coupled to streptavidin-conjugated magnetic beads and incubated with nuclear extracts of TPA-induced BCBL-1 cells or uninduced cells. The bound materials were washed with 150 mM KCl-containing buffer and eluted progressively with 300, 500, and 1,000 mM KCl. Each eluted fraction was assayed by Western blotting with anti-K8 antibody. The results were as follows. (i) With the longest DNA fragment (nucleotides 23024 to 23635; DNA probe 1) which was linked to magnetic beads at the left end, K8 protein could bind to the DNA and be eluted at 500 mM KCl (Fig. 4B, lane 1). (ii) When the DNA was deleted from the right to nucleotide 23555 (DNA probe 2), K8 bound to the DNA probe very weakly and only a very faint band could be detected in the 500 mM KCl elution; when further deleted to nucleotide 23276 (DNA probe 3), K8 could not bind to the DNA at all (Fig. 4B, lanes 2 and 3). (iii) When a 508-bp DNA (nucleotides 23128 to 23635; DNA probe 4) with conjugation to magnetic beads at the right end was mixed with the nuclear extract, K8 could bind to the DNA strongly and be eluted at 500 mM KCl (Fig. 4B, lane 4). (iv) Deletion from the left to nucleotide 23255 (DNA probe 5) greatly impaired the ability of being bound by K8 (Fig. 4B, lane 5). An irrelevant DNA of 400 bp (from the ORF45 coding sequence, designated DNA probe c [the c stands for control]) was included in the experiment as a control and was shown to be unable to be bound by K8 (Fig. 4B, lane 6). The results of this experiment suggest that K8 requires a quite-large DNA fragment (nucleotides 23128 to 23635) for stable binding.
FIG. 4.
Mapping of DNA sequence requirements for K8 binding. (A) Schematic illustration of biotinylated DNA fragments used in this assay, where the biotin label is marked with solid dots. (B) TPA-induced BCBL-1 nuclear extract was incubated with DNA fragments conjugated on magnetic beads, washed, and eluted with D500 elution buffer. Samples were assayed by Western blotting with anti-K8 antibody. (C) The nuclear extract from 293 cells transfected with pCR3.1-K8α was incubated with the DNA fragments conjugated on magnetic beads, washed, and eluted with D500 elution buffer. Samples were assayed by Western blotting with anti-K8 antibody.
To examine whether the binding of K8 protein to DNA requires other viral proteins to be involved, the binding assay was performed with nuclear extract prepared from 293 cells that were transfected with K8α expression vector (pCR3.1-K8α). As illustrated in Fig. 4C, K8 protein in 293 cell extract could bind to probes 1 and 4 efficiently and be eluted at 500 mM KCl, indicating that K8 can bind to DNA in the absence of other viral proteins.
Identification of KSHV ori-Lyt.
Since the EBV counterpart of K8, the Zta protein, binds to EBV ori-Lyt and the binding is essential for viral lytic DNA replication (10), we speculated that K8 is an ori-Lyt binding protein and its binding hot spot may be part of ori-Lyt of KSHV.
In fact, the sequence around the K8 binding site has been described by others as a potential origin of KSHV lytic DNA replication (19). It was found that a 1.7-kb sequence in the KSHV genome between nucleotides 23137 and 24902 has typical features of a herpesvirus lytic DNA replication origin. These features include two perfect AT-rich palindromes that resemble the core loop structures of HSV ori-L and ori-S and are also found in the HHV-6 and HHV-7 lytic origins and an imperfect AT-rich palindrome, multiple short repetitive motifs containing XcaI and PvuII sites that are found in EBV ori-Lyt, two FspI/SphI motifs that are found within cytomegalovirus ori-Lyt, and clustered consensus binding motifs for AP-1, CTF, and ATF. Remarkably, the entire 1.7 kb is duplicated as an almost-identical inverted copy between gene K12 (Kaposin) and ORF71 (vFLIP).
