Abstract
Gammaherpesvirus pathogenesis is dependent on the ability of these viruses to establish a lifelong latent infection and the ability to reactivate from latency. Immediate-early genes of theses viruses are thought to be critical regulators of lytic replication and reactivation from latency. The gene 50-encoded Rta is the only immediate-early gene product that appears to be conserved among all characterized gammaherpesviruses. Previous studies have demonstrated that, in Epstein-Barr virus (EBV), Kaposi's sarcoma-associated virus, and gammaherpesvirus 68 (γHV68, also referred to as murine gammaherpesvirus 68), ectopic expression of Rta in latently infected cell lines can lead to induction of the viral cycle. Recently, studies employing null mutants of EBV have provided a formal demonstration that both Rta and the BZLF1 gene product, Zta, the two EBV immediate-early gene products, are essential for EBV replication. Here we generate and characterize a gene 50-null mutant γHV68 and demonstrate that the gene 50 product Rta is essential for virus replication. Providing γHV68 Rta in trans was sufficient to restore replication of the gene 50-null virus. Notably, Rta expressed from the spliced form of the gene 50 transcript was sufficient to complement growth of the gene 50-null virus. In addition, we provide evidence that loss of Rta expression leads to a complete defect in viral DNA replication and a significant defect in late antigen expression. This work lays the foundation for characterizing the role of Rta in γHV68 chronic infection of mice.
Gammaherpesviruses are lymphotropic viruses that establish a lifelong infection of the host and are associated with cellular transformation and tumor formation in immunosuppressed hosts. Murine gammaherpesvirus 68 (γHV68; also referred to as murine herpesvirus 68) was isolated from a bank vole (3, 4), and the 118-kb γHV68 genome has been sequenced (40). In contrast to other gammaherpesviruses, γHV68 has been shown elsewhere to infect a wide range of species, including inbred and outbred strains of mice (4, 8, 21, 24, 32-34). The ability to infect mice is of great importance for the gammaherpesvirus field, because it provides a genetically tractable system for studying viral pathogenesis. γHV68 infects multiple organs of inbred mice acutely, including the spleen, liver, lung, kidney, adrenal gland, heart, and thymus (4, 24). γHV68 can cause inflammation of the great elastic arteries (42). Additionally, γHV68 can establish a latent infection in the spleen, peritoneal cells, and lymph nodes (5, 32, 43), with B cells, macrophages, and splenic dendritic cells serving as latency sites (11, 33, 44). Additionally, γHV68 can persist in lung epithelial cells (30). Significantly, γHV68 has been associated with lymphoproliferative disease and lymphoma in mice (37). γHV68 readily infects and forms plaques on monolayers of fibroblast cells, and several virus mutants have been generated successfully, demonstrating the utility of this virus for genetic studies (for example, see references 7 and 22).
Latency and reactivation from latency are central aspects of gammaherpesvirus pathogenesis. Latency is thought to be the primary mechanism by which herpesviruses chronically infect their host. In the case of gammaherpesviruses, there is a tight correlation between latent infection and gammaherpesvirus-associated tumors. Reactivation and lytic virus replication are associated with reactivation disease and spread of the virus between hosts. However, the critical question of whether reactivation and subsequent virus replication are required for the maintenance of chronic gammaherpesvirus infection remains unanswered. Additionally, the molecular mechanisms involved in virus reactivation in vivo are poorly understood. Gammaherpesvirus immediate-early genes serve as initiators of the viral lytic cascade, and in the case of Epstein-Barr virus (EBV) reactivation, immediate-early viral gene expression has been shown in vitro to be regulated by signal transduction pathways that trigger reactivation from latency.
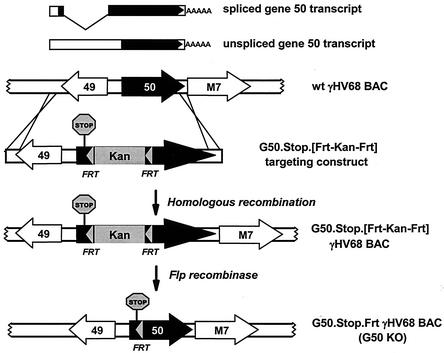
Extensive studies have shown that the EBV immediate-early BRLF1 (also known as gene 50 or rta [replication and transcription activator]) and BZLF1 gene products (Rta and Zta, respectively) act as critical regulators of the viral lytic cascade (9; reviewed in reference 29). The BZLF1 gene does not appear to be well conserved among the gammaherpesviruses. In Kaposi's sarcoma-associated virus and rhesus rhadinovirus the K8 and R8 gene products, respectively, are located in the same position in the viral genome and encode bZIP proteins that appear to be distantly related to Zta (15, 17, 28, 47). Herpesvirus saimiri and γHV68 do not appear to encode homologs of Zta. In contrast, gene 50 is well conserved among all known gammaherpesviruses (31, 39, 40, 45). Both an unspliced form and a spliced form of gene 50 transcript have been characterized (see Fig. 1). Open reading frame (ORF) 49 and gene 50 are encoded on opposite strands and are organized “head-to-head” in the viral genome (Fig. 1). Transcription of both the unspliced and spliced gene 50 transcripts initiates downstream of ORF 49, with transcription extending through ORFs 49 and 50 (Fig. 1). In the case of the spliced transcript, splicing removes the region antisense to ORF 49 and extends the gene 50 ORF. Notably, this organization and transcription of gene 50 are well conserved among the known gammaherpesviruses (12, 18, 20, 31, 38, 45).
