Abstract
Candida albicans, a commensal organism and a pathogen of humans, can switch stochastically between a white phase and an opaque phase without an intermediate phase. The white and opaque phases have distinct cell shapes and gene expression programs. Once switched, each phase is stable for many cell divisions. White–opaque switching is under a1–α2 repression and therefore only happens in a or α cells. Mechanisms that control the switching are unknown. Here, we identify Wor1 (white–opaque regulator 1) as a master regulator of white–opaque switching. The deletion of WOR1 blocks opaque cell formation. The ectopic expression of WOR1 converts all cells to stable opaque cells in a or α cells. In addition, the ectopic expression of WOR1 in a/α cells is sufficient to induce opaque cell formation. Importantly, WOR1 expression displays an all-or-none pattern. It is undetectable in white cells, and it is highly expressed in opaque cells. The ectopic expression of Wor1 induces the transcription of WOR1 from the WOR1 locus, which correlates with the switch to opaque phase. We present genetic evidence for feedback regulation of WOR1 transcription. The feedback regulation explains the bistable and stochastic nature of white–opaque switching.
Keywords: feedback regulation, a1–α2 repression
Candida albicans is associated with humans both as a harmless commensal organism and a pathogen. In most humans, it is a normal part of the microflora in the gastrointestinal tract and in other parts of the body. However, when the host immune system is compromised, it can cause mucosal infections and life-threatening disseminated infections. The ability of C. albicans to undergo a yeast-to-hypha transition and high-frequency phenotypic switching provides the organism with a high degree of phenotypic diversity and the adaptability that is necessary to survive in different host environments (1). One of the high-frequency switching systems is white–opaque switching. White–opaque switching was first identified in a clinical isolate, WO-1, in which cells switched spontaneously between white and opaque phases (2). White phase cells appear round and form hemispherical, white colonies on solid agar, and opaque phase cells are elongated in cell shape with pimples on the surface and form flat, opaque colonies. Several features of white–opaque switching are particularly interesting (1, 3): (i) the transition involves only two phases, and there is not an intermediate phase, (ii) the transition is stochastic, generating heterogeneity in a cell population, and (iii) the phase is stable for many cell divisions, with daughter cells inheriting the phase of their mother cells. White–opaque switching occurs at a frequency of 10−4 to 10−5 per cell division, and opaque–white switching happens at a frequency of 5 × 10−4 per cell generation (4). Opaque cells are stable at 25°C. Upon shifting the temperature to 37°C, they switch en masse to white cells (5).
White–opaque switching is under a1–α2 repression (6). In C. albicans, cell type is determined by the MTL (mating type-like) locus, which contains two alleles, MTLa and MTLα (3, 7, 8). MTLa encodes the a1 and a2 transcriptional regulators, and MTLα encodes the α1 and α2 transcriptional regulators. a2 activates the a-specific genes, α1 activates the α-specific genes, and a1–α2 act together to repress haploid-specific genes, including mating genes and genes required for the white–opaque transition. Miller and Johnson (6) discovered that MTLa/α cells are unable to switch to opaque cells but that deletion of MTLa1 or MTLα2 allows white–opaque transitions.
Opaque cells mate ≈106 times more efficiently than do white cells (6). It is suggested that mating occurs only between opaque cells of opposite mating type and that the low level of mating observed between white cells is due to spontaneously arising opaque cells (3, 6). Genome-wide transcriptional profiling of white and opaque cells has revealed ≈237 genes that are up-regulated in opaque cells and ≈179 genes that are up-regulated in white cells (8, 9). Several opaque genes are in the pheromone-responsive mitogen-activated protein kinase pathway that has been shown to play a role in mating (10–13), further supporting the notion that opaque cells are primed for mating in C. albicans. Many of the differentially up-regulated genes in the opaque phase are implicated in metabolism. Despite the identification of genes that are preferentially expressed in the white and opaque phases, molecular mechanisms for white–opaque switching and the stochastic development of opaque cells in a cell population are not known.
Stochastic cell fate development in a cell population is common in nature. Examples include Xenopus oocyte maturation (14) and Bacillus subtilis competence development (15, 16). Both systems use positive feedback loops in fate development. Theoretical studies have predicted that positive feedback and double-negative feedback regulation can provide a means to support the development of bistable phenotypes among cells within a population (17). Here, we report that WOR1 (white–opaque regulator 1), a gene previously identified as an enhancer of yeast adhesion to polystyrene (EAP2, orf19.4884) (18), is a master regulator of white–opaque switching in C. albicans. WOR1 expression is regulated in an all-or-none manner. We present evidence to suggest that Wor1 activates a feedback regulation of WOR1 transcription and that the feedback regulation controls white–opaque switching.
Results
Wor1 Is Required for White–Opaque Switching.