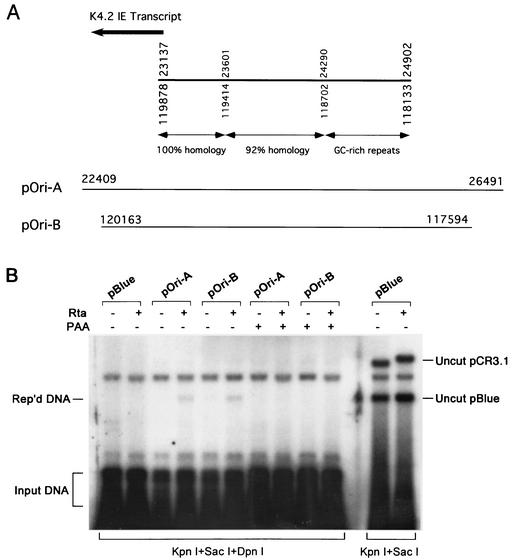
To determine whether the duplicated DNA sequences are indeed lytic origins of KSHV, we examined the duplicated DNA sequences for their abilities to support lytic DNA replication by using a transient-transfection replication assay. Two viral DNA sequences were cloned into pBluescript plasmid as illustrated in Fig. 5A. The resulting plasmids pOri-A and pOri-B encompass an entire potential ori-Lyt from these two loci (spanning nucleotides 23137 to 24902 and nucleotides 119878 to 118133, respectively). These plasmids were introduced into BCBL-1 cells, and the replication of these recombinant DNAs was examined in these cells that can support KSHV lytic DNA replication. Viral lytic replication in these cells was induced by cotransfecting the ORF50 (Rta)-expressing vector, pCR3.1-ORF50. Then, we examined whether any of these KSHV sequences in the plasmid could support replication of the plasmid DNA by using a DNA replication assay. Total DNAs were isolated from the cotransfected cells and digested with KpnI/SacI and KpnI/SacI/DpnI. The replicated pBluescript DNA was distinguished from input DNA by DpnI restriction digestion, which only cleaves E. coli-made DNA because of specific methylation on the DNA. Newly replicated DNA in mammalian cells is resistant to DpnI digestion. The cleaved DNAs were separated on an agarose gel and subjected to Southern hybridization, using pBluescript DNA as a probe. In the cells cotransfected with pCR3.1-ORF50, input methylated pOri-A and pOri-B DNAs were replicated to produce unmethylated copies which were resistant to DpnI digestion (Fig. 5B). The replication of pOri-A and pOri-B plasmids was inhibited when the transfected cells were cultured in the presence of phosphonoacetic acid, a herpesviral DNA polymerase inhibitor (Fig. 5B). The replication was not detected in cells that were in latent stage (cells cotransfected with empty pCR3.1). As a negative control, pBluescript vector cannot be replicated in induced and uninduced BCBL-1 cells. In addition, pCR3.1-ORF50 plasmid in cotransfected cells was not replicated and can be considered as an internal control (Fig. 5B). This result suggests that the origin-like sequence in both loci carries the cis-acting element required for KSHV lytic DNA replication. There are two lytic DNA origins (ori-Lyt) in the KSHV genome.
FIG. 5.
(A) Schematic presentation of the duplicated ori-Lyt region in the KSHV genome. The homology between the two duplicated sequences is indicated. The positions of the inserts in the plasmids pOri-A and pOri-B are shown below the ori-Lyt regions. (B) Replication of KSHV ori-Lyt-containing plasmids in cotransfected BCBL-1 cells receiving ori-Lyt plasmids plus pCR3.1-ORF50. KSHV lytic replication was induced by expression of ORF50 (Rta). Total DNAs were isolated from the transfected cells and used in the replication assay. Replicated DNAs were distinguished from input DNAs by DpnI digestion and detected by Southern blotting with 32P-labeled pBluescript plasmid.
Mapping of right and left boundaries of the ori-Lyt core domain.
