Abstract
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT), like all retroviral RTs, is a versatile DNA polymerase that can copy both RNA and DNA templates. In spite of extensive investigations into the structure-function of this enzyme, the structural basis for this dual template specificity is poorly understood. Biochemical studies with two mutations in HIV-1 RT that affect residues contacting the template-primer now provide some insight into this specialized property. The mutations are N255D and N265D, both adjoining the minor groove-binding track, in the thumb region. The N265D substitution led to a loss of processive polymerization on DNA but not on RNA, whereas N255D drastically reduced processive synthesis on both templates. This differential template usage was accompanied by a rapid dissociation of the N265D variant on DNA but not RNA templates, whereas the N255D variant rapidly dissociated from both templates. Molecular dynamics modeling suggested that N265D leads to a loss of template strand-specific hydrogen bonding, indicating that this is a key determinant of the differential template affinity. The N255D substitution caused local changes in conformation and a consequent loss of interaction with the primer, leading to a loss of processive synthesis with both templates. We conclude that N265 is part of a subset of template-enzyme contacts that enable RT to utilize DNA templates in addition to RNA templates and that such residues play an important role in facilitating processive DNA synthesis on both RNA and DNA templates.
During retroviral replication, the viral genomic RNA is converted to a double-stranded DNA with terminal redundancies not present in the template RNA by the coordinated activities of one viral protein, the reverse transcriptase (RT). Thus, retroviral RTs are versatile DNA polymerases endowed with several activities essential for viral replication: RNA- and DNA-dependent DNA polymerases, RNase H, strand transfer, and strand displacement activities (43). Several efforts have helped map the template-primer and deoxynucleoside triphosphate (dNTP)-binding pockets of human immunodeficiency virus type 1 (HIV-1) RT, leading to a better understanding of structural determinants of its characteristic features pertaining to fidelity (13, 24, 27, 44; K. Bebenek, W. A. Beard, T. A. Darden, L. Li, R. Prasad, B. A. Luton, D. G. Gorenstein, S. H. Wilson, and T. A. Kunkel, Letter, Nat. Struct. Biol. 4:194-197, 1997), selection of dNTPs versus ribonucleotide triphosphates (6, 17, 18), processivity (1, 3, 5, 25), and drug sensitivity (21, 39). To our knowledge, however, there is no information about the structural determinants responsible for the unique ability of this class of DNA polymerases to copy both RNA and DNA templates.
Our current understanding of the interactions between HIV-1 RT and template-primer are derived from structural, biochemical, and genetic analyses. Structural knowledge of RT-nucleic acid interactions have come from three crystal structures of HIV-1 RT in complex with nucleic acid substrates (21, 22, 40). In two of them, the template-primer is duplex DNA, with one being a binary complex (22) and the other a ternary complex with a dNTP at the active site (21). In the third, RT is in a binary complex with an RNA-DNA template-primer (40). Interestingly, HIV-1 RT makes many more contacts with the RNA-DNA template-primer than it does with the DNA duplex, suggesting that the additional contacts may be required for interaction with RNA. However, only a fraction of the additional contacts are specifically with the 2′-hydroxyl of the RNA template (40).
The template-primer duplex contacts each of the subdomains within the polymerase domain of RT, including the fingers, palm, thumb, and the connection. Most of the key contacts in the polymerase domain lie in the palm and thumb subdomains within primer grip (β12-13), template grip (β5a and αE), and the residues in αH and αI helices of the thumb. Biochemical studies suggested that “primer grip” positions the primer 3′ terminus for nucleophilic attack on the incoming dNTP (19, 20, 23). Site-directed mutagenesis of primer grip and biochemical studies of the resulting mutants have revealed its key role in affinity to template-primer (46), discrimination between ribo- and deoxyribo-primers (20, 36) and the fidelity of misinsertion and mispair extension events (45).
Template-primer-RT interactions are also critical for the specific incorporation of dNTP and its analogs. The template grip (β5a) mutation E89G provided the first evidence that multidideoxynucleoside analog resistance (37) can be brought on by a substitution at a template-contacting amino acid residue (22). This effect, an indirect effect of template-primer conformation on the dNTP- or drug-binding pockets, has been termed “template repositioning” (7). The E89G substitution results in resistance to multiple dNTP analog inhibitors, resistance to foscarnet, and loss of divalent cation preference (37). Furthermore, the E89G substitution also leads to increased fidelity of dNTP insertion (13), mispair extension (33, 38), and (−1) frameshift mutations (12). The substitution P157S in the template grip element αE influences site-specific incorporation of nucleoside analogs by HIV-1 RT (28). These results collectively show that the template-primer contacting residues contribute to the dNTP-binding pocket geometry indirectly by determining the template-primer duplex conformation.
Other structural elements in HIV RT determine the affinity of RT to template-primer duplex. Alanine-scanning mutations in the helix clamp (αH) of the thumb subdomain showed that residues that line the template-primer interacting surface of the helix clamp (Q258, G262, and W266) are critical for template-primer affinity (4). Based on a model, generated via molecular simulation modeling, of HIV-1 RT complexed with a double-stranded DNA template-primer, Bebenek et al. proposed that a minor groove DNA-binding track (MGBT) formed by four residues located in the thumb subdomain (Q258, G262, W266, and Q269) and one in β5b (I94) plays a key role in directing the movement of RT along the template-primer duplex (Bebenek et al., letter). In support of this view, alanine-scanning mutagenesis of the αH residues resulted in increased termination probability and a decreased frameshift fidelity (two- to sevenfold) (Bebenek et al., letter).
Residues that interact with the template-primer must make transient contacts that can be repeatedly made and broken during alternating phases of dNMP insertion and RT translocation. Such contacts must avoid strong interactions that may immobilize the template-primer, which in turn would interfere with elongation. In order to utilize both DNA and RNA templates, some amino acid residues may need to interact with both types of nucleic acids, whereas others may be specific to each. A large number of hydrogen bonds between RT residues and the template-primer duplex have been observed in crystal structures (21, 22, 40). Of those contacting the RNA strand, at least six amino acid residues make contact with the 2′-OH of the ribose sugar: W153, E89, Q91, I94, N265, and K353 (40). A majority of the rest of the RNA template-contacting residues interact (as do the DNA template-contacting residues) with other parts of the sugar and the phosphate in the backbone. No genetic or biochemical evidence has thus far implicated any of these residues in dual template utilization (or allowing RT to copy both RNA and DNA templates). During the course of exploring the template-primer binding cleft of HIV-1 RT as a drug target, we obtained evidence that one of those residues, N265, may be involved in the dual recognition of RNA and DNA template strands.
