Abstract
Alphavirus-based expression vectors commonly use a duplicated 26S promoter to drive expression of a foreign gene. Here we describe an expression strategy in which the foreign sequences are linked to the gene encoding the 2A protease of foot-and-mouth disease virus and then inserted in frame between the capsid and E3 genes of Sindbis virus. During replication, the 2A fusion protein is synthesized as a component of the viral structural polyprotein that is then released by intramolecular cleavages mediated by the capsid and 2A proteases. Recombinant Sindbis viruses that expressed fusion proteins composed of 2A linked to the green fluorescent protein (GFP) and to the VP7 protein of bluetongue virus were constructed. Viruses engineered to express GFP and VP7 from a duplicate 26S promoter were also constructed. All four viruses expressed the transgene and grew to similar titers in cultured cells. However, the GFP/2A- and VP7/2A-expressing viruses displayed greater expression stability and were less attenuated in newborn mice than the cognate double-subgenomic promoter-based viruses. By combining the two expression strategies, we constructed bivalent viruses that incorporated and expressed both transgenes. The bivalent viruses grew to lower titers in cultured cells and were essentially avirulent in newborn mice. Groups of mice were vaccinated with each VP7- and VP7/2A-expressing virus, and antibody responses to native VP7 were measured in an indirect enzyme-linked immunosorbent assay. Despite their genetic and phenotypic differences, all viruses induced similarly high titers of VP7-specific antibodies. These results demonstrate that 2A fusion protein-expressing alphaviruses may be particularly well suited for applications that require enduring expression of a single protein or coexpression of two alternative proteins.
Alphaviruses are enveloped, single-stranded, positive-sense RNA viruses that belong to the Togaviridae virus family (Alphavirus genus). Upon entry into the host cell, the viral genome is translated into the four nonstructural proteins (nsp1 to -4) that comprise the viral transcriptase-replicase complex. The viral replicase uses the genomic RNA as a template to synthesize a complementary, full-length, negative-sense RNA. The negative-sense RNA in turn serves as a template for the synthesis of two different positive-sense RNAs. Positive-strand synthesis initiating at the 3′ end of the negative-strand RNA results in the production of full-length genomic RNA. Positive-strand synthesis can also initiate at an internal promoter sequence (26S or subgenomic promoter) to produce a subgenomic mRNA that is colinear with approximately the 3′-terminal one-third of the genomic RNA. Subgenomic mRNAs are translated into the viral structural proteins (40).
Infectious cDNA clones have been constructed for many alphaviruses, including Sindbis virus strain AR339 (15, 21, 31), Venezuelan equine encephalitis virus (6), Ross River virus (17), Semliki Forest virus (19), and South African arbovirus 86 (37). An infectious cDNA clone has also been constructed for rubella virus, the sole member of the Rubivirus genus (45). Alphavirus-based cDNA clones have been modified into expression vectors that are useful for expressing foreign genes in cultured cells and in vivo (29, 36, 42). Most alphavirus-based expression vectors have been constructed according to one of two fundamental designs (3, 11, 35). In the double-subgenomic promoter (DSP) design, the transgene is placed under the transcriptional control of a duplicate 26S promoter inserted within the 3′ nontranslated region of the viral genome or within the short nontranslated region located just upstream of the native 26S promoter. Since DSP-based vectors retain all viral genes, they are capable of multiple rounds of infection and sustained transgene expression. Expression vectors can also be constructed as replicons. In the replicon design, the foreign gene is substituted for the viral structural genes and is expressed under the control of the native 26S promoter. Since replicons lack the structural genes, genome packaging and budding of replicon particles can occur only if the structural proteins are provided in trans, and replicon particles produced in this way are limited to only a single round of infection.
Traditional DSP- and replicon-based vectors are useful for many cell biology and vaccine applications; however, they do have several limitations. First, DSP-based vectors often display poor expression stability, and they can lose transgene expression after relatively few replication cycles (4, 25, 26). The inability of a recombinant vector to sustain transgene expression through repeated replication cycles would be expected to limit its effectiveness as a vaccine vector, because the magnitude of the host immune response to an antigen is likely to be influenced by the duration of antigen exposure (4). The instability of some DSP-based vectors has been attributed to recombination events involving the duplicated 26S promoter sequences (26). Pugachev and coworkers improved the expression stability of a rubella virus-based vector by replacing the duplicate 26S promoter with a picornavirus internal ribosome entry site element (26). Traditional DSP-based vectors are also limited to the expression of a single foreign gene. The primary limitation of replicons is their inability to sustain transgene expression for long periods of time due to the single infection cycle.
Here we describe a strategy for constructing monovalent, live alphavirus expression vectors that display greater expression stability than DSP-based vectors and which can be modified into DSP vectors that coexpress two foreign genes. The design of these vectors is based on a modification of the normal alphavirus structural protein expression strategy. The subgenomic mRNA of alphaviruses is translated into a polyprotein that contains the structural proteins in the order of capsid-PE2-6K-E1 (30). Capsid protein cotranslationally cleaves itself from the polyprotein, and the remainder of the polyprotein is resolved in the rough endoplasmic reticulum by signal peptidase (40). We constructed Sindbis viruses that encode fusion proteins consisting of the green fluorescent protein (GFP) or the VP7 protein of bluetongue virus (BTV) linked to the 2A protease from foot-and-mouth disease virus (FMDV). The 2A protease of FMDV is only 16 amino acids in length (32), and it mediates cis cleavage at its own C terminus (between Gly and Pro) either by using a conventional protease activity (33, 34) or by influencing the translating ribosome to release the 2A-containing polypeptide from the translational complex by promoting hydrolysis of a peptidyl (2A)-tRNAGly ester linkage (8). The recombinant viruses contain the GFP/2A and VP7/2A gene sequences inserted in frame between the capsid and PE2 genes, and they produce structural polyproteins that contain GFP/2A or VP7/2A positioned between capsid and PE2. The 2A fusion protein is then released from the polyprotein by N-terminal and C-terminal cis cleavages performed by capsid and 2A, respectively. DSP-based vectors engineered to express native forms of GFP and VP7 from a duplicated 26S promoter placed within the 3′ nontranslated region also were constructed. By combining the two expression strategies, we constructed bivalent viruses that expressed one protein as a 2A fusion protein and expressed the second protein in native form from the duplicate 26S promoter. Recombinant viruses were compared with respect to levels of transgene expression in cultured cells, expression stability, virulence in newborn mice, and induction of humoral immune responses in adult mice.
MATERIALS AND METHODS
Construction of recombinant viruses.