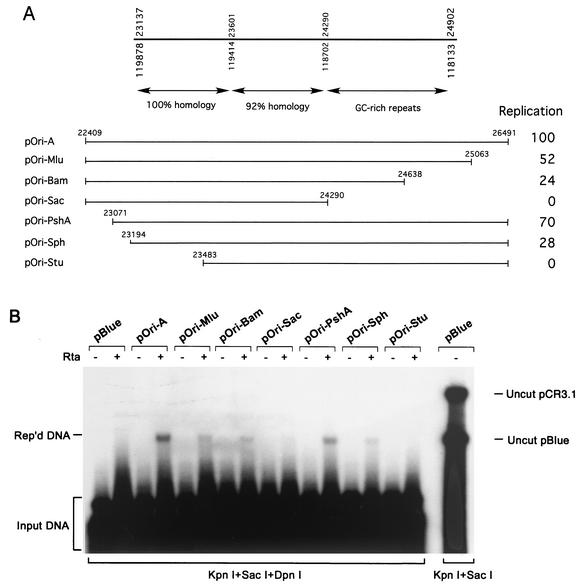
Although both duplicated ori-Lyt domains are able to mediate DNA replication in the viral lytic life cycle, we focused on one locus (between K4.2 and K5) for further analysis. To define the ori-Lyt more closely, we mapped the left and right boundaries of the ori-Lyt domain. Progressive deletions from each end were generated and tested for their abilities to support lytic replication. Four constructs with the right ends of inserts at nucleotides 26491 (PstI), 25063 (MlnI), 24638 (BamHI), and 24290 (SacI) were used to map the right boundary of the ori-Lyt core domain. Similarly, four plasmids with the right boundaries at nucleotides 22409 (EcoRI), 23071 (PshAI), 23194 (SphI), and 23483 (StuI) were also constructed and used to map the left boundary of the ori-Lyt. These deletion constructs were introduced into BCBL-1 cells along with pCR3.1-ORF50 or empty pCR3.1 vector by electroporation. The ability of each deletion mutant to support DNA replication in viral lytic phase was assayed by using a DpnI DNA replication assay. The results showed that deletions up to nucleotide 25063 from the right and deletions up to nucleotide 23071 from the left had limited effects on lytic DNA replication. Any further deletion made replication less efficient or completely abolished it. pOri-Bam has a truncation from the right to nucleotide 24638 and misses half of the GC-rich repeats. The replication of this mutant was apparently impaired. pOri-Sac, which contains the 1.1-kb duplicated DNA sequence and lacks completely the GC-rich repeats, could not support lytic DNA replication at all. These results suggest that the GC-rich tandem repeat domain is needed for lytic DNA replication (Fig. 6). Overall, the 1.7-kb inverted duplicated sequence (nucleotides 23137 to 24902) appears to be necessary and sufficient to act as a cis signal for replication.
FIG. 6.
Mapping of boundaries of KSHV ori-Lyt. (A) A series of deletion mutants were constructed, and the location of the fragments present in different plasmids is indicated with the nucleotides of the KSHV genome (22). (B) Each mutant plasmid was assayed in BCBL-1 cells for its ability to support lytic-phase DNA replication. KSHV lytic replication is induced by expression of ORF50 (Rta). Extrachromosomal DNAs were prepared using the Hirt extraction method and used for the assay. Replicated DNAs were distinguished from input DNAs by DpnI digestion and detected by Southern blotting with 32P-labeled pBluescript plasmid. The replication rate of each mutant relative to that of pOri-A was calculated by measuring intensities of the replicated and the input DNA bands in a phosphorimager. Each number is the average of three independent experiments.
Inspection of the KSHV ori-Lyt DNA sequence.