Previously, we isolated two HIV-1 RT variants displaying resistance (15) to a template analog HIV-1 RT inhibitor, the DNA aptamer, RT1t49, which was previously isolated by systematic evolution of ligands by exponential enrichment (42). Each of these RT1t49-resistant RT variants contained an Asn→Asp substitution in residues in the αH helix (N255 and N265) in the vicinity of the MGBT. In view of the critical importance of this structural element in enzyme translocation, we examined the influence of these mutations on RT's ability to associate with and processively polymerize on RNA and DNA templates. We report here that, in addition to a key role played by αH in RT's interaction with the template-primer, the residue N265 plays a role in the RT's dual recognition of both RNA and DNA templates and that Asn-265 may be one of a set of residues that constitute the determinants of dual template-specificity of HIV-1 RT.
MATERIALS AND METHODS
Proteins.
The wild-type and mutant HIV-1 RT heterodimers were purified as described previously (26). The mutations were present only in the p66 subunit.
RNA- and DNA-dependent polymerase processivity.
Processivity assays were carried out by using a poly(rA)-oligo(dT) enzyme trap to ensure a single round of binding, primer extension, and dissociation. The RNA template (965 nucleotides [nt]) used was derived by in vitro transcription from the pHIV-PBS plasmid (2) (a kind gift of M. A. Wainberg, McGill University) consisting of a 497-bp HIV-1 sequence spanning the R-region long terminal repeat and a portion of the gag region. Processivity on a DNA template was measured by using a single-stranded circular DNA from the phagemid, pKS-HIV, containing a 979-bp BglII-XhoI fragment from pHIV-PBS. RT reactions (20 μl) were performed as follows: the concentrations of KCl used in the reactions were 120 and 40 mM, respectively, for the RNA and DNA templates. The template-primers were prepared by annealing a 27-nt 32P-end-labeled PBS200 primer (5′-GGCTGACCTGATTGCTGTGTCCTGTGT-3′) or Proc2 primer (5′-GATGCACACAATAGAGGGTTGCTACTG-3′) to a twofold molar excess of RNA or DNA template prior to reactions. RT (1 U) and either DNA or RNA template were preincubated 5 min at 37°C. Reactions were initiated by the addition of 50 μM concentrations of dNTPs and a 20-fold molar excess of poly(rA)-oligo(dT) trap and incubated at 37°C for 15 min before being terminated by the addition of 40 μl of stop solution (90% formamide, 10 mM EDTA, and 0.1% each of xylene cyanol and bromophenol blue). Reactions without trap allowed nonprocessive DNA synthesis, and preincubating RT and trap before the addition of template-primer to block all synthesis confirmed the effectiveness of the trap. Reaction products were analyzed by 5% denaturing polyacrylamide gel electrophoresis and phosphorimaging.
RT-template-primer dissociation.
Dissociation rates were determined by a modification of the assay used to measure polymerase processivity. To facilitate binding of RT to the template-primer, a 130 nM concentration of RT and a 130 nM concentration of template-primer were combined and preincubated for 5 min at 37°C in a buffer consisting of 50 mM Tris-Cl (pH 8.0) and 10 mM dithiothreitol. Reactions with an RNA template contained DNA primer VP200 (5′-TAACCTTGCGGCCGTACTCCCC-3′) annealed to 16S rRNA template. Reactions with a DNA template contained M13 sequencing primer (−47) (5′-CGCCAGGGTTTTCCCAAGTCACGAC-3′; New England Biolabs, Beverly, Mass.) annealed to M13 single-stranded DNA (ssDNA) template. The concentrations of KCl used were 80 or 30 mM, respectively, for reactions with RNA or DNA templates. At time zero, 1.0 mg of heparin or 6 μg of poly(rA)-oligo(dT) ml−1 was added into reactions containing RNA or DNA templates, respectively, to trap the free enzyme. At a series of intervals postchallenge, 10-μl aliquots were removed and mixed with 10 μl of 12 mM MgCl2, 100 μM concentrations of dNTPs, and 20 μM [α-32P]dTTP or [α-32P]dATP for RNA and DNA templates, respectively. Reactions were carried out for an additional 10 min at 37°C before they were terminated by the addition of 10 μl of 0.5 M EDTA. Samples were treated, and incorporation was counted as described previously (26). The effectiveness of heparin and poly(rA)-oligo(dT) to limit polymerization on RNA and DNA templates was tested in control reactions in which enzymes were preincubated with trap before the addition of template-primer and radiolabeled dNTP. Rate constants and mean half-lives were determined by fitting the results to a one-phase exponential decay curve by using nonlinear regression (GraphPad Software, Inc., San Diego, Calif.).
Molecular dynamics modeling procedures.
In order to model the N255D or N265D mutations in an unbiased way in the context of an RNA or DNA template, we used a molecular dynamics-free-energy perturbation approach. This approach allows the residue in question (255 or 265) to continuously sample low-energy conformations while its chemical nature is slowly changed from Asn to Asp. More specifically, the empirical force-field parameters for residue 255 or 265 were changed from those of an Asn to those of an Asp by a process of 11 intermediate steps or simulation windows. For example, during the first window the parameters were entirely those of Asn, during the second window they were a mix of one-tenth Asp with nine-tenths Asn, and finally during the eleventh window they were entirely those of Asp. For each window, we performed 10 ps of unconstrained molecular dynamics on the system by using the final coordinates and velocities of the previous simulation window as the initial conditions. The initial conditions for the first window were taken from the appropriate crystal structure (protein data bank entry 1HYS for RT bound to an RNA-DNA hybrid with RNA as a template, 2HMI for RT bound to duplex DNA) after initial molecular dynamics equilibration.