FIG. 1.
Construction of gene 50 recombinant viruses. ORF 50 and the adjacent ORFs are indicated, as are the structures of the spliced and unspliced gene 50 transcripts. The solid black arrow indicates the ORF for each of the transcripts. A schematic representation of the structure of the gene 50 recombinant viruses, G50.Stop.[Frt-Kan-Frt] and G50.Stop.Frt (also designated as G50 KO in the text), is shown below the genome. The kanamycin cassette is indicated, the TAG stop codon mutation (with an associated 2-bp deletion) is represented as a stop sign, and the gray arrowheads represent the FRT sites for Flp-mediated recombination.
The gene 50 product, Rta, has been shown previously to act as a transcriptional activator of downstream viral genes (14, 18, 19, 31; reviewed in reference 25). Additionally, it has roles in DNA replication and cell cycle regulation (10, 16, 27, 35, 50). Most importantly, overexpression of EBV, Kaposi's sarcoma-associated virus, herpesvirus saimiri, and γHV68 gene 50 in cell lines latently infected with the corresponding virus has been shown to lead to disruption of latency (13, 14, 19, 23, 31, 49, 51). Recently, Feederle and colleagues generated EBV null mutants with mutations in the BZLF1 and BRLF1 genes (gene 50) and showed that both genes are essential for EBV replication, thus providing the first genetic evidence for the requirement of these gammaherpesvirus immediate-early genes for virus replication (9). Consistent with an essential role of the γHV68 gene 50 in virus replication, two dominant-negative forms of γHV68 gene 50 have been shown to significantly inhibit viral protein expression and virion production in lytically infected cell lines (48). Taken together, these data strongly indicate that gene 50 is required for the initiation of the viral lytic cycle in vitro and that this requirement is conserved among the characterized gammaherpesviruses.
Here we describe the generation and initial characterization of a gene 50-null mutant γHV68. We show that the gene 50-null mutant is incapable of virion production or viral DNA replication and is attenuated for viral protein synthesis. The defect in gene 50 can be rescued in trans either by a genomic fragment containing gene 50 or by a gene 50 cDNA clone corresponding to the spliced gene 50 transcript. The latter result demonstrates that Rta expressed from the spliced transcript is sufficient to rescue replication of a gene 50-null mutant.
MATERIALS AND METHODS
Viruses and tissue culture.
The γHV68 bacterial artificial chromosome [γHV68(BAC)] (2) was obtained from Heiko Adler and Ulrich Koszinowski (Max von Pettenkofer Institute, University of Munich, Munich, Germany). γHV68(BAC) virus stocks were prepared as follows. NIH 3T12 cells were infected at a multiplicity of infection (MOI) of 0.05 and harvested at 4 days postinfection (p.i.), and samples were homogenized, clarified, and aliquoted for storage at −80°C. Titers of viral stocks were determined by at least two independent plaque assays, as described below. Working gene 50 mutant virus stocks were grown by infecting NIH 3T12-derived gene 50-expressing stable cell lines at an MOI of 0.01, harvested at 7 to 14 days p.i., and otherwise treated the same as described above. NIH 3T12 and the gene 50 stable cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal calf serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 2 mM l-glutamine. Gene 50-expressing cell lines were additionally supplemented with 0.5 mg of active G418/ml. Cells were maintained in a 5% CO2 tissue culture incubator at 37°C.
Plaque assays.
Plaque assays were performed on monolayers of NIH 3T12 or gene 50-expressing stable cell lines under Noble agar overlay. The cells were plated into six-well plates at 3 × 105 cells per well on the day prior to the infection. Infections were performed in an 0.2-ml volume, and plates were rocked every 15 min for 1 h at 37°C. Samples were overlaid with 3 ml of a 1:1 mixture of 1% Noble agar and 2× DMEM supplemented with 10% fetal calf serum and a 2× concentration of antibiotic and l-glutamine. NIH 3T12 monolayers were stained on day 6 or 7 p.i., gene 50-expressing stable cells were stained on day 14 p.i. by the addition of a 2-ml overlay consisting of 0.02% neutral red in a 1:1 mixture of 1% Noble agar and 2× DMEM, and the presence of plaques was scored after 24 h of staining.
Generation of stable cell lines expressing gene 50.
To generate gene 50-expressing stable cell lines, NIH 3T12 cells were transfected with an expression vector (pBK-CMV; Stratagene) containing gene 50 (bp 66642 to 69462 of the γHV68 genome) under the control of the human cytomegalovirus immediate-early promoter. Transfected cells were grown under 1 mg of G418/ml, and G418-resistant colonies were picked, expanded, and tested by reverse transcriptase PCR for the presence of the spliced gene 50 transcript. Positive clones were further tested for their ability to transactivate a γHV68 gene 57 promoter-driven luciferase reporter construct, as previously described (18). One clone (clone 14) exhibited strong activation of the gene 57 promoter, and this line was subcloned by limiting dilution. Subclone 14.18 was used to generate the initial gene 50-null virus stocks. The initial stocks of gene 50 mutant virus were then used to screen multiple subclones of clone 14, and clone 14.29 was chosen for further studies based on its ability to grow gene 50 mutant virus most efficiently.