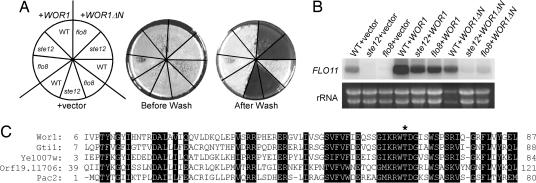
We cloned WOR1 in a high-copy suppressor screen for C. albicans genes that could render a haploid Saccharomyces cerevisiae flo8 mutant invasive on solid media (19). As reported by Li and Palecek (18), we found that WOR1 overexpression increased the expression of FLO11, which encodes a cell surface adhesin that is required for invasive and filamentous growth in S. cerevisiae. WOR1 expression bypassed the requirement of either Flo8 or Ste12 for the expression of FLO11, as shown by invasive growth and Northern blot analysis (Fig. 1 A and B). Wor1 sequence analysis suggests that the N-terminal region (amino acids 6–87) of Wor1 is highly similar to several fungal proteins. The N-terminal region is important for Wor1 function in invasive growth in S. cerevisiae; Wor1 without the region could no longer suppress the flo8 or ste12 mutant in invasive growth (Fig. 1 A and B). Protein sequence alignment of the region of Wor1 with Gti1 and Pac2 of Schizosaccharomyces pombe, YEL007w of S. cerevisiae, and orf19.11706 of C. albicans is shown in Fig. 1C. Overexpression of pac2 inhibits ste11 expression and mating in Sc. pombe (20). Gti1 is required for gluconate uptake upon glucose starvation, which is repressed by the cAMP-dependent protein kinase and requires the Wis1–StyI mitogen-activated protein kinase pathway (21). A potential protein kinase A phosphorylation site is present in all proteins of this family (Fig. 1C). Replacement of T67 with A in Gti1 impaired Gti1 function (21).
Fig. 1.
Identification of Wor1 as a high-copy suppressor of a S. cerevisiae flo8 mutant in invasive growth. (A and B) Invasive assay (A) and FLO11 Northern blot (B) of a haploid S. cerevisiae WT strain (L5528), a flo8 mutant (HLY850) (31), and a ste12 mutant (HLY362) (32) carrying a vector (pVT102-U), WOR1 (pCF68), and WOR1ΔN (pVT-WOR1ΔN). Cells were grown at 30°C on YPD plates for 3 days for invasive growth and in liquid YPD for Northern blot analysis. (C) Protein sequence alignment of the Wor1 N-terminal domain with Sc. pombe Gti1, S. cerevisiae Yel007w, C. albicans Orf19.11706, and Sc. pombe Pac2. Conserved residues are shaded, and a putative protein kinase A phosphorylation site is marked with an asterisk.
To uncover functions of Wor1 in C. albicans, we deleted both copies of WOR1 in an a/α strain, and the deletion was confirmed by Southern blotting (Fig. 6, which is published as supporting information on the PNAS web site). wor1 mutants had no detectable defects in growth and hyphal development (data not shown). Consistent with the lack of phenotypes, WOR1 was not expressed in the a/α strain. A genome-wide transcription profiling study of white and opaque cells of the WO-1 strain showed WOR1 to be differentially expressed in opaque cells (9). Furthermore, genome-wide analyses of gene expression in different cell types and in white and opaque cells have shown that WOR1 is expressed higher in a/− or −/α hemizygote white cells than in a/α cells, and WOR1 expression is ≈47-fold higher in opaque cells than in white cells (8). The expression patterns of WOR1 suggest a potential function in opaque cells. We converted wor1 a/α cells to wor1 a cells by growth selection on sorbose (22). Interestingly, opaque colonies (solid opaque colonies or white colonies with an opaque sector) were never observed with a wor1 cells on any of the media tried, regardless of the length of incubation time, whereas a WT controls consistently gave ≈1–2% opaque colonies (Table 1 and Fig. 2). Heterozygous WOR1/wor1 a cells could form opaque colonies at the same frequency as the WT controls (data not shown). Occasionally, a few dark red colonies on phloxine B-containing plates were observed with the a wor1 strain, but, unlike the elongated morphology of opaque cells, cells from those colonies were abnormally enlarged and round, suggesting that they were not true opaque cells. Consistent with the lack of detectable opaque sectors or colonies, the mating efficiency of the a wor1 strain was consistently low in all mating assays that we carried out and was lower than the mating efficiency of the WT a strains in white phase (Table 2, top three rows). Unlike the a wor1 strain, the mating efficiency of the two WT a strains in white phase varied greatly in different mating assays (data not shown).
Table 1.