All molecular dynamics simulations were carried out by using the Sander module of the AMBER (35) suite of programs. Simulations were performed in explicit solvent under periodic boundary conditions. The periodic unit cells were obtained by including waters up to 9 Å distant from the protein nucleic acid complex, resulting in system sizes of about 140,000 atoms. Neutralizing counterions were added. Simulations were carried out at 300 K with a 2-fs time step. The AMBER parm98 force field (8) was used. Bonds involving hydrogen were constrained by using SHAKE. A cutoff of 8 Å was used for the van der Waals interactions, whereas electrostatic interactions were handled by using the Particle Mesh-Ewald method (14). A careful process of energy minimization, followed by 30 ps of unconstrained dynamics at 300 K, equilibrated the initial structures. Subsequently the above process of chemical mutation was applied.
RESULTS
Mutations in the RT thumb display template-specific functional defects.
We first examined heterodimeric RTs containing N255D, N265D, or both (Dbl) mutations in the p66 subunit only for effects on gross enzymatic activities. Using heteropolymeric RNA or DNA templates annealed to appropriate DNA primers, we measured their RNA- and DNA-dependent DNA polymerase activities. None of the three mutants showed drastic reductions in their catalytic efficiencies (Vmax/Km) compared to the wild type, with the extreme being the Dbl mutant, which showed approximately half the activity as wild type (Vmax/Km) (data not shown).
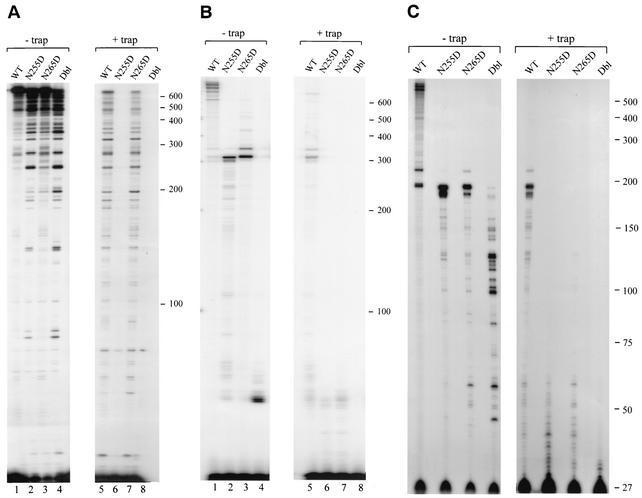
The close proximity of N255 and N265 to the MGBT suggested that these residues play a role in processive DNA synthesis. The processivity of a polymerase refers to its ability to incorporate a series of nucleotides onto a growing primer in a single round of binding, elongation, and dissociation. We tested wild-type and mutant RTs by using a heteropolymeric RNA template annealed to a 5′ P32-labeled oligodeoxynucleotide primer (PBS200). When multiple cycles of RT dissociation and rebinding were allowed, all RTs synthesized similar amounts of the full-length DNA product (Fig. 1A, lanes 1 to 4), indicating no gross defects in polymerization. The N255D and Dbl RTs, however, yielded more products that are of intermediate size (Fig. 1A, lanes 2 and 4). Importantly, under conditions of single-cycle polymerization, only the wild-type HIV-1 RT and N265D variant synthesized full-length DNA products (>600 nt), whereas RTs containing N255D or Dbl displayed dramatically lower processivity, with the longest predominant products being ca. 60 nt in length (Fig. 1A, lanes 5 to 8).
FIG. 1.
Polymerase processivity of wild-type (WT) and mutant RT variants. (A) DNA synthesis on an RNA template. Products synthesized by enzymes in multiple rounds (− trap) or in a single processive cycle (+ trap) are shown. Reaction mixtures contained the pHIV-PBS RNA transcript annealed with the 5′-end-labeled 25-nt DNA primer (PBS200), the RT heterodimer, and dNTPs. (B) DNA synthesis on a DNA template. Reactions were carried out similarly to those with an RNA template. Reactions contained the ssDNA pKS-HIV template annealed to the primer used above. (C) DNA synthesis on a DNA template (ssDNA pKS-HIV) by using the DNA primer Proc 2. The positions of size standards are indicated on the right side of each panel in kilodaltons. All reactions were resolved by denaturing 5% polyacrylamide gel electrophoresis.
In sharp contrast to the results on an RNA template, when an ssDNA template of identical sequence was used in conjunction with the identical DNA primer (PBS200), all mutant RTs displayed severely reduced processivity (Fig. 1B, lanes 5 to 8). Wild-type RT synthesized products of ≥600 nt, compared to ∼40 nt for the N255D and N265D mutant RTs. The Dbl RT displayed nearly wild-type-like activity on a DNA template in the absence of trap; however, it synthesized products of ca. 40 nt when the reactions included a trap (Fig. 1B, lanes 4 and 8). Together, these results demonstrate that the N265D substitution selectively diminishes the processivity of HIV-1 RT on a DNA template while retaining its ability to processively copy an RNA template of identical sequence. On the other hand, the N255 residue in the wild-type RT appears to play a role in processively copying both RNA-DNA and DNA-DNA template-primers.
One may argue that the reason for the inability of the mutants to synthesize more than ∼20 nt lies in some structural or sequence features that force the mutant RTs to stall or dissociate. To address this possibility, we repeated the experiments shown in Fig. 1A and B by using the same RNA or DNA templates but with a primer of identical size that anneals 117 nt downstream of the one used here. Both the RNA-dependent (data not shown) and DNA-dependent (Fig. 1C) processive DNA polymerization experiments with the new primer showed an identical result: on the RNA template, the N265D mutant behaved like the wild type in synthesizing full-length DNA products, whereas on DNA all three mutants synthesized predominantly about 30 nt from the 3′ primer terminus (products of ca. 60 nt; Fig. 1C). Thus, so far, on two sets of template sequences the N265D mutant appears to be unable to processively synthesize DNA on DNA templates. These results support the conclusion that the feature being recognized differentially by the N265D mutant is the backbone sugar (DNA versus RNA) rather than the sequence or a secondary structure.