The parental virus used in these studies is designated TR339 (15, 21). In order to simplify virus constructions, the DSP-based plasmids designated pTR339-26S/GFP and pTR339-26S/VP7 were constructed in the genetic background of a variant of pTR339 that contains two amino acid substitutions (Gln for Arg at nsp3 residue 528 and Val for Ala at E1 residue 72). Neither of these mutations is associated with a phenotype in cell culture or in vivo (16). The construction of pTR339-26S/GFP has been described elsewhere (K. D. Ryman, W. B. Klimstra, and R. E. Johnston, submitted for publication). This plasmid contains the GFP gene and a duplicate 26S promoter placed within the 3′ nontranslated region. pTR339-26S/VP7 was constructed in a similar manner and contains the VP7 gene from BTV in place of GFP. Briefly, the sequences representing gene segment 7 from BTV serotype 10 (BTV-10) were derived from a plasmid designated pBTV10S7. The VP7 gene contained a single XhoI restriction site that was eliminated by using a PCR-based site-directed mutagenesis procedure. Mutagenesis created a noncoding change (T to A) at VP7 nucleotide (nt) 328. The modified VP7 gene was then amplified by PCR with oligonucleotides that incorporated XbaI (5′) and BamHI (3′) restriction sites. The XbaI-containing oligonucleotide also generated a consensus Kozak start site. The amplicon was digested with XbaI and BamHI and ligated into the pH 3′2J1 shuttle vector (12), from which a corresponding XbaI/BamHI fragment had been removed. This transfer placed the VP7 gene downstream of a 26S promoter sequence. The VP7 and 26S promoter sequences were then substituted for the GFP and 26S promoter sequences in pTR339-26S/GFP by transfer of an ApaI/XhoI fragment. Construction of pTR339-GFP/2A and pTR339-VP7/2A was accomplished with a two-step PCR-based cloning strategy essentially as described previously (41). In the first step, sequential PCRs were used to join the coding sequences of capsid to GFP or VP7. The DNA templates used in these PCRs were TR339 (capsid sequence), pTR339-26S/GFP (GFP sequence), and pTR339-26S/VP7 (VP7 sequence). The oligonucleotides used in these reactions were designed so that the final amplicon contained the first three codons of PE2 placed between the capsid and GFP/VP7 sequences and adjacent BglII and StuI restriction sites at the 3′ terminus. The amplified DNA was then digested with AatII (within the capsid sequence) and StuI and ligated into pTR339 from which a corresponding AatII/StuI fragment had been removed. The resulting constructs were designated pTR339-GFP and pTR339-VP7. In the second step, sequential PCRs were used to join the coding sequences of 2A (plus 1D and 2B flanking sequences) and PE2. Sequences representing the FMDV 1D/2A/2B region were contributed entirely by the oligonucleotides used in amplification reactions. The oligonucleotides used in these reactions were designed so that the final amplicon contained a BglII restriction site at its 5′ terminus. The amplified DNA was then digested with BglII and StuI (within PE2) and inserted into pTR339-GFP and pTR339-VP7. The final constructs were designated pTR339-GFP/2A and pTR339-VP7/2A, respectively. Finally, pTR339-GFP/2A/VP7 was produced by transferring a BssII/XhoI fragment from pTR339-26S/VP7 into pTR339-GFP/2A. Similarly, pTR339-VP7/2A/GFP was produced by transferring a BssII/XhoI fragment from TR339-26S/GFP into pTR339-VP7/2A. Consequently, pTR339-GFP/2A/VP7 and pTR339-VP7/2A/GFP contain the phenotypically silent Val-for-Ala substitution at E1 residue 72. All sequences derived from PCRs were confirmed by sequence analysis.
Infectious virus was derived from each cDNA clone as described previously (13). Briefly, cDNA clones were linearized by digestion with XhoI, and runoff transcripts were produced by using SP6 RNA polymerase. RNA transcripts were then electroporated into BHK-21 cells, and virus-containing growth medium was collected at 24 h postelectroporation and frozen at −70°C. BHK-21 cells were obtained from the American Type Culture Collection and were maintained in alpha minimum essential medium (MEM) supplemented with 10% donor calf serum, 10% tryptose phosphate broth, and antibiotics (MEM-complete).
Virus growth in BHK-21 cells.
The kinetics of virus growth was determined for each virus in BHK-21 cells. Cells were electroporated with in vitro viral transcripts as described above. Electroporations were performed in duplicate for each virus, and samples of growth medium were harvested at 6-h intervals postelectroporation. Infectious virus was quantified by standard plaque assay on monolayers of BHK-21 cells. Virus titers were reported as the averages of values obtained for the duplicate samples.
Immunoprecipitation of radiolabeled proteins from cell lysates.
Viral proteins were metabolically radiolabeled with [35S]methionine during growth in BHK-21 cells essentially as described previously (13). BHK-21 cells were grown in 25-cm2 flasks and infected with virus at a multiplicity of infection (MOI) of 5 PFU/cell. Virus was adsorbed to cells for 30 min and then overlaid with MEM-complete. At 6 h postinfection, cells were changed into methionine-free MEM supplemented with 5% fetal bovine serum and 5% tryptose phosphate broth. At 10 h postinfection, [35S]methionine was added to a final concentration of 50 μCi/ml. Cells were maintained in the presence of label for 3 h. Growth medium was removed, and cells were washed four times with ice-cold phosphate-buffered saline (PBS) and then lysed in 700 μl of lysis buffer (PBS containing 0.5% NP-40, 1.0 μM leupeptin, and 1.0 μM pepstatin). Lysates were clarified by microcentrifugation, mixed with protein A-agarose beads (Repligen), and preadsorbed overnight while rocking gently at 4°C. Lysates were analyzed directly by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or used in immunoprecipitation assays as follows. Lysate samples (300 μl) were mixed with 25 μl of protein A-agarose beads (50% suspension in PBS) and 5 μl of antibody in 1.5-ml microcentrifuge tubes. Antibody preparations included polyclonal rabbit antisera to GFP (Clontech), monoclonal antibody (MAb) 290, normal mouse serum, or normal rabbit serum. MAb 290 binds to a highly conserved epitope on VP7 (46). Samples were rocked for 8 h at 4°C, and then beads were washed four times with ice-cold PBS (containing 1.0 μM leupeptin and 1.0 μM pepstatin). Washed beads were mixed with SDS-PAGE sample buffer and boiled for 2 min. Samples were then resolved on SDS-polyacrylamide gels (10% acrylamide) as described previously (18).