Wor1 controls white–opaque switching
| Strain | White to opaque |
Opaque to white |
||||||
|---|---|---|---|---|---|---|---|---|
| Total colonies | Opaque colonies | Opaque sectors | Colonies with opaque cells, % | Total colonies | White colonies | Opaque sectors | Colonies with white cells, % | |
| a (JYC5) | 929 | 0 | 23 | 2.48 | 679 | 0 | 6 | 0.88 |
| a (JYC1) | 1,196 | 3 | 19 | 1.84 | 1,194 | 19 | 0 | 1.59 |
| awor1 | 2,399 | 0 | 0 | 0.00 | N/A | N/A | N/A | N/A |
| awor1 + WOR1 | 1,691 | 0 | 7 | 0.41 | N/A | N/A | N/A | N/A |
| a + WOR1 | >2,000 | All* | 0 | 100 | 1278 | 0 | 0 | 0.00 |
| a/α (CAI4) | 1,266 | 0 | 0 | 0.00 | N/A | N/A | N/A | N/A |
| a/α + WOR1 | 1,191 | 142 | 66 | 17.5 | 2,592 | 0 | 132 | 5.09 |
| a/α wor1+ WOR1 | 2,279 | 0 | 8 | 0.35 | N/A | N/A | N/A | N/A |
Strains used for this experiment are as follows: WT a strains JYC5 and JYC1, a wor1 mutant (CAH3), a wor1 (CAH3) carrying pACT1-WOR1, a (JYC5) carrying pACT1-WOR1, a/α CAI4, a/α (CAI4) carrying pACT1-WOR1, and a/α wor1(CAH2) carrying pACT1-WOR1. For white–opaque switching, 5-day-old solid white colonies from Lee’s medium plates were resuspended and plated onto SC plates with 5 μg/ml phloxine B and grown for 7 days at 23°C. For the opaque–white transition, solid opaque colonies were resuspended and plated onto SC plates as above or onto Lee’s medium plates for a/α + WOR1. The percentage of colonies with opaque or white cells was calculated based on the methods of Miller and Johnson (6). N/A, strains that failed to give any solid opaque colonies for the opaque–white transition.
*No white colonies were observed with this strain.
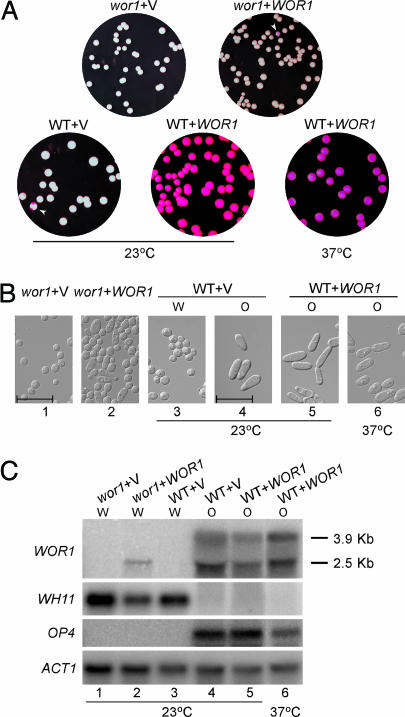
Fig. 2.
Ectopic expression of WOR1 in WT MTLa strains converts all a cells into stable opaque phase. (A) Representative colonies of MTLa wor1 (CAH3) with vector (pACT1) or WOR1 (pACT1-WOR1), and WT MTLa (JYC5) with vector (pACT1) or WOR1 (pACT1-WOR1). Cells of each strain from 5-day-old white colonies on Lee’s medium plates were resuspended and plated on SC plus 5 μg/ml phloxine B plates for 7 days at 23°C or for 3 days at 37°C as indicated. The opaque sectors are marked with white arrowheads. (B) Photographs of cells grown from the white or opaque colonies in A under the indicated conditions. (Scale bars, 20 μm.) (C) Northern blot analysis of WOR1 and phase-specific genes of cells in B. Strains are as follows: lane 1, MTLa wor1/wor1 (CAH3) with vector (pACT1); lane 2, CAH3 with WOR1 (pACT1-WOR1); lanes 3 and 4, MTLa WT (JYC5) with vector from white or opaque colonies, respectively; lanes 5 and 6, JYC5 with WOR1 from opaque colonies. Cells in lanes 1–5 were grown in SC at 23°C overnight. Cells in lane 6 were grown in SC at 23°C overnight and then shifted to 37°C for 8 h. The white or opaque phase of each culture was determined by cell morphology and is indicated above each lane. WH11 and OP4 were used to detect white and opaque phase gene expression. ACT1 was used as a loading control. The exposure times for the probes were 6 h for WOR1, 4 h for OP4, 1 h for WH11, and 3 h for ACT1.
Table 2.
Mating efficiency of the MTLa wor1 mutant
| a strain/phase | Mating efficiency |
|
|---|---|---|
| × α White | × α Opaque | |
| JYC1 white | 8.0 × 10−7 | 1.6 × 10−5 |
| JYC5 white | 3.3 × 10−7 | 5.1 × 10−6 |
| CAH3 white | 1.6 × 10−8 | 3.6 × 10−7 |
| JYC1 opaque | 7.8 × 10−5 | 1.2 × 10−2 |
| JYC5 opaque | 4.8 × 10−5 | 1.6 × 10−1 |
| JYC1 + WOR1 opaque | 8.7 × 10−5 | 4.0 × 10−2 |
The α tester strain used is CHY477 (MTLα/− URA3 HIS1 ade2). JYC1 and JYC2 are WT MTLa ura3 ADE2 strains. CAH3 is the MTLa wor1 ura3 ADE2mutant.JYC1 + WOR1 is MTLa his1 ADE2 pACT1-WOR1. Quantitative mating was performed as described in ref. 6.