Dissociation of RT-template-primer complexes correlates with processivity.
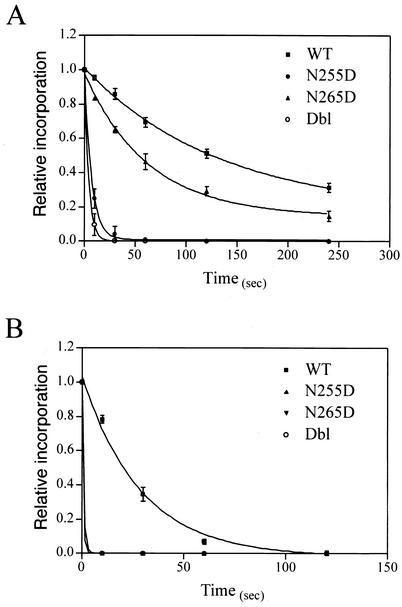
The differential polymerization or processivity of N265D variant with RNA and DNA templates may result indirectly from differential rates of dNTP binding and/or utilization on RNA versus DNA templates. In order to assign a primary role for Asn-265 in dual template recognition by RT, the processivity differences we observed must be associated with differences in the affinity of the mutants to RNA and DNA templates. To test this hypothesis, we determined dissociation rate constants for the template-primer (koff,T-P) and half-life of decay (t1/2) of wild-type and mutant RTs on RNA and DNA templates. The kinetics of dissociation of the RTs on template-primer complexes involving RNA and DNA templates are shown in Fig. 2A and B, respectively. In agreement with the processivity properties, the rates of dissociation on the RNA template are the slowest for the wild type (Fig. 2A). Although the N265D mutant shows a slightly increased rate of dissociation versus the wild type, the N255D and the Dbl mutants dissociate rapidly (∼19- and ∼31-fold, respectively). These results are in agreement with the processivity measurements on RNA template. When the rates of enzyme dissociation were measured on a DNA template, all mutants, including the N265D variant, displayed much higher rates of dissociation than did the wild-type enzyme. The koff,T-P values for the wild-type and mutant RT-template-primer complexes are shown in Table 1. In agreement with previous reports, the koff,T-P for the wild-type RT complex with an RNA template was approximately fivefold lower than with a DNA template (9). When the koff,T-P of N255D, N265D, and Dbl mutant RTs on RNA and a DNA template were compared, a trend similar to that of the overall polymerase processivity (Fig. 1) was observed. The N265D mutant RT displayed a mild effect on the RNA template (2-fold increase in koff,T-P compared to the wild type), whereas it displayed a >30-fold increase in koff,T-P on the DNA template. The N255D and Dbl mutant RTs had a significantly lower affinity (higher koff,T-P, lower t1/2) to both RNA and DNA templates than did wild-type RT (Table 1). It is important to note that in experiments measuring the dissociation of RT from the DNA template, even the earliest time point selected (10 s) showed little bound RT (≤1%) for N255D, N265D, and Dbl RTs, suggesting rapid dissociation. In comparison, >80% of wild-type RT was still bound at this time point. These results indicate, first of all, that the N265D mutation had a dissimilar influence on RT's affinity to template-primers comprised of RNA or DNA templates and, second, that the N255D mutation led to a much lower affinity of RT for both RNA and DNA templates. Taken together, these results indicate that single mutations can dramatically affect RT association with the template-primer at the level of initial binding or during enzyme translocation events. Consequently, one mutation affects processive DNA synthesis severely on both RNA and DNA templates (N255D). More interestingly, a single mutation (N265D) appears to selectively affect stable association with and processive polymerization on DNA but not on RNA templates.
FIG. 2.
Dissociation of wild-type (WT) and mutant RT complexes with template-primers. (A) Dissociation kinetics on an RNA template. (B) Dissociation kinetics on a DNA template. Equal amounts (130 nM) of RT and template-primer were preincubated for 5 min at 37°C. At time zero, preformed complexes were combined with a competing trap, and the dissociation of RT from the template-primer was monitored by measuring the incorporation of radiolabeled dNTP as described in Materials and Methods. The results presented are the means ± the standard errors of the mean from at least three separate experiments. The curves represent nonlinear least-squares fits of the data to an equation for one-phase exponential decay.
TABLE 1.
Dissociation rate constants of wild-type and mutant RTs on RNA and DNA template-primersa
| Enzyme type | RDDP activityb
|
DDDP activityc
|
||
|---|---|---|---|---|
| koff,T-P (s−1) | Half-life (s) | koff,T-P (s−1) | Half-life (s) | |
| WT | 0.0075 ± 0.001 | 92 | 0.0390 ± 0.002 | 18 |
| N255D | 0.1406 ± 0.008 | 4.9 | 1.179 ± 0.01 | ≤0.6 |
| N265D | 0.0161 ± 0.001 | 43 | 1.179 ± 0.01 | ≤0.6 |
| Dbl | 0.2347 ± 0.03 | 2.9 | 1.179 ± 0.01 | ≤0.6 |
Kinetic constants were determined as described in Materials and Methods. The koff,T-P values were obtained from at least three independent measurements.
Reactions were carried out by using DNA primer VP200 annealed to 16S rRNA template.
Reactions were carried out by using M13 DNA primer (−47) annealed to ss-DNA template.
Molecular modeling of the RT-template-primer complex.