The 1.7-kb ori-Lyt is composed of two domains: a 1.1-kb duplicated sequence which is almost identical between the two ori-Lyt's; and high-GC-content tandem repeats that are represented as 20-bp and 30-bp tandem arrays. The 1.1-kb domain has many prominent structural motifs that were either found in ori-Lyt's of other herpesviruses or implicated to have roles in ori-Lyt function. The most remarkable motifs in the domain are two long AT palindromes (16-bp AT-Pal, AAAAATTATAATTTTT, nucleotides 23275 to 23290; 18-bp AT-Pal, TATATATATATATATAAT, nucleotides 23572 to 23590). Such AT palindromes resemble the core loop structures of HSV ori-L and ori-S (18) and are also found in the HHV-6 and HHV-7 lytic origins (8). AT palindromes are often seen in both viral and cellular DNA replication origins, and initiation of replication generally involves local unwinding at these AT-rich sequences (6). In addition, there is an additional imperfect 14-bp AT-rich palindrome (14-bp AT-Pal, TTTATGTGCATAAA, nucleotides 23159 to 23172) in the 1.1-kb domain. Other interesting motifs include six copies of a short repetitive motif containing XcaI and PvuII sites which resemble features of EBV ori-Lyt, two Fsp/SphI motifs that are found within CMV ori-Lyt, and numerous other features that are present in the domain, including three TATA boxes (CATAAAA, nucleotides 23164 to 23158; TATAATTAA, nucleotides 23711 to 23719; and TATAATA, nucleotides 23894 to 23900) and clustered consensus binding motifs for known transcription factors such as AP-1, ATF, SP-1, and C/EBP.
An AT-palindrome sequence in the ori-Lyt is required for lytic DNA replication as well as K8 binding.
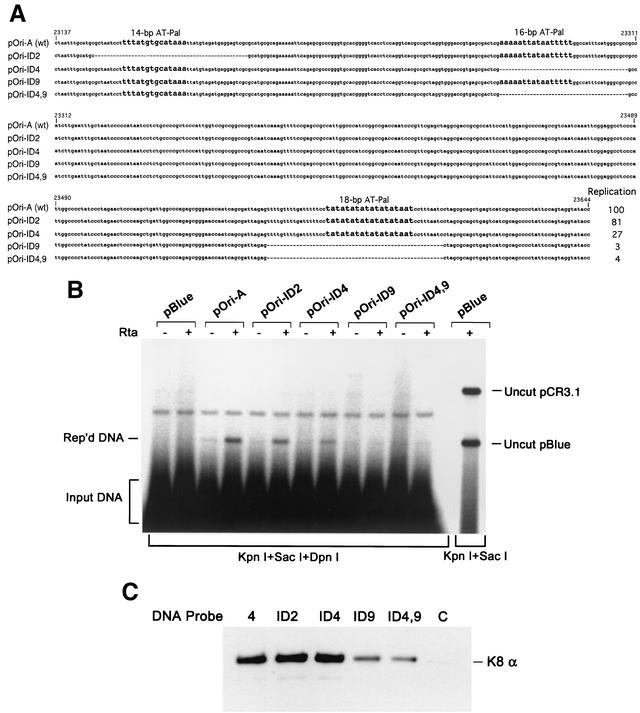
In the ori-Lyt domain, some conserved motifs have been implicated to have roles in ori-Lyt function (19). Among them, three AT palindromes are extremely intriguing. AT palindromes are often found in eukaryotic cellular DNA replication origins, and initiation of replication generally involves local unwinding at an AT-palindromic sequence (6). To examine roles of these AT palindromes in KSHV ori-Lyt in DNA replication, several internal deletion mutants were constructed. These deletion mutants, namely, pOri-ID2 (Δ23149-23198), pOri-ID4 (Δ23275-23309), pOri-ID9 (Δ23555-23599), and pOri-ID4,9 (a double mutant), were designed to delete each or two of the AT palindromes (Fig. 7A). These internal deletion constructs were used for a transient-transfection replication assay in BCBL-1 cells. BCBL-1 cells were cotransfected with each of these internal deletion plasmids along with pCR3.1-ORF50. At 72 h posttransfection, extrachromosomal DNAs were isolated from the cotransfected cells by using the Hirt DNA extraction method and subjected to Southern analysis for replicated (DpnI-resistant) DNAs as described above. As shown in Fig. 7B, deletion of the imperfect 14-bp AT-rich palindrome (pOri-ID2) had little effect on ori-Lyt-mediated DNA replication. pOri-ID4, which had the 16-bp AT-Pal deleted, was still able to replicate, though with a reduced efficiency (27% of the replication rate of wild-type ori-Lyt). In contrast, deletion of the 18-bp AT-Pal (pOri-ID9) completely abolished DNA replication. This result suggested that the 18-bp AT palindrome is essential for KSHV ori-Lyt-dependent DNA replication.