Ectopic Expression of WOR1 from the ACT1 Promoter in WT a Strains Converts All Cells to the Opaque Phase.
WOR1 is preferentially expressed in opaque cells, and its expression in white cells is very low (8, 9) and is not detectable by Northern blotting (Fig. 2). The low level of WOR1 could be a limiting factor in white–opaque switching. To determine whether ectopic expression of WOR1 can promote white cells to switch to opaque cells, we placed WOR1 under the ACT1 promoter and transformed pACT1-WOR1 into a strains. Expression of WOR1 from the ACT1 promoter promoted white–opaque switching. All transformants grew up as opaque colonies (Fig. 2A and Table 1), and cells showed a uniformly elongated cell shape (Fig. 2B) (a characteristic of opaque cells), the expression of the opaque-specific gene OP4, and the absence of WH11 transcript (Fig. 2C, lanes 4 and 5). No white colonies or colonies with a white sector were observed when cells from an opaque colony were plated (Table 1). Cells remained in the opaque phase with the expression of opaque-specific genes even after growth at 37°C (Fig. 2), a condition that has been reported to shift opaque cells en masse to white cells (5). Therefore, the ectopic expression of WOR1 from the ACT1 promoter promotes white–opaque switching and locks cells in the opaque phase.
Unlike WT a cells, ectopic expression of WOR1 from the ACT1 promoter in a wor1 cells did not generate solid opaque colonies (Fig. 2A and Table 1). Approximately 0.4% of the colonies had an opaque sector (Fig. 2A and Table 1, wor1 + WOR1). Although some cells from the opaque sector seemed elongated (Fig. 2B), opaque-specific gene expression was not detected from cells grown from an opaque sector (Fig. 2C, lane 2). Therefore, Wor1 expressed from the ACT1 promoter is not sufficient to drive cells into a stable opaque phase. To determine whether the inability to complement the wor1 mutant is a dosage effect, we placed C-terminal HA-tagged WOR1 under the MAL2 promoter. The expression of Wor1-HA in a wor1 cells was able to convert most wor1 cells into elongated opaque cells after 15 h of growth at 25°C in liquid SC-Ura (synthetic complete medium without uracil) plus maltose (data not shown).
Ectopic Expression of WOR1 Allows the Formation of Opaque Cells in a/α Cells.
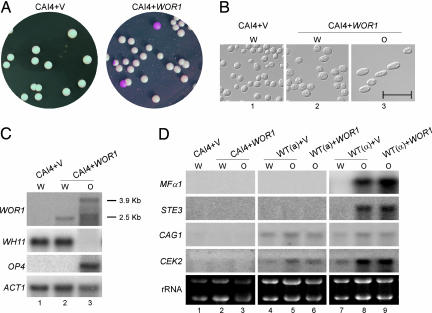
Miller and Johnson (6) showed that a/α cells are unable to undergo white–opaque switching because of the repression of a haploid-specific gene(s) by a1–α2. WOR1 was identified by Tsong et al. (8) as one of the a1–α2-repressed genes. Promoter sequence analysis identified an a1–α2-binding consensus sequence (GCGTGTTAAAAATCACATCA) at −5,640 to −5,621 bp upstream of the WOR1 coding sequence (23). Two additional sequences at −7,966 to −7,947 and −6,993 to −6,974 bp could potentially be a1–α2-binding sites. If WOR1 is the master control of white–opaque switching under a1–α2 repression, ectopic WOR1 expression should be able to promote white–opaque switching in a/α cells. As expected, the ectopic expression of WOR1 produced opaque a/α cells (Fig. 3). Plating cells from a white colony gave rise to 17% opaque colonies (Table 1 and Fig. 3A). The cells from the opaque colonies were elongated in cell shape and expressed the opaque-specific gene OP4 (Fig. 3 B and C). In contrast to ectopic WOR1 expression in a cells, the a/α opaque cells were unstable. They became white cells after an overnight growth at 37°C (data not shown). In addition, colonies with white sectors were observed when cells from an opaque a/α colony were replated (Table 1).
Fig. 3.
Ectopic expression of WOR1 promotes opaque cell formation in a/α cells. (A) Representative white and opaque colonies of CAI4 (MTLa/α) with vector (pACT1) (Left) or with WOR1 (pACT1-WOR1) (Right). (B) Cells from white and opaque colonies in A were grown in SC at 23°C overnight and photographed. (Scale bar, 20 μm.) (C) Logarithmic-phase cultures of white and opaque cells from CAI4 with vector or WOR1 were harvested, and RNA was extracted for Northern blot analysis. Lane 1, white cells of CAI4 with vector; lane 2, white cells of CAI4 with WOR1; lane 3, opaque cells of CAI4 with WOR1. WOR1, WH11, OP4, and ACT1 probes were used for hybridization as described in Materials and Methods. (D) Northern blot analysis of genes required for mating. Lanes 1–3, same as lanes 1–3 in C; lanes 4 and 5, MTLa WT (JYC5) with vector from white or opaque colonies; lane 6, JYC5 with WOR1; lanes 7 and 8, MTLα WT (CHY257) with vector from white or opaque colonies; lane 9, CHY257 with WOR1. WOR1, WH11, OP4, MFα1, STE3, CAG1, CEK2, and ACT1 probes were used for hybridization as described in Materials and Methods.