To generate stable cell lines expressing only the spliced form of gene 50 (Fig. 1), the spliced gene 50 transcript was cloned by reverse transcriptase PCR with RNA prepared from the gene 50-expressing cell line clone 14.29 (see above). The spliced gene 50 cDNA was generated with the SuperScript First-Strand synthesis system (Invitrogen), and the resulting cDNA was amplified with gene 50 primers (forward primer 36.1s, 5′-CTCTGACAGCCCGGGCATGGCCTCTGACTCGGAT-3′, and reverse primer 36.3as, 5′-GAGGGGCGAAGCTTTGAGGGTTTTATAGCGTCAC-3′). The forward primer contains a SrfI site, and the reverse primer contains a HindIII site (underlined). The forward primer also contains a translation initiation codon for the spliced gene 50 transcript (boldface). The resultant cDNA was cloned downstream of the FLAG epitope sequence into the SrfI and HindIII sites of pCMV-Tag 2B (Stratagene) to generate sG50-FLAG. Sequencing analysis confirmed that the cloned fragment corresponded to the sequence for the spliced gene 50 transcript previously described (18). sG50-FLAG was transfected into NIH 3T12 cells, the cells were grown under selection with 1 mg of G418/ml, and the bulk stable cells were subcloned by limiting dilution and screened by infection with gene 50 mutant virus. Subclone s27 was chosen based on its ability to most efficiently support growth of the gene 50 mutant virus.
Generation of recombinant viruses.
For the generation of recombinant viruses by using the γHV68(BAC), we essentially followed the previously reported procedure with minor modifications (2). As outlined in Fig. 1, we initially targeted a translation stop codon and kanamycin resistance cassette into gene 50. The gene 50 cDNA clone 50-1 (18) was cloned into the EcoRV and XmaI sites of pMECAΔ55 (pMECA [36], in which all restriction endonuclease cleavage sites between NheI and HindIII were eliminated by digestion with NheI and HindIII, followed by filling in the overhangs with T4 DNA polymerase, and blunt-end ligation) to generate g50cDNA/pMecaΔ55. A PCR fragment spanning the region of gene 50 from the BglII site at bp 67745 to the BclI site at bp 67984 was generated which incorporated a deletion of bp 67971 (5′-T) and 67972 (5′-A) and changed bp 67976 to 67978 from 5′-CAC to 5′-AGA, resulting in the generation of an in-frame TAG stop codon at bp 67975 and an XbaI site at bp 67973. The incorporation of these changes into the resulting PCR product was confirmed by sequence analysis. The mutated fragment of gene 50 was subsequently cloned into the BglII and BclI sites of g50cDNA/pMecaΔ55 to generate g50cDNA.Stop/pMecaΔ55. Since cleavage at the BclI site is blocked by methylation, all steps that involved cloning with BclI were done in Escherichia coli strain SCS110, which is defective in dam methylation. Finally, the kanamycin resistance cassette, flanked by Flp recombinase recognition target sites, was cloned from pCP15 (6) by PCR (sense primer, 5′-CTCTAATTGCTAGCGTACCCGGGGATCTTGAAG-3′, and antisense primer, 5′-CTCTGATTGCTAGCTTCAAAAGCGCTCTGAAG-3′; note that the NheI site is underlined), digested with NheI, and cloned into the inserted XbaI site of g50cDNA.Stop/pMecaΔ55.
To generate G50.Stop.[Frt-Kan-Frt] γHV68(BAC), E. coli JC8679 (recBC sbcA) carrying the γHV68(BAC) and a recA expression plasmid (p2650; gift of Wolfgang Hammerschmidt, Institute of Clinical Molecular Biology and Tumor Genetics, Munich, Germany) was electroporated with a twice-agarose-gel-purified G50.Stop.[Frt-Kan-Frt] fragment in which the vector backbone was removed by digestion with XmnI and ScaI. The cells were incubated at 37°C for 2 h, plated onto agar plates containing 17 μg of chloramphenicol/ml and 50 μg of kanamycin/ml, and incubated overnight at 42°C. The BAC vector contains the gene encoding chloramphenicol resistance while the recA expression plasmid (p2650) has a temperature-sensitive origin of replication and cannot replicate at 42°C. Cam+ Kan+ colonies were picked and restreaked onto plates containing chloramphenicol and kanamycin or plates containing ampicillin to confirm the loss of the recA expression plasmid. DNA was isolated from Cam+ Kan+ Amp− colonies and analyzed by Southern blotting to detect the presence of the mutation in the viral genome. To generate G50.Stop.Frt γHV68(BAC), E. coli DH10B cells carrying one of the correct G50.Stop.[Frt-Kan-Frt] γHV68(BAC) clones were electroporated with an Flp recombinase expression plasmid (pCP20) (6) and grown overnight at 30°C on plates containing chloramphenicol and ampicillin, and colonies were restreaked onto chloramphenicol plates and grown overnight at 42°C to cure the cells of the Flp recombinase expression plasmid (pCP20), which contains a temperature-sensitive origin of replication. Colonies were again restreaked onto plates containing either chloramphenicol or kanamycin, and DNA from clones that were Cam+ Kan− was subjected to Southern blot analyses. The gene 50 region from one of the correct clones was sequenced to confirm the correct structure of the mutation (see Fig. 2B) and was transfected into a gene 50-expressing cell line to generate virus stocks of G50.Stop[Frt] γHV68(BAC) [which will subsequently be referred to as G50 KO(BAC)].
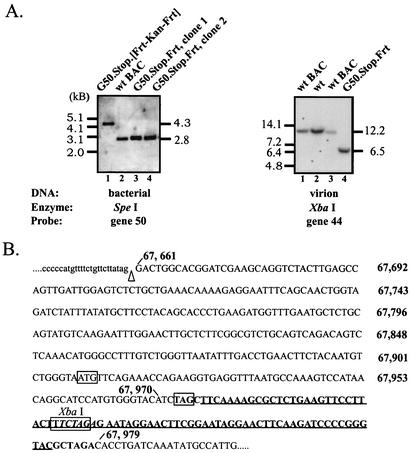
FIG. 2.