The level of transgene expression by monovalent viruses was quantified by phosphorimaging analysis. Briefly, vector-expressed proteins were metabolically radiolabeled with [35S]methionine, and samples of infected-cell lysates were resolved by SDS-PAGE as described above. Dried gels were analyzed with a VersaDoc model 5000 imaging system (Bio-Rad). This experiment was repeated twice, and results were reported as an average.
Stability of transgene expression.
BHK-21 cells grown in 60-mm-diameter dishes were infected with selected stock viruses at an MOI of 0.1 PFU/cell. Virus was adsorbed to cells for 30 min, and unadsorbed virus was then removed by repeated washes with PBS. Cells were overlaid with MEM-complete and placed at 37°C. At 20 h postinfection, the growth medium was harvested and frozen at −70°C. Samples were quick thawed, and infectious virus was quantified by plaque assay on BHK-21 cells. These virus samples were then used to infect fresh BHK-21 cells as before. This process was repeated for a total of 10 serial passages. Each virus sample in the passage series was then used to generate radiolabeled infected-cell lysates as described above except that lysates were not preadsorbed with protein A-agarose beads. An equal volume (8 μl) of each lysate was then mixed with SDS-PAGE sample buffer, boiled for 2 min, and resolved in SDS-polyacrylamide gels (10% acrylamide).
Virulence studies in newborn CD-1 mice.
Neonatal (12 to 24 h of age) CD-1 mice were infected subcutaneously (s.c.) in the ventral thorax with 103 BHK-21 PFU of each virus. Viruses were diluted in PBS-1% donor calf serum. The mice were then observed twice daily for 21 days, and average survival times (AST) and percent mortality were calculated. AST values were used to analyze differences in virulence by using a conservative two-sample t test. Animal care and experimental procedures were performed in accordance with the Institutional Animal Care and Use Committee guidelines.
Purification of BTV virions.
Twelve 175-cm2 flasks of BHK-21 cells were infected with BTV-10 at an MOI of 1.0 PFU/cell for 42 h. Cells were harvested with a cell scraper, consolidated into six centrifuge tubes, and spun at 1,250 × g for 15 min. Supernatants were discarded, and each pellet was resuspended in 1.0 ml of 0.002 M Tris-HCl (pH 8.8)-0.5% Triton X-100 and homogenized in a glass tissue grinder (Kontes). The material was placed into a 1.5-ml microcentrifuge tube and spun at 15,000 rpm for 10 min. The liquid from the six samples was then pooled and placed on ice. The six pellets where then pooled together in 4 ml of 0.002 M Tris-HCl (pH 8.8)-0.5% Triton X-100, vortexed for 4 min, and pelleted as before. Supernatants were collected and pooled with that set aside earlier. The sample was extracted three times with 6.0 ml of 1,1,2-trichlorotrifluoroethane and then extracted three times with 6.0 ml of diethyl ether. The extracted sample was overlaid onto a 6.0 ml-discontinuous sucrose gradient (66 and 40% sucrose in 0.002 M Tris-HCl [pH 8.8]) and centrifuged at 26,000 rpm for 3 h at 4°C. The virus band that formed at the interface of the sucrose solutions was collected and dialyzed overnight against 0.2 M Tris-HCl (pH 8.0). The sample was then collected and digested with chymotrypsin (40 μg/ml) for 1 h at 37°C. The digested sample was overlaid onto a 3.0-ml 20% sucrose cushion and centrifuged at 26,000 rpm for 3 h at 4°C. The virus pellet was collected in 0.002 M Tris-HCl (pH 8.8) and used as the antigen in enzyme-linked immunosorbent assay (ELISA).
Serological responses in mice infected with VP7-expressing viruses.
Groups of 3-week-old female CD-1 mice (Charles River Laboratories) (four mice per group) were immunized with 104 PFU of TR339 and each VP7-expressing recombinant virus in both rear footpads 1 day after a tail vein prebleed. Four mice were mock infected with PBS. Viruses used for vaccination were partially purified from electroporation supernatants. Specifically, supernatants were clarified of cell debris by low-speed centrifugation and then pelleted through 20% sucrose cushions. Mice were boosted with 104 PFU of virus at 21 and 42 days after the initial immunization. Blood samples were then collected 14 days after the final boost. Serum was collected by using Microtainer serum separators (Becton Dickinson).
Antibody responses to VP7 were analyzed by testing each serum sample in an indirect ELISA. ELISA plates were coated with purified BTV-10 (200 ng/well) in carbonate buffer (pH 9.6) overnight at room temperature. Wells were washed twice with PBS then blocked for 1 h with 3% bovine serum albumin (BSA) in PBS (PBS-BSA). The wells were washed twice with PBS, and then 100 μl of each serum sample (diluted 1:20 in PBS-BSA) was added to duplicate top wells of a 96-well ELISA plate (Falcon). Serial twofold dilutions were then performed with a multichannel pipette. Serum samples were incubated for 1 h at room temperature, and then the plates were washed four times with PBS. A horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G secondary antibody (diluted 1:1,000 in PBS-BSA) was then added to each well, and plates were incubated for 1 h. The plates were then washed four times with PBS. One hundred microliters of substrate (o-phenylenediamine dihydrocholoride) was then added to each well, and the optical density at 450 nm (OD450) was measured 30 min later. Antibody titers were calculated as the inverse of the serum dilution yielding OD450 readings of ≥0.2 above background.
RESULTS
The proteolytic activity of 2A is influenced by residues immediately N terminal of 2A (1D region), and requires a proline residue immediately downstream of its cleavage site (33). Using a PCR-based cloning strategy and a cDNA clone of Sindbis virus strain AR339, we linked the gene encoding the GFP (714 nt) or VP7 (1,047 nt) upstream of a 2A gene construct consisting of the sequences encoding the three C-terminal residues of 1D, the 16 residues of 2A, and the N-terminal proline of 2B (60 nt total). These sequences were then inserted in frame between the sequences encoding capsid and PE2 (Fig. 1). The cloning procedure also created a unique BglII restriction site (encoding Arg-Ser) between GFP or VP7 and the FMDV sequences. The enzymatic activity of capsid protein is not strictly dependent on downstream sequences (11, 38); however, to enhance capsid-mediated cleavage, the three N-terminal residues of PE2 (Ser-Ala-Ala) were included between the capsid and the transgene sequences. The recombinant plasmids were designated pTR339-GFP/2A and pTR339-VP7/2A, and viruses derived from these plasmids were designated TR339-GFP/2A and TR339-VP7/2A, respectively (Fig. 2). Two DSP-based expression vectors that express the GFP or VP7 genes from a duplicated 26S promoter placed within the 3′ nontranslated region also were constructed. Viruses derived from these constructs were designated TR339-26S/GFP and TR339-26S/VP7, respectively (Fig. 2). Finally, two bivalent viruses in which the two expression strategies were combined were constructed. These viruses were designated TR339-GFP/2A/VP7, and TR339-VP7/2A/GFP (Fig. 2).