FIG. 7.
AT palindromes are essential for KSHV ori-Lyt replication and K8 binding. (A) The nucleotide sequence of wild-type DNA and mutants in which each of the AT-rich palindromes in the ori-Lyt domain was deliberately deleted. (B) Each mutant plasmid was assayed in BCBL-1 cells for its ability to support lytic-phase DNA replication. KSHV lytic replication is induced by expression of ORF50 (Rta). Extrachromosomal DNAs were prepared using the Hirt extraction method and used for the assay. Replicated DNAs were distinguished from input DNAs by DpnI digestion and were detected by Southern blotting with 32P-labeled pBluescript plasmid. The replication rate of each mutant relative to that of pOri-A was calculated by measuring the intensities of the replicated and the input DNA bands. Each number is the average of two independent experiments. (C) The AT-palindrome deletion mutants were assayed for their capabilities of being bound by K8 protein. Biotinylated DNA probe 4 (from nucleotides 23128 to 23635; see Fig. 4) and its deletion derivatives (probe ID2, probe ID4, probe ID9, and probe ID4,9) were prepared by PCR. These DNA fragments were conjugated on magnetic beads and incubated with TPA-induced BCBL-1 nuclear extract. After washing, bound proteins were eluted with D500 elution buffer. Samples were assayed by Western blotting with anti-K8 antibody. C, control with an irrelevant DNA.
These AT-palindrome deletion mutants were also used to examine the contribution of each of the palindrome sequences to K8 binding to ori-Lyt DNA. Biotinylated DNA fragments were synthesized using PCR with pOri-A DNA and AT-palindrome deletion mutants (pOri-ID2, pOri-ID4, pOri-ID9, and pOri-ID4,9) as templates. The DNAs were coupled to streptavidin-conjugated magnetic beads and then incubated with nuclear extract of TPA-induced BCBL-1 cells. The bound materials were washed with 150 mM KCl-containing buffer and eluted with 500 mM KCl. Each eluted fraction was assayed on Western blotting with anti-K8 antibody. The result, as illustrated in Fig. 7C, showed that deletion of 14-bp and 16-bp palindromes (pOri-ID2 and pOri-ID4) did not impair the binding of K8 to these DNAs, whereas removal of the 18-bp AT palindrome caused a dramatic reduction in the ability of the DNA to be bound by K8 protein. Thus, the 18-bp AT-palindrome sequence was proven to be critical for K8 binding and ori-Lyt-dependent viral DNA replication as well.
DISCUSSION
In this study, we showed that two regions in the KSHV genome, between K4.2 and K5 and between K12 and ORF71, are able to serve as origins for lytic cycle-specific DNA replication (ori-Lyt). The two ori-Lyt domains share an almost identical 1,153-bp sequence and a 600-bp downstream GC-rich repeat sequence. A mapping analysis suggests that these 1.7-kb DNA sequences are sufficient to act as a cis signal for KSHV lytic replication.
In addition, a virally encoded bZip protein, namely K8, was found to bind to a KSHV sequence within the ori-Lyt by using a DNA binding site selection. The binding of K8 to this region was confirmed in cells by using the ChIP method. Further analysis showed that stable K8 binding to DNA requires an extended DNA sequence that is 100% conserved between two KSHV ori-Lyt's. The K8 protein displays significant similarity to the Zta protein of EBV, which is known to be an OBP of EBV. Therefore, it is suggested that K8 may function as an OBP of KSHV.
KSHV ori-Lyt.