Interestingly, opaque a/α cells were unable to mate with a or α cells (data not shown). The few colonies obtained from the crosses were due to the loss of one MTL locus, as determined by quantitative PCR (data not shown), suggesting that additional genes required for mating are likely repressed by a1–α2. Indeed, MFα1 (α factor) and STE3 (a factor receptor) transcripts were readily detected in α opaque cells but not in a or a/α opaque cells (Fig. 3D). CAG1 (Gα) and CEK2 (MAPK) were detected only in a and α cells, not in a/α cells (Fig. 3D). This result is consistent with the genome-wide transcriptional analysis that identified CAG1 and CEK2 as being repressed by a1–α2 and identified MFα1 and STE3 as α-specific genes (8). Pheromone-inducible genes FIG1 and HWP1 were not detectable in any of the samples (data not shown). Therefore, a1–α2 controls the expression of WOR1 for white–opaque switching and controls the expression of genes in the pheromone-responsive MAPK pathway for mating. In addition, mating occurs primarily in opaque cells and therefore also requires WOR1.
Wor1 Induces the Expression of WOR1 from the WOR1 Locus.
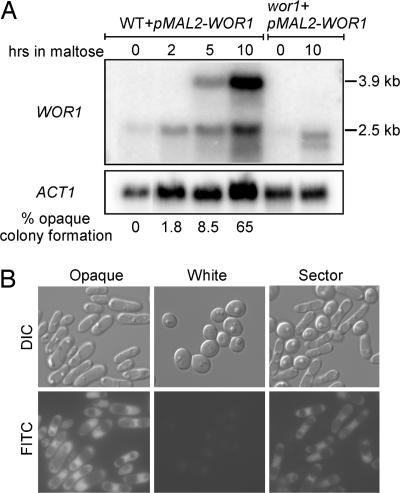
Northern blot analysis showed that WOR1 is not detectable in white cells and has two transcripts of 2.5 kb and 3.9 kb in opaque cells (Fig. 2C, lanes 3 and 4). Both transcripts were observed in opaque wor1/WOR1 heterozygous cells (data not shown) and therefore were transcribed from the same allele of WOR1. It is likely that the 3.9-kb WOR1 transcript has a long 5′ UTR, because the Candida genome identifies only one ORF in this locus, and WOR1 expressed in the wor1 mutant from the ACT1 promoter showed only the 2.5-kb transcript (Fig. 2C, lane 2). Interestingly, the WOR1 expression level from the ACT1 promoter is significantly lower than that of the 3.9-kb WOR1 transcript in the opaque cells (compare lane 2 to lanes 4 and 5 in Fig. 2C), and the former was not sufficient to establish the opaque transcriptional program (Fig. 2C, lane 2). It is possible that the Wor1 expressed from the ACT1 promoter, directly or indirectly, induced the expression of the 3.9-kb WOR1 from the WOR1 locus, which then promoted the switch to the opaque phase. To examine whether Wor1 can induce the expression of the 3.9-kb WOR1 transcript from the WOR1 locus, we transformed the pMAL2-WOR1-HA construct into WT a cells and induced Wor1 expression from the MAL2 promoter in a maltose medium. Induction of WOR1 from the MAL2 promoter was evident by 2 h, and the 3.9-kb WOR1 transcript was not visible yet at this time point (Fig. 4A). By 5 h, the 3.9-kb WOR1 transcript from the WOR1 locus was expressed. The level was significantly higher by 10 h. The increase in the 3.9-kb WOR1 transcript correlated closely with the rise in the percentage of opaque colony formation. Change in cell morphology was not observed by 10 h, but most cells became elongated after 15 h of growth (data not shown). All cells from the 15-h induction produced opaque colonies (data not shown). The MAL2 promoter was not completely off in SC (synthetic complete medium) (Fig. 4A), which is consistent with the relatively frequent appearance of opaque sectors in old colonies of the pMAL2-WOR1-HA transformants on SC plates (data not shown). Therefore, Wor1 can stimulate the expression of the 3.9-kb WOR1 transcript from the WOR1 locus, and the expression correlates with the commitment of cells to the opaque phase.
Fig. 4.
Wor1 induces its own transcription and bistable expression of WOR1 in single cells. (A) Expression of Wor1 from the inducible MAL2 promoter promotes the expression of WOR1 from its own promoter and the transition to opaque phase. MTLa cells (JYC5) transformed with the pMAL2-WOR1-HA construct from a fresh white colony were grown to logarithmic phase in SD, washed five times, and resuspended into SC/maltose at 25°C. Cells were harvested at the indicated times for Northern blot analysis and plated to determine the percentages of opaque colony formation. The percentage of opaque colonies on SD plates with 5 μg/ml phloxine B is shown for each time point. MTLa wor1 transformed with the pMAL2-WOR1-HA construct was included to show the WOR1 transcribed from the MAL2 promoter. (B) Expression of WOR1 shows an on-and-off pattern in single cells. Differential interference contrast microscopy (DIC) and FITC images were taken with cells of MTLa pWOR1-GFP (JYC1 plus pWOR1-GFP) collected from a white colony, an opaque colony, and a sector with both white and opaque cells. GFP expression is under the control of the WOR1 promoter in this strain.