FIG. 1.
Expression of 2A fusion proteins as cleavable components of the structural polyprotein. The alphavirus subgenomic mRNA is translated into a polyprotein containing the viral structural proteins in the order of capsid-PE2-6K-E1. The recombinant viruses TR339-GFP/2A and TR339-VP7/2A express polyproteins containing GFP/2A or VP7/2A inserted between capsid and PE2. The three N-terminal residues of PE2 (S-A-A) were retained downstream of capsid to facilitate capsid-mediated cleavage of the polyproteins. The residues that comprise 2A and the flanking residues (derived from FMDV proteins 1D and 2B) that were included to enhance 2A activity are highlighted. Two additional residues (R-S) located N terminal to the FMDV residues were contributed by the codons comprising a BglII restriction site that was generated in the cloning process.
FIG. 2.
Genomic structure of TR339 and recombinant Sindbis viruses. Recombinant viruses expressing GFP/2A or VP7/2A fusion proteins as cleavable components of the viral structural polyprotein and/or native GFP or VP7 proteins from a duplicated 26S promoter placed within the 3′ nontranslated region were constructed as shown.
The rationale for using GFP and VP7 in this study was based on the size of their respective genes, which must be considered due to genome packaging constraints, and on the phenotypic properties of the expressed proteins. Specifically, GFP was chosen because of the ease with which GFP expression can be detected within cells. VP7 was chosen because it is highly immunogenic for mice and thus could be used as a model antigen for inducing humoral immune responses in mice infected with the recombinant vectors. In addition, we have developed well-characterized immunological reagents and reliable assays for detecting VP7 and VP7-specific antibodies, and thus the use of VP7 facilitated several aspects of vector characterization.
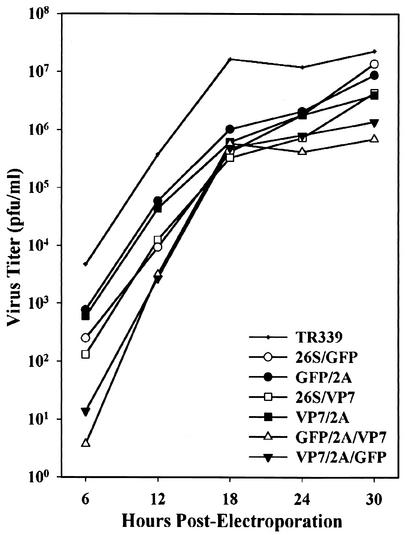
The growth characteristics of each virus were compared in BHK-21 cells (Fig. 3). The peak titer obtained for each monovalent recombinant virus was within 1 log10 unit of the peak titer of TR339. The peak titers of TR339-GFP/2A/VP7 and TR339-VP7/2A/GFP were approximately 33- and 17-fold lower, respectively, than that of TR339. The kinetics of virion production were similar for each virus (Fig. 3). Cytopathic effects (CPE), measured as cell lysis and visible plaque formation, developed most rapidly in cells infected with TR339-GFP/2A and TR339-VP7/2A. CPE developed at a lower rate in cells infected with TR339-26S/GFP and TR339-26S/VP7 and developed most slowly in cells infected with TR339-GFP/2A/VP7 and TR339-VP7/2A/GFP.
FIG. 3.
Growth kinetics of TR339 and recombinant viruses in BHK-21 cells. BHK-21 cells were electroporated with in vitro transcripts as described in Materials and Methods. Virus released into the growth medium was collected at 6-h intervals and quantified by plaque assay. Infections were performed in duplicate, and virus titer values from the duplicate samples were averaged.
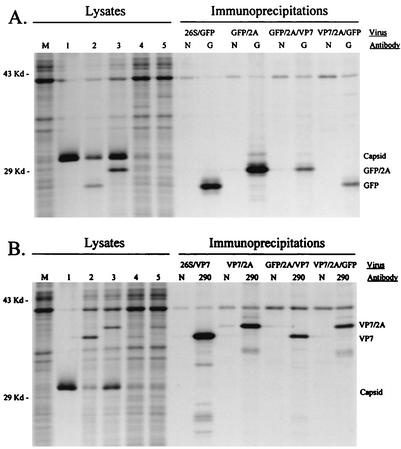
Transgene expression by the six recombinant viruses was then evaluated in cell culture. Cells infected with each GFP- or GFP/2A-expressing virus displayed cytoplasmic fluorescence when viewed under a fluorescence microscope (data not shown). Phenotypic comparisons between TR339-GFP/2A and TR339-26S/GFP, and between TR339-VP7/2A and TR339-26S/VP7, were used to evaluate potential differences between the two expression strategies. First, infected-cell lysates were analyzed by SDS-PAGE (Fig. 4). Polyproteins containing the GFP/2A and VP7/2A fusion proteins were resolved normally, indicating that the intramolecular cleavages mediated by the capsid and 2A proteases occurred as predicted. The relative mobilities of the GFP/2A and VP7/2A fusion proteins were decreased compared to those of GFP and VP7, respectively, due to the three additional residues of PE2 located at the fusion protein N terminus and the C-terminal addition of 21 residues contributed by the BglII site (2 residues) and FMDV 1D/2A sequences (19 residues). The identity of the expressed proteins was confirmed by immunoprecipitation of radiolabeled proteins from infected-cell lysates by using a rabbit polyclonal antiserum to GFP (Fig. 4A) and a MAb to VP7 (Fig. 4B). These experiments confirmed that each virus expressed the appropriate transgene(s) during growth in cell culture. Differences in the levels of transgene expression were observed between the monovalent viruses; however, these differences did not necessarily correlate with the mechanism used for transgene expression. Based on phosphorimaging analysis of cell lysate proteins resolved in SDS-polyacrylamide gels, the level of GFP/2A expression was approximately 2.0 ± 0.8 times higher than that of GFP, and the level of VP7 expression was approximately 2.0 ± 0.5 times higher than that of VP7/2A. Therefore, neither expression strategy appeared to be consistently superior to the other with respect to level of transgene expression.
FIG. 4.