Genomic structure of gene 50 recombinant viruses. (A) Southern blot analysis. (Left) wt, G50.Stop.[Frt-Kan-Frt], and G50.Stop.Frt BAC viral genomes were purified from E. coli DH10B cells, digested with SpeI, electrophoresed, blotted, and hybridized with a biotin-labeled gene 50 probe (bp 66642 to 69462). The biotin-labeled gene 50 probe was generated by using the Detector random primer DNA biotinylation kit (Kirkegaard & Perry Laboratories), and the Southern blot was developed by using the DNA Detector genomic Southern blotting kit (Kirkegaard & Perry Laboratories) according to the manufacturer's instructions. (Right) wt γHV68 or wt BAC and G50.Stop.Frt BAC (G50 KO in the text) viral genomes were purified from virions isolated from NIH 3T12 and gene 50-expressing stable cell line 14.29, respectively, digested with XbaI, electrophoresed, blotted, and hybridized with a 32P-labeled gene 44 probe (bp 61444 to 62183). On both panels, the fragment sizes of the molecular size markers are shown to the left of each blot (1-kb DNA ladder for the left panel, lambda DNA-BstEII digest for the right panel; New England Biolabs). To the right of each blot are shown the predicted sizes of the viral DNA fragments detected by the respective probes in each blot. (B) Nucleotide sequence of the region containing the mutation in the G50.Stop.Frt BAC. The genome coordinates are to the right of the nucleotide sequence. The nucleotide sequence shown in lowercase letters denotes the gene 50 intron of the spliced gene 50 transcript, and the sequence in uppercase letters denotes the second gene 50 exon of the spliced gene 50 transcript. The splice acceptor site is denoted with an arrowhead. The ORF 50 ATG is boxed. The mutation in G50.Stop.Frt BAC is depicted in boldface, the introduced TAG stop codon is boxed, the FRT site is underlined, and the XbaI site within the FRT site is boxed and italicized.
Immunoblot analysis.
NIH 3T12 or 14.29 cells (4 × 105 to 5 × 105) were either mock infected or infected with wild-type (wt) γHV68(BAC) virus or G50 KO(BAC) virus at 1 PFU per cell in the presence or absence of phosphonoacetic acid (PAA). Total cell lysates were harvested at 24 h p.i., and 1/20 of the sample was run under reducing conditions on a sodium dodecyl sulfate-12.5% polyacrylamide gel. Immunoblotting was performed with a 1:500 dilution of rabbit polyclonal anti-γHV68 antiserum (42), followed by detection employing a 1:1,000 dilution of donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories), and blots were developed with ECL chemiluminescent reagent (Amersham) according to the manufacturer's protocol.
Viral DNA analysis with quantitative real-time PCR.
NIH 3T12 cells were infected with wt γHV68(BAC) or G50 KO(BAC) at 1 PFU per cell and harvested at various times p.i. (see Fig. 6). Data were collected from two independent experiments. DNA from cell pellets was isolated with the DNeasy tissue kit (Qiagen) and quantified with a VersaFluor fluorometer (Bio-Rad), and 1 ng of each sample was used per PCR with the QuantiTect SYBR Green PCR kit (Qiagen) and gene 50 primers (sense primer, 5′-GGCCGCAGACATTTAATGAC; antisense primer, 5′-GCCTCAACTTCTCTGGATATGCC) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers (sense primer, 5′-TGCACCACCAACTGCTTAG; antisense primer, 5′-GGATGCAGGGATGTTC) in a 30-μl reaction mixture volume. All PCRs were performed in duplicate on a Bio-Rad iCycler real-time PCR instrument, and results were analyzed with iCycler 3.0 software. The starting viral DNA amounts (time zero) were similar between G50 KO and wt BAC at 1.34 × 104 and 5.76 × 103 viral DNA copies per ng of DNA, respectively. GAPDH copy numbers per reaction did not vary significantly, ranging from 5.5 × 104 to 1.15 × 105 copies per ng of DNA.
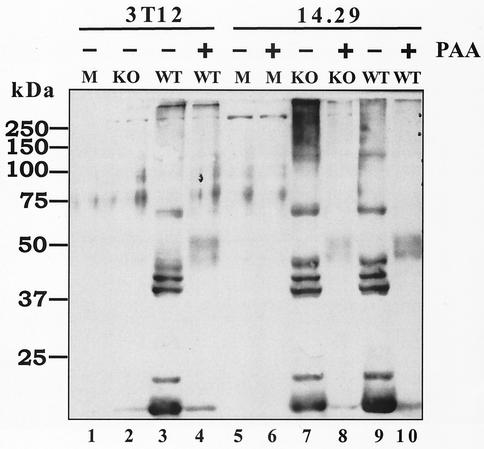
FIG. 6.
G50 KO is defective for late viral protein expression. NIH 3T12 or 14.29 cells were mock infected or infected with wt γHV68(BAC) virus or G50 KO virus at 1 PFU per cell in the presence or absence of PAA. Total cell lysates were harvested at 24 h p.i., and samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblot analysis was carried out with a rabbit polyclonal anti-γHV68 antiserum raised in γHV68-infected rabbits (42) as described in Materials and Methods. Molecular mass markers are shown to the left of the gel.