WOR1 Expression Shows an All-or-None Pattern in Single Cells.
To visualize WOR1 expression in individual cells, GFP was cloned under a 3-kb WOR1 upstream sequence, and the construct was integrated into the WOR1 promoter at the WOR1 locus. The GFP integration site was confirmed by PCR, and all transformants still had one copy of WOR1 under the endogenous promoter. Cells from a fresh opaque colony all had an elongated cell shape and cytoplasmic GFP, whereas cells from a white colony of the same transformant did not give a detectable GFP signal (Fig. 4B). Northern blot analysis confirmed that the GFP was expressed from the WOR1 locus with a long 5′ UTR (data not shown). After cells from an opaque colony were grown overnight at 37°C, they all switched to white phase cell morphology, and the GFP signal was not detectable anymore (data not shown), further supporting the observation that the GFP signal was expressed specifically in opaque cells. Cells from a colony with an opaque sector gave a mixed cell morphology (Fig. 4B). As expected, a cytoplasmic GFP signal was detected only in the elongated cells, not in round cells, demonstrating that the expression of WOR1 has an all-or-none pattern at the single-cell level, consistent with the established opaque-specific expression pattern of WOR1.
Discussion
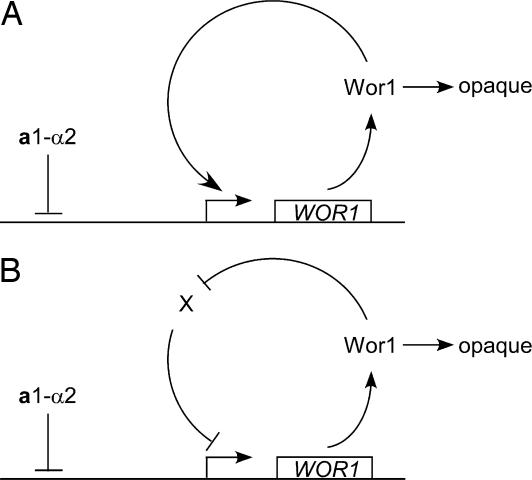
White–opaque switching is a bistable transition that happens stochastically, and the phase is stable for many cell divisions after switching. Mechanisms that control the switching are unknown. In this study, we have identified Wor1 as a master regulator of white–opaque switching. Deletion of WOR1 blocks opaque cell formation. The ectopic expression of WOR1 converts all white cells to stable opaque cells in MTL homozygotes. In addition, the ectopic expression of WOR1 in a/α cells is sufficient to induce opaque cell formation, placing white–opaque switching under the control of a1–α2 (8). Importantly, WOR1 expression displays an all-or-none pattern, and the ectopic expression of Wor1 induces the transcription of WOR1 from the WOR1 locus. We propose that a feedback regulation of WOR1 expression supports the bistability of WOR1 expression and provides a mechanism for white–opaque switching (Fig. 5). This model also explains the stochastic nature of white–opaque switching and phase stability.
Fig. 5.
Model of feedback loops for WOR1 expression and white–opaque switching. A simplified diagram depicts the idea that either a positive feedback loop (A) or a double-negative feedback loop (B) can establish the bistable expression pattern of WOR1. Once expressed, Wor1 activates opaque cell formation.
The expression of WOR1 shows bistability. A GFP fusion to the WOR1 promoter is not detectable in cells from a colony in white phase and is highly expressed in cells from an opaque phase colony (Fig. 4B). Consistent with the study in single cells, WOR1 transcription in white cells is not detectable by Northern blot but is increased ≈47-fold in opaque cells (Fig. 2C) (8). Theoretical studies have shown two feedback configurations that can explain bistability (17). First, a feedback system that contains positive feedback loops with a cooperative response to an activator can display bistability. In this scenario, random cells that happen to have a high concentration of the activator can initiate the positive feedback loop. Second, a system with two mutually repressing negative feedback loops can also lead to bistability. In this case, random cells with a high concentration of the first repressor will have a decreased amount of the second repressor, leading to phase-specific target gene expression. Artificial bistable systems of these two configurations have been demonstrated in Escherichia coli and S. cerevisiae (24, 25). Here, we propose that the bistable WOR1 expression is also driven by feedback regulation (Fig. 5). WOR1 expression from the ACT1 promoter or the MAL2 promoter in a cells induces the expression of WOR1 from its own promoter to generate a high level of Wor1 that promotes the white–opaque transition. The ectopic expression of WOR1 converts all cells into stable opaque cells in MTL homozygotes. In a/α cells, the ectopic expression of WOR1 from the ACT1 promoter also promotes white–opaque switching, but at greatly reduced efficiency, and the opaque phase is not stable. This reduced efficiency indicates a weakened feedback loop in a/α cells, which is likely due to the existence of a1–α2 repression on the WOR1 promoter (Fig. 5). We speculate that the stability of the opaque phase may depend on the strength of the feedback loop, which is proportional to the amount of active Wor1 in the opaque phase. Further research is needed to distinguish between a positive feedback loop versus a double-negative feedback loop. Additional regulators may also be part of the feedback loop.