Protein expression by recombinant Sindbis viruses. Subconfluent monolayers of BHK-21 cells were infected with virus at an MOI of 5 PFU/cell. Proteins were metabolically radiolabeled with [35S]methionine, and infected-cell lysates were collected and processed as described in Materials and Methods. Cell lysates were loaded directly onto gels (left sides of gels) or used in immunoprecipitation assays (right sides of gels). (A) Lysates from mock-infected cells (lane M) and cells infected with TR339 (lane 1), TR339-26S/GFP (lane 2), TR339-GFP/2A (lane 3), TR339-GFP/2A/VP7 (lane 4), and TR339-VP7/2A/GFP (lane 5). Immunoprecipitations were performed with infected-cell lysates and antibody preparations as shown at the top. (B) Lysates from mock-infected cells (lane M) and cells infected with TR339 (lane1), TR339-26S/VP7 (lane 2), TR339-VP7/2A (lane 3), TR339-GFP/2A/VP7 (lane 4), and TR339-VP7/2A/GFP (lane 5). Immunoprecipitations were performed with infected-cell lysates and antibody preparations as shown at the top. Lanes N, normal mouse serum; lanes G, rabbit polyclonal antiserum to GFP; lanes 290, VP7-specific MAb.
To compare the stability of transgene expression between the DSP-based viruses and those engineered to express the protein as a cleavable component of the structural polyprotein, each virus was serially passaged 10 times at a low MOI (0.1 PFU/cell) in BHK-21 cells, and the relative levels of transgene expression were compared between viruses collected after each passage (Fig. 5). TR339-26S/GFP and TR339-GFP/2A expressed detectable levels of GFP and GFP/2A, respectively, throughout the 10-passage series. No visible decrease in the levels of GFP/2A expression by TR339-GFP/2A was observed through the serial passage; however, the level of GFP expression by TR339-26S/GFP began to decline noticeably after four passages. Expression of VP7/2A by TR339-VP7/2A appeared to be stable through six serial passages. Expression of VP7/2A declined in subsequent passages but was still detectable after the 10th passage. In contrast, expression of VP7 by TR339-26S/VP7 decreased dramatically after only two passages and was not observed after the fourth passage.
FIG. 5.
Stability of transgene expression following serial, low-multiplicity virus passage in BHK-21 cells. Viruses were serially passaged 10 times at a low MOI (0.1 PFU/cell) in BHK-21 cells as described in Materials and Methods. The original stock virus (lanes P0) and virus obtained after each passage (lanes P1 to P10) were then used to infect BHK-21 cells. Infected cells were radiolabeled with [35S]methionine, and cell lysates were collected at 12 h postinfection. An equal volume of each cell lysate was then mixed with sample buffer and analyzed directly on SDS-polyacrylamide gels.
Prior to testing of the vaccine potential of each recombinant virus in adult CD-1 mice, the virulence of each virus was assessed in newborn mice. Groups of 30 newborn mice (12 to 24 h old) were inoculated s.c. with 103 PFU of each virus. Mice were observed daily for 21 days, and then values for percent mortality and AST were calculated (Table 1). Infection with TR339 resulted in 100% mortality with an AST of 1.5 ± 0.5 days. Infection with TR339-GFP/2A, TR339-26S/GFP, and TR339-VP7/2A also resulted in 100% mortality; however, AST values were extended. TR339-26S/VP7 caused 80% mortality, with the longest AST (5.5 ± 2.6 days) of any monovalent recombinant virus. The bivalent viruses were nearly avirulent. TR339-GFP/2A/VP7 caused 6.7% mortality, with an AST of 11 ± 4.2 days, and TR339-VP7/2A/GFP caused 3.3% mortality, with an AST of 15 days. All recombinant viruses were significantly less virulent than TR339 (P ≤ 0.0001) based on comparisons of AST values, and the monovalent DSP-based viruses were significantly less virulent than the cognate viruses engineered to express 2A fusion proteins. Specifically, TR339-26S/GFP was less virulent than TR339-GFP/2A (P ≤ 0.002), and TR339-26S/VP7 was less virulent than TR339-VP7/2A (P ≤ 0.001).
TABLE 1.
Virulence of viruses in newborn CD-1 mice
| Virus | No. of infected mice | % Mortality | ASTa (days) |
|---|---|---|---|
| TR339 | 30 | 100 | 1.5 ± 0.5 |
| TR339-GFP/2A | 30 | 100 | 4.0 ± 1.0 |
| TR339-26S/GFP | 30 | 100 | 4.8 ± 0.9 |
| TR339-VP7/2A | 30 | 100 | 3.6 ± 0.7 |
| TR339-26S/VP7 | 30 | 80 | 5.5 ± 2.6 |
| TR339-GFP/2A/VP7 | 30 | 6.7 | 11 ± 4.2 |
| TR339-VP7/2A/GFP | 30 | 3.3 | 15b |
AST for mice that succumbed to virus infection ± standard deviation.
Data based on the death of a single mouse.
Groups of 3-week-old female CD-1 mice were vaccinated s.c. with each VP7- and VP7/2A-expressing virus as described in Materials and Methods. Groups of mice were also infected with TR339 or mock infected with PBS. Serological responses to VP7 or VP7/2A were quantified for each animal in an indirect ELISA with purified BTV-10 virions as the antigen. Each serum sample was assayed twice, and results are reported as the average (Fig. 6). All prevaccination serum samples and all serum samples obtained from mice receiving PBS were negative for VP7-specific or cross-reacting antibodies (OD450 values were 0.1 or less at lowest dilution tested [1:20]). Two of four mice vaccinated with TR339 yielded minimal positive results (both had titers of 20). These results probably reflect the presence of antibodies to BHK-21 cell proteins, as virions used in vaccinations were grown in BHK-21 cells and only partially purified prior to use in vaccinations. In addition, the purified BTV virions used to coat ELISA plates were grown in BHK-21 cells, and it is unlikely that our purification procedure removed all cellular proteins (22). One animal from each of the TR339-26S/VP7- and TR339-GFP/2A/VP7-vaccinated groups registered postboost titers of 20 or lower and were essentially nonresponders. This is not an unexpected result, as CD-1 is an outbred mouse strain. Postboost serum antibody titers from all other animals infected with the recombinant viruses were markedly higher, ranging from 640 to 10,240 (Fig. 6). Neither expression strategy appeared to be superior to the other with respect to induction of humoral responses to vector-expressed proteins. In addition, the bivalent viruses were as effective at inducing antibody responses to VP7 and VP7/2A as their monovalent counterparts.
FIG. 6.