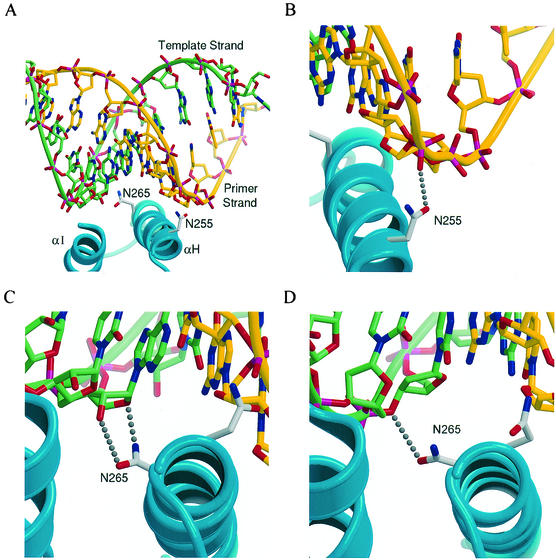
X-ray crystallographic analysis of HIV-1 RT in complex with RNA or DNA templates indicates that the αH helix of the thumb subdomain is positioned in the minor groove of the template-primer duplex (11, 21, 40). The N255 and N265 residues are on the side of the helix that faces the minor groove of the template-primer duplex (Fig. 3A). The side chain of N255 is close to the primer strand, which is DNA in all three structures (1HYS structure [40] includes RNA-DNA duplex, whereas in the 2HMI [11] and 1RTD [21] structures HIV-1 RT is complexed with duplex DNA). In the 1HYS structure, N255 forms a hydrogen bond with a phosphate group of the primer strand. In the 2HMI structure, N255 is pointed away from the backbone, although a simple side chain rotation yields an H bond to a phosphate analogous to that in 1HYS, with no resulting steric conflicts. In the 1RTD structure, the N255 side chain is pointed toward a phosphate but does not interact strongly (4.2 Å). The side chain of N265, in the 1HYS structure, is pointed toward the RNA template strand, forming hydrogen bonds to the O2′ and O4′ of the ribose. In the 2HMI and 1RTD structures the N265 side chain points toward the DNA template strand, forming hydrogen bonds to the O3′ of the deoxyribose.
FIG. 3.
Interactions formed between the αH helix of the thumb subdomain with the minor groove of the template-primer. (A) Initial structure for the simulation of the N255D and N265D mutations with an RNA template. Helices H and I are shown as ribbons, with the side chains of N255 and N265 also indicated. The template and primer strands are also shown as ribbons in addition to a ball-and-stick representation. The backbone ribbon and carbons of the template strand are shown in light green, while those of the primer strand are shown in yellow. (B to D) Views of the RT-substrate interactions in the crystal structures of the wild-type RT with an RNA (40) and DNA (10) template, showing the N255 side chain hydrogen bonded to a phosphate on the DNA primer strand (B), the N265 side chain hydrogen bonded to O4′ and O2′ of a template ribose on the RNA template (C), and Asn-265 hydrogen bonded to O3′ of a template deoxyribose (D). Figures were generated by using MOLSCRIPT (29) and Raster3D (32).
From the above structural evidence, we hypothesized that N265 makes functionally important contacts with the RNA template and that the N265D mutation disrupts hydrogen bonding on DNA template but not on RNA template, since an Asp being isosteric with Asn could still interact strongly with the O2′ of the ribose as a hydrogen bond acceptor. Although the structural evidence for the role of N255 is less obvious, the N255D mutation would prevent hydrogen bonding to the phosphate group on either RNA or DNA primers and would cause the local electrostatic potential to be more negative, i.e., less favorable for DNA binding. It could also cause local distortions of the RT structure due to close proximity of N255 to residues D256, K259, and Q258.
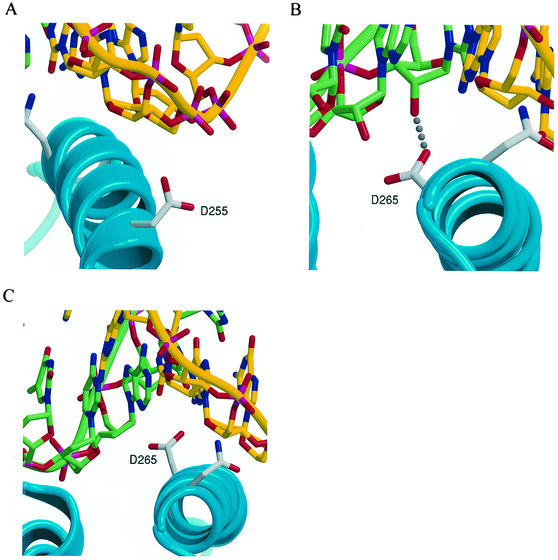
To examine these hypotheses, we performed molecular dynamics modeling of HIV-1 RT in complex with both RNA-DNA and DNA-DNA template-primers. Both N255D and N265D mutants were simulated in complex with each template-primer, giving four simulations in all. Interestingly, although the side chain of Asn-255 in the 2HMI structure is pointed away from the primer strand, this side chain quickly reoriented to the previously mentioned rotamer during the initial model building process, forming a hydrogen bond to the phosphate that is similar to that seen in the 1HYS structure. The subsequent N255D simulations with the DNA-DNA (as well as with RNA-DNA) template-primer resulted in D255 pointing away from the phosphate backbone (Fig. 4A). Since, as noted above, the orientation of N255 varies among the available crystal structures, it is difficult to ascribe a functional significance to the loss of this hydrogen bond. However, superposing the final structures of the four simulations upon their respective starting crystal structures, the N terminus of the αH helix (including residues 255 to 258) was found to be significantly displaced from the primer-template in both N255D simulations. No such displacement occurred in the two N265D simulations. The N-terminal helix displacement, as well as the residue 255 side chain reorientation, is likely due to electrostatic repulsion between the negatively charged aspartate and the phosphate backbone.
FIG. 4.
Molecular modeling of N255D and N265D mutations. (A) A close-up view showing the D255 side chain having lost its contact with the DNA primer backbone due to loss of hydrogen bonding and electrostatic repulsion. (B) View showing D265 residue that loses the hydrogen bond to O4′ and O2′ of one ribose while gaining a hydrogen bond to O2′ of a neighboring template ribose. (C) View showing the D265 residue, which loses both H bonds and is repelled from the template DNA strand. The results of the N255D simulation with a DNA template were essentially the same as for the RNA template and are not shown.
Since all of our polymerase assays (activity, processivity, and dissociation rates) used a DNA primer annealed to either RNA or DNA templates, the absence of hydrogen bonding and disruption of minor groove contact near the N terminus of helix αH is consistent with the loss of activity for N255D mutant on both RNA-DNA and DNA-DNA template-primers. The model suggests that the template nonspecific loss of RT-template-primer association of the N255D variant is due to the disruption of its contact with the primer strand (Fig. 4).