RESULTS
Targeted disruption of gene 50 with the γHV68 into a BAC.
Recently, Adler et al. (2) reported the cloning of the entire γHV68 genome as a BAC [wt γHV68(BAC)], which they generously provided to us. The wt γHV68(BAC) is maintained in recombination-deficient bacteria and to date appears to be stable under these conditions. The presence of the BAC sequences does not affect viral growth in vitro (2) but does influence lytic replication and reactivation in vivo (1). We first aimed to disrupt the γHV68 gene 50 by inserting a translation stop codon (bp 67975), along with a kanamycin resistance cassette (flanked by FRT sites), to allow selection for the desired mutant in E. coli {G50.Stop.[Frt-Kan-Frt] γHV68(BAC)} (Fig. 1). The incorporation of FRT sites allowed for removal of the kanamycin cassette by Flp recombinase-mediated excision. In addition to incorporation of a translation stop codon, a 2-bp deletion was introduced that generated a frameshift to ensure that any protein resulting from stop codon read-through or reversion would not encode a functional gene 50 product.
The G50.Stop.[Frt-Kan-Frt] γHV68(BAC) mutant was generated as described previously (2) through homologous recombination in E. coli JC8679 cells by using a plasmid containing the mutation and ca. 1.4 kb of homology to γHV68 sequence on each side of the mutation (Fig. 1). Southern blot analysis of G50.Stop.[Frt-Kan-Frt] γHV68(BAC) by digestion with SpeI and hybridization with a biotin-labeled gene 50 probe demonstrated, as predicted, the presence of a 2.8-kb restriction fragment in the wt γHV68(BAC) and a 4.3-kb fragment in G50.Stop.[Frt-Kan-Frt] γHV68(BAC) consistent with the insertion of the kanamycin cassette (Fig. 2A, left panel, lanes 1 and 2). Additionally, this analysis demonstrated that the insertion is in the gene 50 genomic locus, because one of the SpeI sites in the γHV68 genome lies outside the region used for homologous recombination.
One of the correct G50.Stop.[Frt-Kan-Frt] γHV68(BAC) clones was then subjected to Flp-mediated recombination (see Materials and Methods) to generate G50.Stop.Frt γHV68(BAC) (Fig. 1). The excision of the kanamycin cassette was demonstrated by Southern blot analysis (Fig. 2A, left panel, lanes 3 and 4). Additionally, the region around the mutation in G50.Stop.Frt was sequenced to confirm that the desired mutation had been introduced into the viral genome (Fig. 2B). For ease, the G50.Stop.Frt γHV68(BAC) will be referred to as G50 KO(BAC).
Isolation of a gene 50-null γHV68 in gene 50-expressing stable cells.
Viral stocks of G50 KO were generated by transfection of the gene 50-expressing cell line 14.18 with G50 KO(BAC) DNA purified from E. coli. Notably, no infectious virus could be isolated upon multiple cycles of reinfection of NIH 3T12 cells with G50 KO(BAC), demonstrating a dependence on gene 50 expression for growth of G50 KO(BAC) virus. Subsequent screening of multiple subclones of the gene 50 stable cell line clone 14 for growth of the G50 KO virus identified clone 14.29 as the most efficient in supporting growth of the mutant virus. Homologous recombination between the G50 KO viral DNA and gene 50 inserted into the cellular genome can result in the generation of revertant virus containing the wt gene 50 locus. Nonhomologous recombination is less likely, but also possible. To assess the reversion frequency in the viral stocks, they were analyzed by plaque assay on NIH 3T12 cells, which do not express gene 50. This method accounts for virus resulting from both homologous and nonhomologous recombination, provided that the resultant virus has wt growth characteristics. The reversion frequency of the G50 KO(BAC) virus stocks generated on the gene 50-expressing cell line 14.29 was estimated to be 1 PFU of revertant in 3 × 104 to 1 × 105 PFU of G50 KO virus. To ensure that the genetic lesion in the G50 KO remains unaffected by growth in tissue culture, G50 KO DNA was isolated from virions and analyzed by Southern analysis employing the presence of an additional XbaI site in the FRT sequence of the G50 KO genome (Fig. 2A). Southern analyses of XbaI-digested G50 KO virion DNA, with a 32P-labeled gene 44 probe, demonstrated hybridization of the probe, as expected, to a 12.2-kb fragment in the wt γHV68 and in the wt γHV68(BAC) and a 6.5-kb fragment in the G50 KO (Fig. 2A, right panel, lanes 1 through 4). Additionally, this analysis also demonstrated that the mutation is contained within the gene 50 genomic locus, because XbaI generates cuts in the γHV68 genome outside the region used for homologous recombination.
Gene 50 is necessary and sufficient for G50 KO(BAC) virus production in NIH 3T12 cells.
The hypothesis on which these studies are based is that gene 50 is essential for initiation and progression of the γHV68 lytic cycle. To test the essentiality of gene 50 in a quantitative manner, we infected cell monolayers of NIH 3T12 and the gene 50-expressing cell line 14.29 with G50 KO(BAC) and wt γHV68(BAC) virus at 0.1 PFU per cell, and total virus (cells and supernatants) was collected at different times postinfection. Importantly, the G50 KO(BAC) virus stock used had less than 25 PFU of revertant virus/ml (below the limit of detection). Mock-infected samples did not display any cytopathic effect (CPE) up to 14 days p.i. Both NIH 3T12 and 14.29 cells infected with wt γHV68(BAC) virus reached 100% CPE at 4 days p.i. Notably, NIH 3T12 cells infected with G50 KO(BAC) virus did not display any CPE, similar to mock-infected cells, up to 14 days p.i. In contrast, G50 KO(BAC) virus-infected 14.29 cells showed a progression from small plaques without cleared centers starting at ca. 4 days p.i. to plaques with cleared centers (typical for wt γHV68 infection), appearing at day 8 p.i., to 100% CPE by 14 days p.i.