Signals that promote white–opaque switching are not yet known. It seems that a higher proportion of cells becomes opaque after colonies have been on solid media for a prolonged period. During the long incubation time, the Wor1 basal level may fluctuate and, in some random cells, may accumulate to a level high enough for the initiation of feedback activation of WOR1 transcription. Once the feedback loop is activated, WOR1 will continue to increase and trigger the white–opaque transition. Therefore, noise, or variation, in gene expression may contribute to the formation of some opaque cells in a population of white cells, as has been suggested for phenotypic variability in many systems (26). The feedback loop is also an integral part of a mechanism that explains the stochastic nature of white–opaque switching.
In conclusion, we show that Wor1 is a master regulator of white–opaque switching. We present evidence to support feedback regulation of WOR1 transcription to explain the bistable and stochastic natures of the phase transition. We speculate that feedback regulation could be used commonly by C. albicans in phenotypic switching, providing C. albicans with phenotypic diversity in a cell population and adaptability to adverse conditions, thereby increasing its fitness in hosts.
Materials and Methods
Strains and Culture Conditions.
The C. albicans strains used in this study are listed in Table 3, which is published as supporting information on the PNAS web site. For the routine growth of yeast strains, YPD (1% yeast extract/2% peptone/2% glucose) and SC were used. Media for the invasive growth of S. cerevisiae and the mating of C. albicans were as described in refs. 10 and 19.
Cloning of WOR1.
The plasmid pCF68 was isolated from a C. albicans genomic library, based on its ability to suppress the invasive growth of a S. cerevisiae flo8 mutant (HLY850) on SC-Ura (SC without uracil) (19). pCF68 contains a 4.3-kb insert containing the whole WOR1 ORF. The nucleotide sequence has been deposited in GenBank (accession nos. AY245453 and orf19.4884).
Construction of C. albicans wor1 Strains.
WOR1 was deleted in C. albicans by a PCR product-directed disruption strategy (27) (Fig. 6A). Primers WOR5DR and WOR3DR were used to amplify C. albicans HIS1 and ARG4 from plasmids pGEM-HIS1 and pRS-ARG4ΔSpeI, respectively (27). The PCR products were transformed into BWP17 sequentially, generating the WOR1/wor1 heterozygous strain CAH1 and the wor1/wor1 null mutant strain CAH2. wor1 deletions were confirmed by PCR and Southern blot analysis (Fig. 6B). C. albicans MTLa strains were isolated by streaking the MTLa/α strains on YEPS (yeast extract-peptone plus 2% sorbose) medium and incubating the plates at 37°C for 1 week (22). Colonies from Sou+ plates were identified by PCR with primer pairs for MTLa and MTLα as described in ref. 22. Primers used in the deletion are listed in Table 4, which is published as supporting information on the PNAS web site.
Plasmid Construction.
pACT1 was constructed by PCR amplification of a 1-kb promoter sequence of C. albicans ACT1 from CAI4 genomic DNA with primers ACT1pF (AACTGCAGCCTCGTTTATAATAAACTTAGTC) and ACT1pR (CCGATATCCATTTTGAATGATTATATTTTTTTAA) and by subcloning the PCR fragment into the PstI/EcoRV site of the vector BES116 (28).
pACT1-WOR1 was constructed by PCR amplification of the WOR1 ORF and the 660-bp 3′ UTR with primers WOR1F (CCGATATCTCTAATTCAAGTATAGTCCCTAC) and WOR1R (TGAAGCTTAGAAAAGAAATTTGCATCGCC) and by subcloning the PCR fragment into the EcoRV/HindIII site of pACT1.
pWOR1-GFP was constructed by cloning a 3-kb upstream region of WOR1 with PCR primers CCATCGATTTATTCAAAATCTTGATACCCTATT and CCCAAGCTTGCTTAATATTGAATTGAATTATAC into the ClaI/HindIII site upstream of the GFP coding sequence in plasmid HLP471 (29). The plasmid pWOR1-GFP was then digested with BglII and integrated into the WOR1 upstream sequence at the BglII site by homologous recombination.
pMAL2-WOR1-HA was constructed by amplifying the WOR1 ORF with PCR primers TGCTCTAGAGGATCCATGTCTAATTCAAGTATAGTCCC and CGGGGTACCGCATGCCTAAGCGTAATCTGGAACATCGTATGGGTATCCAGTACCGGTGTAATACGACCC. The HA sequence was included in the C-terminal primer and in-frame with WOR1. The PCR product was digested with XbaI and KpnI and subcloned into a pMAL2 plasmid derived from BES119 (28).