The final structure of the N265D-RNA-DNA simulation showed that, although the side chain of D265 had lost the hydrogen bonds to O4′ and O2′ of one ribose, it had gained a single, but stronger hydrogen bond to O2′ of a neighboring ribose (Fig. 4B). In the superposition of this structure upon 1HYS, the backbone and side chain of D265 (as well as the rest of helix αH) remain close to those of N265 in the crystal structure. This suggests that the D265 residue in the mutant RT can bind RNA-DNA template-primers with near-wild-type affinity, which would account for its ability to processively polymerize on RNA templates. The hydrogen of the O2′ on the ribose carries a positive partial charge that complements the negative charge of the D265 resulting in a lack of electrostatic repulsion on RNA templates. In contrast, the final structure of the N265D-DNA-DNA simulation showed that the side chain of D265 no longer formed any hydrogen bonds to the template strand (Fig. 4C). In this case, the negative charge on the D265 is not compensated for, leading to electrostatic repulsion between the side chain and the deoxyribose backbone. Unlike the N255D mutation, the disruption appears to be quite local, e.g., interactions involving W266 are undisturbed. The electrostatic repulsion due to D265 is likely less than that due to D255, given the smaller partial charges on the sugar ring compared to the phosphate group. However, the hydrogen bond involving N255 and the ribose or deoxyribose (or N265 and the ribose) is buried and likely to be functionally significant. Thus, the modeling predicts that the D265 mutation leads to loss of favorable interactions with a DNA template, thus explaining the loss of processive polymerization on DNA templates.
DISCUSSION
The knowledge that the MGBT plays a key role in RT's affinity to template-primer (3, 4) and that it serves as a railing on which the template-primer duplex tracks along during DNA polymerization by HIV-1 RT (5, 16, 30) has led to a better understanding of the dynamics of RT-template-primer interactions. However, structural determinants of RT responsible for its ability to use both RNA and DNA templates have thus far not been addressed (34). Here, we have presented evidence that the residue N265 plays an important role in HIV-1 RT's ability to use both RNA and DNA as templates for DNA synthesis. In addition, our results show that residues 265 and 255 in the αH of the thumb subdomain in HIV-1 RT play an important role in processive DNA synthesis since N→D substitutions at each of these residues resulted in dramatic reductions in polymerase processivity.
It is important to note that the defect of N255D and Dbl mutants on both DNA and RNA and that of N265D on DNA is unusual. In reactions where rebinding of RT is permitted (distributive synthesis), the activity of N255D and N265D RT mutants is similar to that of wild-type RT. However, in single cycle reactions, they display a dramatic loss of processive DNA synthesis on specific templates, stopping abruptly after synthesizing 20 to 40 nt. Thus, these residues may be critical for the recognition of template strand or part of a set of dynamic contacts transiently formed during successive cycles of enzyme translocation. We note that, although many mutations in HIV-1 RT have been described to affect processivity, very few show this severe phenotype.
It is known that residues in alpha helix that form the MGBT aid in the translocation of RT. Thus, these residues interact with the minor groove of the template-primer duplex as a guide during the translocation. Since the residue D265 is adjacent to one of these residues (W266), it is possible that D265 may be sensitive to changes in the groove geometry. It is known that runs of nucleotides in DNA or RNA can influence the minor groove by either compressing or widening the groove. However, inspection of the sequence around the points where the N255D or Dbl mutant terminate on RNA and that around the points where all three mutants terminate on DNA did not reveal such runs of nucleotides. Our attempt to find sequences with potential to form secondary structures was also unsuccessful. Thus, the termination of N265D on DNA but not on RNA appears to be independent of template sequence.
There have been other reports of HIV-1 RT mutations differentially affecting template-primer utilization. The mutation Q151A led to a 15- to 100-fold reduction in kcat on RNA templates, while causing a mere fivefold reduction on DNA templates (41). This effect was found to be due to template-specific differences in an event subsequent to dNTP binding and not due to a defect in a primary event such as initial recognition and binding of templates. Furthermore, the Q151A mutant displayed severely reduced polymerase activity even in the absence of an enzyme trap, which was further reduced on RNA templates in that the polymerization stopped after the addition of a single dNTP. In contrast, both of the mutants described here appear to be capable of forming full-length DNA similar to the wild type when the rebinding of RT is allowed (nonprocessive conditions). The N265D mutant is novel in that it selectively affects processivity and template-primer affinity on a DNA but not RNA template. Furthermore, most mutations in HIV-1 RT reported to reduce processivity reduce the length of the longest product by 10 to 50%, reduce the amount of the longest product, or increase the pausing at various intermediate positions. Both of the mutations reported here are unique in that they are severely defective for processive DNA synthesis, whereas their distributive synthesis appears largely intact (Fig. 1B). Attempts to examine the ability of such mutant RTs to contribute to viral replication by creating molecular clones of HIV with each of these mutations led to replication-defective viruses (15). These results indicate the essential role of processive polymerization for viral replication.
The biochemical experiments (processivity and template-primer affinity) and molecular modeling reported here highlight the importance of hydrogen bonding and charge repulsion in RT-template-primer interactions. It appears that the residue N265 plays an important part in HIV RT stably associating with both RNA and DNA template strands during processive DNA synthesis. The retention of processive synthesis on RNA templates observed for N265D RT suggests that contacts between such anchor residues and the template strand are functionally essential.
The N265 residue can be substituted by at least one other residue (Asp) without affecting processive RNA-dependent DNA synthesis, but not processive DNA-dependent DNA synthesis. Both N255 and N265 are partially conserved among retroviral RTs: N255 is conserved among all lentiviruses and deltaretroviruses, whereas N265 is conserved among immunodeficiency viruses (31). We believe that residues forming hydrogen bonds with template-primer in a sequence-nonspecific manner may not need to be highly conserved since other residues may be able to play this role equally well. Furthermore, we recognize that the ability to use both RNA and DNA templates also requires the template-primer cleft to be flexible in order to accommodate both types of nucleic acid duplexes, requiring adjustments to the overall template-primer binding cleft. Residues that facilitate this versatile ability to adapt to overall changes in the conformation may or may not contact one or both of the templates and are yet to be identified.