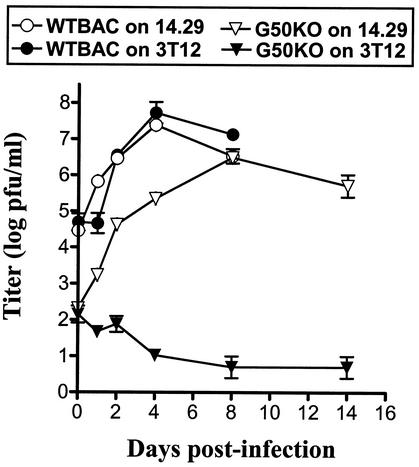
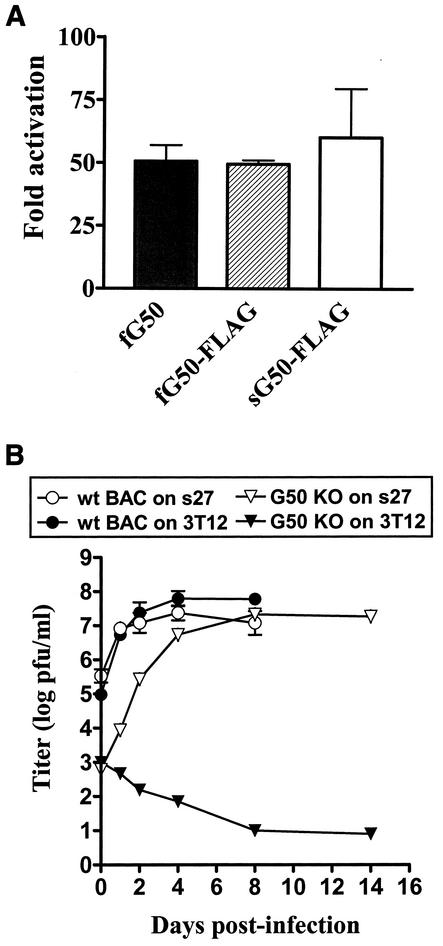
Viral titers of samples recovered from infection of NIH 3T12 and 14.29 fibroblasts were determined on the gene 50-expressing cell line 14.29 (Fig. 3). Prior to these experiments, we established that γHV68 grew with comparable kinetics and had a comparable growth output on both NIH 3T12 and 14.29 cells (data not shown), and therefore wt γHV68(BAC) viral titers were also determined on 14.29 cells. wt γHV68(BAC) reached a maximum titer of ca. 2 × 107 to 5 × 107 PFU/ml at day 4 p.i. on either NIH 3T12 or 14.29 cells. Titers of virus recovered from G50 KO(BAC) virus-infected NIH 3T12 cells slowly declined with time, demonstrating an absence of virus growth on cells lacking gene 50 expression. In contrast, G50 KO(BAC) virus growth on the 14.29 complementing cell line was readily detectable, reaching a maximum titer of 3 × 106 PFU/ml at day 8 p.i. In addition, samples from G50 KO virus-infected 14.29 cells were plaqued on NIH 3T12 cells to test for the presence of revertant virus, and none was detected (data not shown), indicating that there is <25 PFU of revertant virus/ml present in these samples. Therefore, the high levels of virus production detected upon infection of the 14.29 cell line with G50 KO(BAC) virus can be attributed to G50 KO(BAC) virus growth, and not virus that has restored expression of wt gene 50.
FIG. 3.
G50 KO virus is defective for virus replication in NIH 3T12 fibroblasts and can be rescued by expression of gene 50 in trans. NIH 3T12 fibroblasts or a stable NIH 3T12-derived cell line expressing gene 50 (14.29 cells; see Materials and Methods) was infected with wt BAC or G50 KO at 0.1 PFU per cell. Samples were harvested at the indicated times. All samples were quantified by plaque assay on the 14.29 cell line. Samples from G50 KO-infected 14.29 cells were additionally quantified by plaque assay on NIH 3T12 cells to determine the presence of revertant virus, and none was detected by this assay. Data were compiled from two independent experiments.
Since the 14.29 cell line was created with a fragment of the γHV68 genome containing the entire ORF 49, in addition to gene 50, it is formally possible that the 14.29 cell lines expresses both Rta and the ORF 49 product. ORF 49 is a putative gene with no known function that is conserved among the sequenced gammaherpesviruses (40) and thus is likely to be expressed during virus infection. To exclude the possibility that defects in the expression of other genes in the gene 50 locus, specifically ORF 49, are responsible for the inability of G50 KO virus to grow on NIH 3T12 fibroblasts, we designed a strategy to generate a gene 50-expressing cell line that can express only Rta. To accomplish this, we cloned the spliced gene 50 transcript, which lacks ORF 49 (Fig. 1). This approach was based on the assumption that it is the spliced transcript that encodes the complete Rta protein. This cDNA was cloned into an expression vector and used to generate stable NIH 3T12 cell lines expressing only the spliced form of the gene 50 transcript (see Materials and Methods).