Serological responses to VP7. Three-week-old CD-1 mice (four per group) were immunized with 104 PFU of TR339 and each VP7-expressing recombinant virus in each rear footpad. Four mice were mock infected with PBS. Mice were boosted with 104 PFU of virus 21 and 42 days later. Serum samples were collected 14 days after the final boost. Antibody responses to VP7 were evaluated by using an indirect ELISA. ELISA plates were coated with purified BTV-10 virions (200 ng/well). All prevaccination serum samples were negative for VP7-specific or cross-reacting antibodies (OD450 values were ≤0.1 at the lowest dilution tested [1:20]). *, OD450 of <0.2 at a 1:20 dilution. ×, no serum available due to death of the animal. Each bar represents the serum titer for a single animal.
DISCUSSION
The FMDV 2A protease possesses two properties that make it particularly useful for protein expression applications. First, 2A mediates cleavage at its own C terminus and therefore can remove itself and all upstream residues from polyproteins in which it is incorporated in a site-specific manner. This property of 2A has been exploited previously to construct recombinant picornavirus-based (20, 24, 44) and influenza A virus-based (23) expression vectors and alphavirus-based (14, 39) and Kunjin virus-based (43) replicons. Here we used the 2A protease to construct live Sindbis virus expression vectors that express GFP/2A and VP7/2A fusion proteins as cleavable components of the viral structural polyprotein. Removal of the fusion protein was achieved by N-terminal and C-terminal cis cleavages mediated by capsid and 2A, respectively. Second, the FMDV 2A protease is very small, which helps to minimize its effect on the functional and antigenic properties of fusion proteins containing 2A as the C-terminal sequence. A number of proteins have been shown to retain their function in the context of a 2A fusion protein, including chloramphenicol acetyltransferase (7, 23, 34), β-glucuronidase (9), GFP (8, 9), and bovine interleukin-12 (5). We confirmed that fusion of 2A to GFP does not affect the functional properties of GFP in cell culture or in vivo, as BHK-21 cells and newborn mice infected with TR339-GFP/2A actually displayed brighter fluorescence than did cells and mice infected with TR339-26S/GFP (data not shown). Similarly, GFP and VP7 appeared to retain their antigenic properties when fused to 2A, as GFP/2A and VP7/2A were efficiently immunoprecipitated from infected-cell lysates with a polyclonal rabbit antiserum and a MAb, respectively. In addition, antibodies raised to VP7/2A in mice bound to native VP7 in ELISA. These results suggest that recombinant Sindbis viruses can be designed to express a wide range of functional 2A fusion proteins.
Phenotypic comparisons between TR339-GFP/2A and TR339-26S/GFP and between TR339-VP7/2A and TR339-26S/VP7 did not reveal obvious differences between the viruses with respect to growth rates in cultured cells. However, CPE developed most rapidly in cells infected with TR339-GFP/2A and TR339-VP7/2A, and these viruses were more virulent in newborn mice than TR339-26S/GFP or TR339-26S/VP7, respectively. These differences probably reflect the attenuating effects of the second active subgenomic promoter present in the DSP viruses. Incorporation of a second subgenomic promoter can slow virus replication by competing with the native subgenomic promoter for viral transcriptase complexes (28). Transcription from the native 26S promoter did appear to be reduced in viruses containing a second active 26S promoter. First, the level of capsid protein synthesized by TR339-26S/GFP and TR339-26S/VP7 was visibly decreased compared to that synthesized by TR339-GFP/2A and TR339-VP7/2A, respectively (Fig. 4). Second, the expression of capsid protein by TR339-26S/VP7 increased noticeably upon serial passage, and this increase correlated with diminished expression of VP7 from the downstream 26S promoter (Fig. 5).
The stability of transgene expression has been a concern with DSP-based expression vectors, as stable expression is important in cell culture and vaccine applications. For instance, a recombinant rubella virus engineered to express GFP in a manner similar to that for TR339-26S/GFP lost virtually all GFP expression after only three serial passages in Vero cells (26). Similarly, Sindbis virus-based vectors engineered to express protective antigens of Japanese encephalitis virus from duplicate 26S promoters placed upstream of the native 26S promoter or within the 3′ nontranslated region did not produce detectable levels of Japanese encephalitis virus proteins after five serial passages in cell culture (25). The instability of transgene expression by some DSP-based vectors may result from deletions arising through recombination events involving the duplicated 26S promoter sequences (26). Loss of transgene expression by DSP-based vectors may also result from deleterious events other than genetic recombination. Inactivating mutations arising within the transgene would not be deleterious to the virus, and we would not expect these nonexpressing mutants to be selected against. Furthermore, mutations that inactivate the duplicate 26S promoter may actually enhance the replication of the nonexpressing mutant due to increased use of the native 26S promoter. Under these circumstances, nonexpressing mutants would be expected to increase within the virus population over time. In contrast, alphavirus vectors based on the design of TR339-GFP/2A and TR339-VP7/2A should be under pressure to retain the foreign gene in functional form. Since the transgene is placed in frame with the viral structural genes, lesions within the transgene that alter the reading frame, or produce nonsense mutations, should be lethal, and mutants harboring such mutations should be selected against (23).
To address the issue of expression stability directly, we compared the stability of transgene expression between TR339-26S/GFP and TR339-GFP/2A and between TR339-26S/VP7 and TR339-VP7/2A during 10 serial passages in cell culture. Our results demonstrated that the expression stability of viruses designed to express their foreign protein as a cleavable component of the viral structural polyprotein was greater than the expression stability of the cognate DSP-based viruses. Expression stability also appeared to be influenced by properties of the transgene itself. Specifically, the expression stability of native GFP and GFP/2A was greater than that of native VP7 and VP7/2A, respectively. These results suggest that expression stability may be influenced by factors such as the transgene size, sequence, and/or secondary structures that these sequences may assume. Interestingly, TR339-26S/VP7 and TR339-VP7/2A induced similar levels of VP7-specific antibodies in vaccinated mice despite their differences in expression stability. This result may indicate that the differences in expression stability between the DSP and 2A fusion protein-expressing viruses are not significant enough to influence immune responses to the expressed proteins in vivo, that the lower expression stability was compensated for by the higher level to which VP7 was expressed compared to VP7/2A, or that these differences in expression stability were not reproduced in vivo.