Other key anchor sites important for differentiating between RNA and DNA templates could be located in other parts of template-primer cleft, such as the template and primer grip regions in the palm subdomain, the β3-β4 hairpin of fingers domain, and the MGBT of the thumb subdomain. The crystal structure of HIV-1 RT bound to the RNA template-DNA primer shows several amino acid residues forming hydrogen bonds with the 2′-OH of the RNA template strand. In addition to N265, these include E89, Q91, K154, A284, and K353 residues in the polymerase domain and R448 and Q475 residues in the RNase H domain (40). Although we have demonstrated the role of N265, a systematic analysis to delineate the contribution of the remaining residues to the dual recognition of RNA or DNA templates by HIV-1 RT is currently in progress. Such studies, along with crystal structures of RTs from additional retroviruses in complex with RNA and DNA, should aid us in understanding the mechanisms by which these multifunctional polymerases use both substrates.
Acknowledgments
We thank William Drosopoulos, Ganjam Kalpana, Scott Garforth, and Ian Willis for critically reading the manuscript.
This study was supported by a Public Health Service grant to V.R.P. (RO1 AI30861). T.S.F. acknowledges support from an institutional predoctoral training grant (NIH T32-GM07491).
REFERENCES
- 1.Arion, D., G. Borokov, Z. Gu, M. A. Wainberg, and M. A. Parniak. 1996. The K65R mutation confers increased DNA polymerase processivity to HIV-1 reverse transcriptase. J. Biol. Chem. 271:19860-19864. [DOI] [PubMed] [Google Scholar]
- 2.Arts, E. J., X. Li, Z. Gu, L. Kleiman, M. A. Parniak, and M. A. Wainberg. 1994. Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem. 269:14672-14680. [PubMed] [Google Scholar]
- 3.Beard, W. A., K. Bebenek, T. A. Darden, L. Li, R. Prasad, T. A. Kunkel, and S. H. Wilson. 1998. Vertical-scanning mutagenesis of a critical tryptophan in the minor groove binding track of HIV-1 reverse transcriptase. Molecular nature of polymerase-nucleic acid interactions. J. Biol. Chem. 273:30435-30442. [DOI] [PubMed]
- 4.Beard, W. A., S. J. Stahl, H. R. Kim, K. Bebenek, A. Kumar, M. P. Strub, S. P. Becerra, T. A. Kunkel, and S. H. Wilson. 1994. Structure-function studies of human immunodeficiency virus type 1 reverse transcriptase: alanine scanning mutagenesis of an alpha-helix in the thumb subdomain. J. Biol. Chem. 269:28091-28097. [PubMed] [Google Scholar]
- 5.Bebenek, K., J. Abbotts, S. H. Wilson, and T. A. Kunkel. 1993. Error-prone polymerization by HIV-1 reverse transcriptase. J. Biol. Chem. 268:10324-10334. [PubMed] [Google Scholar]
- 6.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2000. Analysis of mutations at postions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 97:3056-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, P. L., C. Tantillo, A. Jacobo-Molina, R. G. Nanni, J. Ding, E. Arnold, and S. H. Hughes. 1994. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc. Natl. Acad. Sci. USA 91:4882-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheatham, T. E., III, P. Cieplak, and P. A. Kollman. 1999. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 16:845-862. [DOI] [PubMed] [Google Scholar]
- 9.DeStefano, J. J., R. A. Bambara, and P. J. Fay. 1993. Parameters that influence the binding of human immunodeficiency virus reverse transcriptase to nucleic acid structures. Biochemistry 32:6908-6915. [DOI] [PubMed] [Google Scholar]
- 10.Ding, J., K. Das, Y. Hsiou, S. G. Sarafianos, A. D. Clark, A. Jacobo-Molina, C. Tantillo, S. H. Hughes, and E. Arnold. 1998. Structure and functional implication of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 284:1095-1111. [DOI] [PubMed] [Google Scholar]
- 11.Ding, J., S. H. Hughes, and E. Arnold. 1997. Protein-nucleic acid interactions and DNA conformation in a complex of human immunodeficiency virus type 1 reverse transcriptase with a double-stranded DNA template-primer. Biopolymers 44:125-138. [DOI] [PubMed] [Google Scholar]
- 12.Drosopoulos, W. C., and V. R. Prasad. 1998. The increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall rates measured in vitro. J. Virol. 72:4224-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drosopoulos, W. C., and V. R. Prasad. 1996. Increased polymerase fidelity of E89G, a nucleoside analog-resistant variant of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase. J. Virol. 70:4834-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Essmann, U. A. 1995. A smooth particle mesh method. J. Chem. Phys. 103:8577-8593. [Google Scholar]
- 15.Fisher, T. S., P. Joshi, and V. R. Prasad. 2002. Mutations that confer resistance to template-analog inhibitors of human immunodeficiency virus (HIV) type 1 reverse transcriptase lead to severe defects in HIV replication. J. Virol. 76:4068-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forgacs, E., G. Latham, W. A. Beard, R. Prasad, K. Bebenek, T. A. Kunkel, S. H. Wilson, and R. S. Lloyd. 1997. Probing structure/function relationships of HIV-1 reverse transcriptase with styrene oxide N2-guanine adducts. J. Biol. Chem. 272:8525-8530. [DOI] [PubMed] [Google Scholar]
- 17.Gao, G., and S. P. Goff. 1998. Replication defect of Moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides. J. Virol. 72:5905-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, G., M. Orlova, M. M. Georgiadis, W. A. Hendrickson, and S. P. Goff. 1997. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl. Acad. Sci. USA 94:407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh, M., P. S. Jacques, D. W. Rodgers, M. Ottman, J.-L. Darlix, and S. F. J. Le Grice. 1996. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 35:8553-8562. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, M., J. Williams, M. D. Powell, J. G. Levin, and S. F. Le Grice. 1997. Mutating a conserved motif of the HIV-1 reverse transcriptase palm subdomain alters primer utilization. Biochemistry 36:5758-5768. [DOI] [PubMed] [Google Scholar]