To study the functionality of the spliced gene 50 cDNA, FLAG epitope-tagged spliced gene 50 (sG50-FLAG) and FLAG epitope-tagged full-length gene 50 (fG50-FLAG) expression vectors were cotransfected with the gene 57 promoter-driven luciferase (57pLuc) reporter construct into NIH 3T12 cells and analyzed for luciferase activity (Fig. 4A). The fG50-FLAG expression vector displayed approximately the same levels of activation of the gene 57 promoter as did the untagged full-length gene 50 (fG50) expression vector, indicating that addition of the FLAG epitope does not affect the function of gene 50 as previously reported (48). Notably, the sG50-FLAG expression vector could also achieve a similar level of activation of the gene 57 promoter as the full-length gene 50 constructs, consistent with the hypothesis that it is the spliced transcript that gives rise to functional Rta (note that the full-length gene 50 constructs encode both the spliced and unspliced gene 50 transcripts). This observation is consistent with previous findings that the spliced form, but not the unspliced form, of the herpesvirus saimiri gene 50 is capable of activating the gene 57 promoter (46). However, this analysis does not rule out a function for a gene 50 product expressed from the unspliced gene 50 transcript.
FIG. 4.
Gene 50-null mutant growth deficiency can be rescued by expression of only the spliced gene 50 transcript. (A) The spliced form of gene 50 can transactivate the γHV68 early gene 57 promoter. With respect to the activation of the gene 57 promoter by the full-length gene 50 expression vector (fG50), NIH 3T12 cells were cotransfected with 1 μg of pBK-CMV (Stratagene) containing gene 50 (see Materials and Methods) and 1 μg of either pGL2-Basic (Promega) or pGL2-Basic containing the gene 57 promoter (G57pLuc) (18). Alternatively, for the amino-terminal FLAG epitope-tagged full-length gene 50 (fG50-FLAG) and the amino-terminal FLAG epitope-tagged spliced gene 50 (sG50-FLAG) expression vectors, NIH 3T12 cells were cotransfected with 1 μg of either pCMV-Tag 2B (Stratagene) or pCMV-Tag 2B containing the relevant gene 50 construct and 1 μg of either pGL2-Basic or G57pLuc. Transfections were performed with the lipid-based transfection reagent LT-1 (Mirus), according to the manufacturer's protocol. Cell lysates were prepared 48 h posttransfection and assayed for luciferase activity. Data are presented as fold activation of the gene 57 promoter (G57pLuc) in the presence and absence of each of the gene 50-expressing vectors (fG50, fG50-FLAG, and sG50-FLAG). Data were compiled from two independent experiments. (B) NIH 3T12 or a stable NIH 3T12-derived cell line expressing the spliced form of gene 50 (s27 cells) was infected with wt γHV68(BAC) virus or G50 KO virus at 0.1 PFU per cell, and samples were harvested at the times indicated. The virus dilutions that were used for the infections were quantified by plaque assay on 14.29 cells to determine actual input virus titers (see Results). All samples were quantified by plaque assay on the 14.29 cell line. Samples from G50 KO-infected 14.29 cells were additionally quantified by plaque assay on NIH 3T12 cells to determine the presence of revertant virus, and none was detected by this assay. Data are compiled from three independent experiments.
The sG50-FLAG construct was transfected into NIH 3T12 cells, the cells were grown under G418 selection, and the bulk stable cells were tested for the ability to support G50 KO growth. Since G50 KO virus did not cause any CPE on the bulk stable cells, multiple subclones were generated by limiting dilution and again tested for G50 KO(BAC) virus growth. One clone, s27, was chosen for further studies based on its ability to support G50 KO(BAC) growth most efficiently. Cell monolayers of NIH 3T12 and s27 cells were infected with G50 KO(BAC) virus and wt γHV68(BAC) virus at 0.1 PFU of virus per cell. Total virus was collected at different times p.i. and analyzed for virus production by plaque assay on 14.29 cells (Fig. 4B). Mock-infected samples did not display any CPE up to 14 days p.i. wt γHV68(BAC) reached 100% CPE by 2 days p.i. on s27 cells and by 4 days on NIH 3T12 cells, with a maximum titer of ca. 2 × 107 to 6 × 107 PFU/ml by 4 days p.i. for both cell lines. G50 KO(BAC)-infected NIH 3T12 cells did not display any CPE, and there was no detectable virus production by day 14 p.i. Importantly, G50 KO(BAC)-infected s27 cells reached 100% CPE and maximal viral titers of ca. 2 × 107 PFU/ml, similar to wt γHV68(BAC) levels, by 8 days p.i. This represents a ca. 300-fold increase compared to input G50 KO(BAC) virus in an assay that measures multiple rounds of infection. Plaque assay of G50 KO(BAC)-infected s27 samples on NIH 3T12 cells failed to detect the presence of any virus that had restored wt gene 50 in the G50 KO(BAC) virus, thus demonstrating that the high levels of virus production in the gene 50-expressing s27 cells were due to trans-complementation of the G50 KO(BAC) defect by gene 50. This experiment provides unambiguous genetic evidence that the growth defect observed with the G50 KO(BAC) can be attributed to disruption of γHV68 gene 50. Thus, the latter analyses demonstrate that the expression of Rta from the spliced gene 50 transcript is both necessary and sufficient for growth of the G50 KO(BAC) virus.