TR339-GFP/2A/VP7 and TR339-VP7/2A/GFP grew to markedly lower titers than TR339 (33- and 17-fold lower, respectively), expressed their transgenes at lower levels than the monovalent viruses in cell culture, and were essentially avirulent in newborn mice. The degree to which these viruses were attenuated in cell culture and in vivo is not surprising, as each contains approximately 2,000 nucleotides of inserted sequence, a second active 26S promoter, and a significantly altered structural polyprotein. These genetic modifications could be expected to influence a wide range of processes related to virus growth, including but not limited to genome replication rate, genome packaging and virion morphogenesis, expression and processing of viral proteins, and induction of antiviral host cell responses. However, despite their genetic modifications, TR339-GFP/2A/VP7 and TR339-VP7/2A/GFP induced titers of VP7-specific antibodies in vaccinated mice that were equivalent to those induced by the monovalent vectors. This result may indicate that the magnitude of humoral immune responses to the vector-expressed antigens does not increase significantly once antigen levels have reached a threshold level that the monovalent and bivalent viruses are both able to achieve and/or that the lower levels of antigen delivered by the bivalent viruses is compensated for by their ability to express antigens for a longer duration due the decreased rate at which they kill their host cell.
BTV does not cause disease in adult mice, and antibodies to VP7 do not neutralize BTV infectivity. Therefore, it was not possible to perform meaningful challenge experiments with the immunized mice or to perform cell culture-based neutralization assays with immune sera. However, the immunization experiments that were performed suggest that monovalent and bivalent vectors designed to express a foreign protein as a cleavable component of the structural polyprotein can be used successfully as vaccine vectors. Specifically, it was shown that fusion of 2A to VP7 and GFP did not significantly alter the antigenic properties of these proteins and that each VP7/2A-expressing vector expressed the antigen to levels that were sufficient to induce high-titer antibody responses to native VP7.
Bivalent alphavirus-based replicons have been described previously (1, 2, 27); however, as far as we are aware, TR339-GFP/2A/VP7 and TR339-VP7/2A/GFP represent the first published examples of bivalent, live alphavirus vectors. The results obtained in the immunization experiments suggest that bivalent viruses based on the design of TR339-GFP/2A/VP7 and TR339-VP7/2A/GFP should be particularly useful for vaccine applications that require the expression of two interacting proteins or expression of two proteins with different immunological functions. For example, induction of protective immune responses to some pathogens may require coexpression of two gene products that assemble into a single functional unit. Expression of both of the major envelope glycoproteins (GL and M) of equine arteritis virus (EAV) was required to induce neutralizing antibodies against EAV in mice and in horses (1, 2). The GL and M glycoproteins form heterodimers on the surface of EAV virions, and neutralizing epitopes of EAV appear to be dependent on heterodimer formation. Macaques vaccinated individually with vaccinia viruses engineered to express the G1 or G2 glycoprotein of Lassa virus were not protected when challenged with Lassa virus. However, animals that received both the G1- and G2-expressing vaccinia viruses at different sites were fully protected, and this protection was primarily cell mediated (10). Bivalent vectors may also be engineered to express one antigen that is important for eliciting protective antibodies and a second antigen that contains critical T-cell epitopes. It may be most appropriate to express the antigen targeting T-cell responses as a 2A fusion protein, because some proteins may lose B-cell epitopes when fused to 2A; however, most T-cell epitopes derived from the 2A fusion proteins should be processed and presented normally by major histocompatibility complex I antigens on the surface of infected cells. Finally, bivalent vectors may be particularly well suited for coexpressing an antigen of interest along with a protein, such as a cytokine, which possesses adjuvant or immune-modulating properties.
Acknowledgments
We thank N. James MacLachlan (University of California, Davis) for providing MAb 290 and an isolate of BTV-10, and we thank William C. Wilson (ABADRL, Laramie, Wyo.) for providing cDNA clone pBTV10S7. We thank Ming-Ying Leung for assistance with statistical analysis and Jacqueline Williams for helping with the construction of recombinant viruses.
This work was supported by grants R01 AI22186 and R29 AI40937 from the National Institutes of Health, grant S06 GM08194 from the National Institutes of Health-Minority Biomedical Research Service, and a grant from the San Antonio Area Foundation.
REFERENCES
- 1.Balasuriya, U. B. R., H. W. Heidner, J. F. Hedges, J. C. Williams, N. L. Davis, R. E. Johnston, and N. J. MacLachlan. 2000. Expression of the two major envelope proteins of equine arteritis virus as a heterodimer is necessary for induction of neutralizing antibodies in mice immunized with recombinant Venezuelan equine encephalitis virus replicon particles. J. Virol. 74:10623-10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasuriya, U. B. R., H. W. Heidner, N. L. Davis, H. M. Wagner, P. J. Hullinger, J. F. Hedges, J. C. Williams, R. E. Johnston, W. D. Wilson, I. K. Liu, and N. J. MacLachlan. 2002. Alphavirus replicon particles expressing the two major envelope proteins of equine arteritis virus induce high level protection against challenge with virulent virus in vaccinated horses. Vaccine 20:1609-1617. [DOI] [PubMed] [Google Scholar]
- 3.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caley, I. J., M. R. Betts, N. L. Davis, R. Swanstrom, J. A. Frelinger, and R. E. Johnston. 1999. Venezuelan equine encephalitis virus vectors expressing HIV-1 protein: vector design strategies for improved vaccine efficacy. Vaccine 17:3124-3135. [DOI] [PubMed] [Google Scholar]
- 5.Chaplin, P. J., E. B. Camon, B. Villarreal-Ramos, M. Flint, M. D. Ryan, and R. A. Collins. 1999. Production of interleukin-12 as a self-processing 2A polypeptide. J. Interferon Cytokine Res. 19:235-241. [DOI] [PubMed] [Google Scholar]
- 6.Davis, N. L., L. V. Willis, J. F. Smith, and R. E. Johnston. 1989. In vitro synthesis of infectious Venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology 171:189-204. [DOI] [PubMed] [Google Scholar]
- 7.Donnelly, M. L. L., D. Gani, M. Flint, S. Monaghan, and M. D. Ryan. 1997. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J. Gen. Virol. 78:13-21. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly, M. L. L., G. Luke, A. Mehrotra, X. Li, L. E. Hughes, D. Gani, and M. D. Ryan. 2001. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal “skip.” J. Gen. Virol. 82:1013-1025. [DOI] [PubMed] [Google Scholar]
- 9.Donnelly, M. L. L., L. E. Hughes, G. Luke, H. Mendoza, E. ten Dam, D. Gani, and M. D. Ryan. 2001. The ‘cleavage' activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like' sequences. J. Gen. Virol. 82:1027-1041. [DOI] [PubMed] [Google Scholar]