In an initial effort of searching for a lytic origin of KSHV, we attempted to map the entire genome of KSHV for the DNA sequence that supports DNA replication in the lytic life cycle of KSHV. Six overlapping cosmid clones, which represent the whole genome of KSHV (27), were introduced individually into BCBL-1 cells, along with the KSHV ORF50 (Rta) expression vector. Three days after transfection, DNA was isolated from the cells and analyzed for cosmid DNA replication. Unfortunately, we were not able to detect any replicated cosmid DNA, probably because of the insensitivity of this assay. After K8 was found to bind to a region between K4.2 and K5 by a DNA binding site selection, we focused on this region and its duplicated region between K12 and ORF71 in the KSHV genome and showed that these two regions can serve as an origin of lytic-phase DNA replication. Is there any possibility of the existence of an additional ori-Lyt in the KSHV genome? We think that is unlikely. First of all, the two ori-Lyt regions display typical features of a herpesviral lytic DNA replication origin. Such features are not found in other regions of the KSHV genome. Second, inspection of the whole KSHV genome revealed that the regions between ORF K4.2 and ORF K5 and between ORF K12 and ORF71 are only two large noncoding intergenic domains (>3 kb) in the viral genome. Thus, it is highly likely that KSHV possesses two ori-Lyt's in its genome, just as EBV does. There are two ori-Lyt's in the EBV genome in most strains, even though one is sufficient for EBV lytic replication (13). When this article was under review, these two regions were also reported to be KSHV ori-Lyt by others (2). The results presented here, together with those of AuCoin et al. (2), located two regions in the KSHV genome that serve as origins of lytic-phase DNA replication.
An ori-Lyt for rhesus macaque rhadinovirus (RRV) was identified in the RRV genome between ORF69 and ORF71 (20). Since the RRV is a close relative of KSHV and its ori-Lyt is located in the same relative position as the right ori-Lyt of KSHV, we compared the nucleotide sequences between these two ori-Lyt's. To our surprise, besides the GC-rich tandem repeat sequences, our sequence analysis did not find any significant homology between RRV and KSHV ori-Lyt's. However, further analysis revealed the presence of an analogous sequence of KSHV ori-Lyt in the genome of RRV. The homologous sequence in the RRV genome is located between ORF69 and ORF71 but not inside the ori-Lyt, which Pari et al. (20) have mapped to the region from nucleotides 112979 to 116210 (nucleotide numbers of the RRV genome according to Searles et al. [23]). The most significant homology was found between the KSHV ori-Lyt conserved sequence from nucleotides 119663 to 119414 and an RRV region around nucleotides 117785 to 117537. There is 68% nucleotide sequence homology between these two regions. Furthermore, some KSHV ori-Lyt motifs, including the critical 18-bp AT palindrome, are present in the RRV homologous region (TATATATATATATAT, nucleotides 117551 to 115537).
Is K8 an OBP?
K8 is thought to be an analogue of Zta of EBV, based on its limited sequence homology to Zta (both are proteins of the bZip family), colinear genomic localization, and similar splicing pattern. Using an in vitro binding site selection assay, K8 bZip protein was found to bind a hot spot located within the KSHV ori-Lyt core domain. It was reported that the K8 protein is associated with a viral DNA replication complex which is composed of at least six other viral replication proteins (e.g., SSB, POL, PAF, HEL, PRI, and PPF) (26). Given the role of Zta in EBV replication as an OBP, we speculate that K8 may play a role in KSHV lytic DNA replication similar to that of Zta in EBV DNA replication. In other words, K8 may be an OBP of KSHV.
However, the DNA binding property of K8 appears to be unusual. First of all, in contrast to Zta, which recognizes and binds to Zta-responsive element motifs of an only-6-bp sequence, our results showed that stable binding of K8 to DNA required an extended DNA sequence (400 to 500 bp in length). Since we were very surprised by this observation, we repeated the experiment three times and found that the results were reproducible. Second, in a bZip protein, the basic amino acid domain located immediately adjacent to the leucine repeat is known to serve as a DNA binding domain (11, 14, 25). Inspection of the amino acid sequence of K8 revealed that a couple of the basic residues that are critical for Zta and other bZip proteins to bind DNA are missing in K8, even though the overall basic domain of K8 is very rich in basic amino acids. Based on these observations, we speculate that a single K8 molecule (or a single K8 homodimer) and a single binding site may not result in stable K8-DNA binding; K8 recognizes and binds to the ori-Lyt DNA sequence through synergy of several K8 molecules binding to multiple K8 binding sites or synergistic action of K8 with another DNA binding protein(s). Thus, multiple binding motifs scattered in an extended DNA sequence are required for stable DNA binding of K8 protein.