Northern Blot Analysis.
Northern blot analysis was performed as described in ref. 10. PCR products for C. albicans WOR1, WH11, OP4, ACT1, MFα1, STE3, CAG1, CEK2, and S. cerevisiae FLO11 were used for probe labeling. Primers used for PCR are listed in Table 4.
White–Opaque Switching and Mating Assays.
C. albicans white–opaque switching was performed as reported in refs. 6 and 30. The strains were streaked on Lee’s medium, and colonies were resuspended, plated onto Lee’s medium plus 2% mannitol plates supplemented with 5 μg/ml phloxine B, and incubated at 23°C for 7 days. Quantitative mating analysis was performed as described by Miller and Johnson (6). YPD medium, Lee’s medium, or SC was used for mating between C. albicans MTLa and MTLα strains. The strains were mixed and incubated at 23°C for 6 days. The mating efficiency = conjugants/(limiting parent + conjugants) = the greater of (-Ade-Ura-His-Arg)/(-Ade) or (-Ade-Ura-His-Arg)/ (-Ura-His-Arg).
Supplementary Material
Acknowledgments
We thank Shelley Lane for technical assistance and for reading the manuscript. This work was supported by Chinese National Natural Science Foundation Grants 30330010 and 30028010, Chinese National 863 Grants 2004AA223120 and CAS2004-2-8 (to J.C.), and National Institutes of Health Grant GM55155 (to H.L.).
Abbreviation
- SC
synthetic complete medium.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY245453).
See Commentary on page 12659.
References
- 1.Soll D. R., Lockhart S. R., Zhao R. Eukaryot. Cell. 2003;2:390–397. doi: 10.1128/EC.2.3.390-397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slutsky B., Staebell M., Anderson J., Risen L., Pfaller M., Soll D. R. J. Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett R. J., Johnson A. D. Annu. Rev. Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 4.Rikkerink E. H., Magee B. B., Magee P. T. J. Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srikantha T., Soll D. R. Gene. 1993;131:53–60. doi: 10.1016/0378-1119(93)90668-s. [DOI] [PubMed] [Google Scholar]
- 6.Miller M. G., Johnson A. D. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 7.Hull C. M., Johnson A. D. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 8.Tsong A. E., Miller M. G., Raisner R. M., Johnson A. D. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 9.Lan C. Y., Newport G., Murillo L. A., Jones T., Scherer S., Davis R. W., Agabian N. Proc. Natl. Acad. Sci. USA. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J., Chen J., Lane S., Liu H. Mol. Microbiol. 2002;46:1335–1344. doi: 10.1046/j.1365-2958.2002.03249.x. [DOI] [PubMed] [Google Scholar]
- 11.Magee B. B., Legrand M., Alarco A. M., Raymond M., Magee P. T. Mol. Microbiol. 2002;46:1345–1351. doi: 10.1046/j.1365-2958.2002.03263.x. [DOI] [PubMed] [Google Scholar]
- 12.Panwar S. L., Legrand M., Dignard D., Whiteway M., Magee P. T. Eukaryot. Cell. 2003;2:1350–1360. doi: 10.1128/EC.2.6.1350-1360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett R. J., Uhl M. A., Miller M. G., Johnson A. D. Mol. Cell. Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrell J. E., Jr, Machleder E. M. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 15.Maamar H., Dubnau D. Mol. Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smits W. K., Eschevins C. C., Susanna K. A., Bron S., Kuipers O. P., Hamoen L. W. Mol. Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- 17.Ferrell J. E., Jr Curr. Opin. Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 18.Li F., Palecek S. P. Biotechnol. Prog. 2005;21:1601–1609. doi: 10.1021/bp050236c. [DOI] [PubMed] [Google Scholar]
- 19.Cao F., Lane S., Raniga P. P., Lu Y., Zhou Z., Ramon K., Chen J., Liu H. Mol. Biol. Cell. 2006;17:295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunitomo H., Sugimoto A., Wilkinson C. R., Yamamoto M. Curr. Genet. 1995;28:32–38. doi: 10.1007/BF00311879. [DOI] [PubMed] [Google Scholar]
- 21.Caspari T. J. Cell Sci. 1997;110:2599–2608. doi: 10.1242/jcs.110.20.2599. [DOI] [PubMed] [Google Scholar]
- 22.Magee B. B., Magee P. T. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 23.Jin Y., Zhong H., Vershon A. K. Mol. Cell. Biol. 1999;19:585–593. doi: 10.1128/mcb.19.1.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becskei A., Seraphin B., Serrano L. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner T. S., Cantor C. R., Collins J. J. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 26.Raser J. M., O’Shea E. K. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson R. B., Davis D., Mitchell A. P. J. Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Q., Summers E., Guo B., Fink G. J. Bacteriol. 1999;181:6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazan I., Sepulveda-Becerra M., Liu H. Mol. Biol. Cell. 2002;13:134–145. doi: 10.1091/mbc.01-03-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson J. M., Soll D. R. J. Bacteriol. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H., Styles C. A., Fink G. R. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H., Styles C. A., Fink G. R. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.