- 10.Fisher-Hoch, S. P., L. Hutwagner, B. Brown, and J. B. McCormick. 2000. Effective vaccine for Lassa fever. J. Virol. 74:6777-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolov, I., T. A. Hoffman, B. M. Pragai, S. A. Dryga, H. V. Huang, S. Schlesinger, and C. M. Rice. 1996. Alphavirus-based expression vectors: strategies and applications. Proc. Natl. Acad. Sci. USA 93:11371-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn, C. S., Y. S. Hahn, T. J. Braciale, and C. M. Rice. 1992. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc. Natl. Acad. Sci. USA 89:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidner, H. W., K. L. McKnight, N. L. Davis, and R. E. Johnston. 1994. Lethality of PE2 incorporation into Sindbis virus can be suppressed by second-site mutations in E3 and E2. J. Virol. 68:2683-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heise, M. T., D. A. Simpson, and R. E. Johnston. 2000. Sindbis-group alphavirus replication in periosteum and endosteum of long bones in adult mice. J. Virol. 74:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klimstra, W. B., K. D. Ryman, K. A. Bernard, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 1999. Infection of neonatal mice with Sindbis virus results in a systemic inflammatory response syndrome. J. Virol. 73:10387-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn, R. J., H. G. Niesters, Z. Hong, and J. H. Strauss. 1991. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182:430-441. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Liljeström, P., L. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattion, N. M., E. C. Harnish, J. C. Crowley, and P. A. Reilly. 1996. Foot-and-mouth disease virus 2A protease mediates cleavage in attenuated Sabin 3 poliovirus vectors engineered for delivery of foreign antigens. J. Virol. 70:8124-8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKnight, K. L., D. A. Simpson, S. C. Lin, T. A. Knott, J. M. Polo, D. F. Pence, D. B. Johannsen, H. W. Heidner, N. L. Davis, and R. E. Johnston. 1996. Deduced consensus sequence of Sindbis virus strain AR339: mutations contained in laboratory strains which affect cell culture and in vivo phenotypes. J. Virol. 70:1981-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mertens, P. P. C., J. N. Burroughs, and J. Anderson. 1987. Purification and properties of virus particles, infectious subviral particles, and cores of bluetongue virus serotypes 1 and 4. Virology 157:375-386. [DOI] [PubMed] [Google Scholar]
- 23.Percy, N., W. S. Barclay, A. García-Sastre, and P. Palese. 1994. Expression of a foreign protein by influenza A virus. J. Virol. 68:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percy, N., W. S. Barclay, M. Sullivan, and J. W. Almond. 1992. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J. Virol. 66:5040-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugachev, K. V., P. W. Mason, R. E. Shope, and T. K. Frey. 1995. Double-subgenomic Sindbis virus recombinants expressing immunogenic proteins of Japanese encephalitis virus induce significant protection in mice against lethal JEV infection. Virology 212:587-594. [DOI] [PubMed] [Google Scholar]
- 26.Pugachev, K. V., W. P. Tzeng, and T. K. Frey. 2000. Development of a rubella virus vaccine expression vector: use of a picornavirus internal ribosome entry site increases stability of expression. J. Virol. 74:10811-10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pushko, P., J. Geisbert, M. Parker, P. Jahrling, and J. Smith. 2001. Individual and bivalent vaccines based on alphavirus replicons protect guinea pigs against infection with Lassa and Ebola viruses. J. Virol. 75:11677-11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raju, R., and H. V. Huang. 1991. Analysis of Sindbis virus promoter recognition in vivo, using novel vectors with two subgenomic RNA promoters. J. Virol. 65:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rayner, J. O., S. A. Dryga, and K. I. Kamrud. 2002. Alphavirus vectors and vaccination. Rev. Med. Virol. 12:279-296. [DOI] [PubMed] [Google Scholar]
- 30.Rice, C. M., and J. H. Strauss. 1981. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc. Natl. Acad. Sci. USA 28:2062-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutations. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson, B. H., M. J. Grubman, G. N. Weddell, D. M. Moore, J. D. Welsh, T. Fischer, D. J. Dowbenko, D. G. Yansura, B. Small, and D. G. Kleid. 1985. Nucleotide and amino acid sequence coding for polypeptides of foot-and-mouth disease virus type A12. J. Virol. 54:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan, M. D., A. M. Q. King, and G. P. Thomas. 1991. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 72:2727-2732. [DOI] [PubMed] [Google Scholar]
- 34.Ryan, M. D., and J. Drew. 1994. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 13:928-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlesinger, S. 2000. Alphavirus expression vectors. Adv. Virus Res. 55:565-577. [DOI] [PubMed] [Google Scholar]
- 36.Schlesinger, S., and T. W. Dubensky. 1999. Alphavirus vectors for gene expression and vaccines. Curr. Opin. Biotechnol. 10:434-439. [DOI] [PubMed] [Google Scholar]
- 37.Simpson, D. A., N. L. Davis, S. C. Lin, D. Russell, and R. E. Johnston. 1996. Complete nucleotide sequence and full-length cDNA clone of S.A.AR86, a South African alphavirus related to Sindbis virus. Virology 222:464-469. [DOI] [PubMed] [Google Scholar]
- 38.Sjöberg, E. M., M. Suomalainen, and H. Garoff. 1994. A significantly improved Semliki Forest virus expression system based on the translational enhancer segments from the viral capsid gene. Bio/Technology 12:1127-1131. [DOI] [PubMed] [Google Scholar]
- 39.Smerdou, C., and P. Liljeström. 1999. Two-helper RNA system for production of recombinant Semliki Forest virus particles. J. Virol. 73:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan, K. F. 1999. Enlightening mitosis: construction and expression of green fluorescent fusion proteins. Methods Cell Biol. 61:113-135. [DOI] [PubMed] [Google Scholar]
- 42.Tubelukas, I., P. Berglund, M. Fleeton, and P. Liljeström. 1997. Alphavirus expression vectors and their use as recombinant vaccines. Gene 190:191-195. [DOI] [PubMed] [Google Scholar]
- 43.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable, high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 74:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vignuzzi, M., S. Gerbaud, S. van der Werf, and N. Escriou. 2002. Expression of a membrane-anchored glycoprotein, the influenza virus hemagglutinin, by dicistronic replicons derived from the poliovirus genome. J. Virol. 76:5285-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, C. Y., G. Dominguez, and T. K. Frey. 1994. Construction of rubella virus genome-length cDNA clones and synthesis of infectious RNA transcripts. J. Virol. 68:3550-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whetter, L. E., N. J. MacLachlan, D. H. Gebhard, H. W. Heidner, and P. F. Moore. 1989. Bluetongue virus infection of bovine monocytes. J. Gen. Virol. 70:1663-1676. [DOI] [PubMed] [Google Scholar]