- 21.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 22.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, et al. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacques, P. S., B. M. Wohrl, M. Ottman, J.-L. Darlix, and S. F. J. Le Grice. 1994. Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem. 269:26472-26478. [PubMed] [Google Scholar]
- 24.Jonckheere, H., E. De Clercq, and J. Anne. 2000. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino acid sequence at residues Leu74, Glu89, Tyr115, Tyr183, and Met184. Eur. J. Biochem. 267:2658-2665. [DOI] [PubMed] [Google Scholar]
- 25.Kew, Y., L. Olsen, A. Japour, and V. R. Prasad. 1998. Insertions into the β3-β4 hairpin loop of HIV-1 reverse transcriptase reveal a role for fingers subdomain in processive polymerization. J. Biol. Chem. 273:7529-7537. [DOI] [PubMed] [Google Scholar]
- 26.Kew, Y., Q. Song, and V. Prasad. 1994. Subunit selective mutagenesis of Glu89 residue in human immunodeficiency virus reverse transcriptase. J. Biol. Chem. 269:15331-15336. [PubMed] [Google Scholar]
- 27.Kim, B., T. R. Hathaway, and L. A. Loeb. 1998. Fidelity of mutant HIV-1 reverse transcriptases: interaction with the single-stranded template influences the accuracy of DNA synthesis. Biochemistry 37:5831-5839. [DOI] [PubMed] [Google Scholar]
- 28.Klarmann, G. J., R. A. Smith, R. F. Schinazi, T. W. North, and B. D. Preston. 2000. Site-specific incorporation of nucleoside analogs by HIV-1 reverse transcriptase and the template grip mutant P157S. Template interactions influence substrate recognition at the polymerase active site. J. Biol. Chem. 275:359-366. [DOI] [PubMed] [Google Scholar]
- 29.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24:946-950. [Google Scholar]
- 30.Latham, G. J., E. Forgacs, W. A. Beard, R. Prasad, K. Bebenek, T. A. Kunkel, S. H. Wilson, and R. S. Lloyd. 2000. Vertical-scanning mutagenesis of a critical tryptophan in the “minor groove binding track” of HIV-1 reverse transcriptase: major groove DNA adducts identify specific protein interactions in the minor groove. J. Biol. Chem. 275:15025-15033. [DOI] [PubMed] [Google Scholar]
- 31.Li, M. D., D. L. Bronson, T. D. Lemke, and A. J. Faras. 1995. Phylogenetic analyses of 55 retroelements on the basis of the nucleotide and product amino acid sequences of the pol gene. Mol. Biol. Evol. 12:657-670. [DOI] [PubMed] [Google Scholar]
- 32.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics, p. 505-524. In C. W. Carter, Jr. (ed.), Macromolecular crystallography, part B, vol. 277. Academic Press, Inc., New York, N.Y. [DOI] [PubMed]
- 33.Oude Essink, B. B., N. K. T. Back, and B. Berkhout. 1997. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 25:3212-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel, P. H., and L. A. Loeb. 2001. Getting a grip on how DNA polymerases function. Nat. Struct. Biol. 8:656-659. [DOI] [PubMed] [Google Scholar]
- 35.Pearlman, D. A. 1995. AMBER, a package of computer programs for applying molecular mechanics, normal mode of analysis, molecular dynamics, and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 91:1-41. [Google Scholar]
- 36.Powell, M. D., M. Ghosh, P. S. Jacques, K. J. Howard, S. F. LeGrice, and J. G. Levin. 1997. Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J. Biol. Chem. 272:13262-13269. [DOI] [PubMed] [Google Scholar]
- 37.Prasad, V. R., I. Lowy, T. De Los Santos, L. Chiang, and S. P. Goff. 1991. Isolation and characterization of a dideoxyguanosine triphosphate-resistant HIV-1 reverse transcriptase expressed in bacteria. Proc. Natl. Acad. Sci. USA 88:11363-11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinek, T., M. Bakhanashvili, R. Taube, O. Avidan, and A. Hizi. 1997. The fidelity of 3′ misinsertion and mispair extension during DNA synthesis exhibited by two drug-resistant mutants of the reverse transcriptase of human immunodeficiency virus type 1 with Leu74Val and Glu89Gly. Eur. J. Biochem. 247:238-247. [DOI] [PubMed] [Google Scholar]
- 39.Sarafianos, S. G., K. Das, J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Chem. Biol. 6:R137-R146. [DOI] [PubMed] [Google Scholar]
- 40.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarafianos, S. G., V. N. Pandey, N. Kaushik, and M. J. Modak. 1995. Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Biochemistry 34:7207-7216. [DOI] [PubMed] [Google Scholar]
- 42.Schneider, D. J., J. Feigon, Z. Hostomsky, and L. Gold. 1995. High-affinity ssDNA inhibitors of the reverse transcriptase of type 1 human immunodeficiency virus. Biochemistry 34:9599-9610. [DOI] [PubMed] [Google Scholar]
- 43.Telesnitsky, A., and S. P. Goff. 1998. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 44.Wainberg, M. A., W. C. Drosopoulos, H. Salomon, M. Hsu, G. Borkow, M. Parniak, Z. Gu, Q. Song, J. Manne, S. Islam, G. Castriota, and V. R. Prasad. 1996. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 271:1282-1285. (Comments, 275:228-231, 1997.) [DOI] [PubMed]
- 45.Wisniewski, M., C. Palaniappan, Z. Fu, S. F. Le Grice, P. Fay, and R. A. Bambara. 1999. Mutations in the primer grip region of HIV reverse transcriptase can increase replication fidelity. J. Biol. Chem. 274:28175-28184. [DOI] [PubMed] [Google Scholar]
- 46.Wohrl, B. M., R. Krebs, S. H. Thrall, S. F. LeGrice, A. J. Scheidig, and R. S. Goody. 1997. Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66: implications for DNA synthesis and dimerization. J. Biol. Chem. 272:17581-17587. [DOI] [PubMed] [Google Scholar]