In the 1.7-kb ori-Lyt domain, the first 465 nucleotides are identical between these two KSHV ori-Lyt's. K8 protein appears to need the entire identical duplicated region for stable binding. One may think of the possibility that the binding of K8 to the ori-Lyt DNA is only stable when the whole replication complex (including at least six other viral proteins) forms on the ori-Lyt DNA. However, the observation that K8 protein expressed in 293 cells is also able to bind to ori-Lyt DNA disagrees with this hypothesis. Our results suggested that K8 protein can bind to ori-Lyt DNA without involvement of any other viral proteins. However, we cannot rule out the possibility that some cellular protein may facilitate K8 binding to KSHV ori-Lyt DNA. The detailed mechanism underlying K8 binding to DNA will be investigated through a fine mapping of K8 binding motifs in the ori-Lyt domain and mutagenesis studies.
Overall, we report here the identification of two ori-Lyt's in the KSHV genome and the observation that the viral bZip protein K8 binds to the ori-Lyt. This work makes possible a number of future studies on mechanism of KSHV lytic DNA replication. One of these studies, which is ongoing in our laboratory, is to address if K8 bZip protein is obligatory for origin-dependent DNA replication and whether K8 is a key viral component in initiating KSHV ori-Lyt-dependent DNA replication.
Acknowledgments
We thank Jae Jung at the New England Regional Primate Research Center for providing us with anti-K8 polyclonal antibody. We thank Paul Lieberman at the Wistar Institute and Rolf Renne at Case Western Reserve University for helpful discussions.
This work was supported by research grants from the National Institutes of Health (R01CA86839 and R01AI52789) to Y.Y.
REFERENCES
- 1.Ambroziak, J. A., D. J. Blackbourn, B. G. Herndier, R. G. Glogau, J. H. Gullett, A. R. McDonald, E. T. Lennette, and J. A. Levy. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582-583. [DOI] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 76:7890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 6.Challberg, M. D., and T. J. Kelly, Jr. 1989. Animal virus DNA replication. Annu. Rev. Biochem. 58:671-717. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Dewhurst, S., D. M. Krenitsky, and C. Dykes. 1994. Human herpesvirus 6B origin: sequence diversity, requirement for two binding sites for origin-binding protein, and enhanced replication from origin multimers. J. Virol. 68:6799-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentz, R., F. Rausher, C. Abate, and T. Curran. 1989. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science 243:1695-1699. [DOI] [PubMed] [Google Scholar]
- 12.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 14.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 15.Lin, S.-F., D. R. Robinson, G. Miller, and H.-J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, G. 1990. The switch between latency and replication of Epstein-Barr virus. J. Infect. Dis. 161:833-844. [DOI] [PubMed] [Google Scholar]
- 17.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockshorn, D., and D. A. Galloway. 1988. Sequence and structural requirements of a herpes simplex viral DNA replication origin. Mol. Cell. Biol. 8:4018-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholas, J., J. Zong, D. Alcendor, D. M. Ciufo, L. Poole, R. T. Sarisky, C. Chiou, X. Zhang, X. Wan, H. Guo, M. Reitz, and G. S. Hayward. 1998. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 23:79-88. [DOI] [PubMed] [Google Scholar]
- 20.Pari, G. S., D. AuCoin, K. Colletti, S. A. Cei, V. Kirchoff, and S. W. Wong. 2001. Identification of the rhesus macaque rhadinovirus lytic origin of DNA replication. J. Virol. 75:11401-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 22.Russo, J. J., R. A. Bohenzky, M.-C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 91:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, and F. Sigaux. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 25.Turner, R., and R. Tjian. 1989. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science 243:1689-1694. [DOI] [PubMed] [Google Scholar]
- 26.Wu, F. Y., J. Ahn, D. J. Alcendor, W. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]