It should be noted that the analyses of G50 KO(BAC) virus growth on NIH 3T12, 14.29, or s27 cell lines revealed that the titer of mutant virus after 1 h of incubation with the cell monolayer (Fig. 3 and 4B, 0-day time point) was nearly 100-fold lower than the titer of wt virus at this same time point. We have carefully assessed the amount of input virus in these assays by again determining the titers of the virus stocks used, as well as determining the titers of the virus dilutions used to infect the cell monolayers (data not shown). None of these analyses revealed the source of this difference between mutant and wt virus titers at this early time point. It is formally possible that mutant virus grown on gene 50-complementing cell lines has altered kinetics of adsorption, penetration, and/or uncoating. Future characterization of the G50 KO(BAC) mutant may reveal the basis for this discrepancy.
Absence of viral DNA replication and late gene expression in cells infected with gene 50-null γHV68.
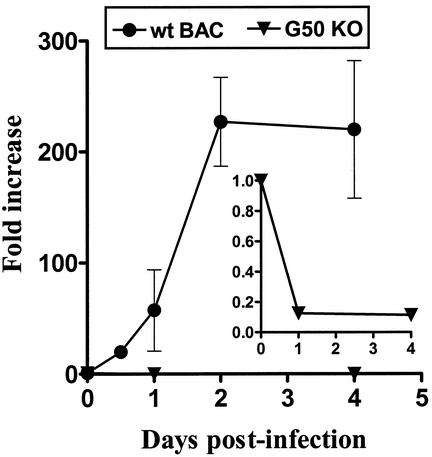
To gain insights into the basis of the replication defect in the G50 KO(BAC) virus, we assessed viral DNA replication in NIH 3T12 cells infected with either G50 KO(BAC) or wt γHV68(BAC) virus. Cells were either mock infected or infected at an MOI of 1.0, and DNA was isolated from cell pellets at different times p.i. To assess the amount of viral DNA present in each sample, we developed a quantitative real-time PCR assay with primers specific for the viral genome (gene 50) or a cellular gene (GAPDH) (Fig. 5). GAPDH levels were monitored to ensure that the DNA amounts analyzed by real-time PCR were consistent between time points and between the wt γHV68(BAC) and G50 KO(BAC) viruses (data not shown). The levels of viral DNA present at 1 h p.i. (Fig. 5, 0-day time point) were very similar between G50 KO(BAC) and wt γHV68(BAC) viruses (1.34 × 104 and 5.76 × 103 viral DNA copies per ng of DNA, respectively). Additionally, agarose gel electrophoresis demonstrated that the PCR products were of the expected sizes (data not shown). Real-time PCR analyses revealed that, as expected, wt viral genome copy numbers increased as infection progressed, reaching a maximum increase of >200-fold (compared to time zero) at 2 days p.i., which coincided with 100% CPE of the cell monolayer. In contrast, we could not detect replication of the G50 KO(BAC) virus over the 4-day time course of the experiment, and viral DNA copy numbers actually decreased ca. 10-fold by 24 p.i. (Fig. 5, inset). Thus, as expected, the G50 KO(BAC) virus is deficient in viral DNA replication.
FIG. 5.
G50 KO is deficient in viral DNA replication. The figure shows quantitation of viral DNA replication in NIH 3T12 cells infected with wt γHV68(BAC) virus or G50 KO virus at 1 PFU per cell. Cells were harvested at the indicated times, and the copy number of viral genomes was determined by a real-time PCR assay, as described in Materials and Methods. Copy number was normalized per nanogram of whole-cell DNA. Data are represented as the fold increase in viral copy number per nanogram of DNA compared to time zero (which was arbitrarily defined as 1.0). The inset shows on an expanded scale the decrease in viral DNA in NIH 3T12 cells infected with G50 KO virus. Data are compiled from two independent experiments.
We extended the above study by analyzing viral protein expression in NIH 3T12 and 14.29 cells infected with G50 KO(BAC) or wt γHV68(BAC) virus at 1 PFU per cell in the presence or absence of PAA, an inhibitor of herpesvirus DNA polymerase. Expression of viral proteins was analyzed by immunoblotting of cell lysates recovered at 24 h p.i., with a rabbit polyclonal anti-γHV68 antiserum generated by infecting rabbits with γHV68 (Fig. 6). In wt γHV68(BAC)-infected cells, the antiserum detected several lytic antigens (Fig. 6, lanes 3 and 9), whose expression was dependent on viral replication (Fig. 6, lanes 4 and 10), indicating that they are late viral proteins. There was no detectable expression of these late viral proteins in G50 KO(BAC) virus-infected 3T12 cells (Fig. 6, lane 2), but their expression was apparent in G50 KO(BAC) virus-infected 14.29 cells (Fig. 6, lanes 7 and 8). Notably, there was no detectable viral protein expression observed in G50 KO(BAC)-infected NIH 3T12 cells upon overexposure of the blot, or with longer periods of infection (up to 8 days; data not shown). This analysis is consistent with the observed viral DNA replication defect and suggests that there is a global defect in late viral gene expression associated with the loss of gene 50 function.
DISCUSSION
Previously, Wu et al. reported that γHV68 expression of gene 50 is sufficient to drive γHV68 reactivation in the latently infected S11 B-lymphoma cell line (49). Recently, these researchers have also shown that dominant-negative forms of γHV68 gene 50 can inhibit virus replication (48). The analyses presented here confirm and extend these observations. The genetic proof for the requirement of a gene in a particular process is provided by analysis of the process when the gene is disrupted completely (i.e., a null gene mutant). We show here that a genetic lesion in γHV68 gene 50 renders the virus completely defective for virus production in NIH 3T12 cells and that complementation of this defect in trans by γHV68 Rta restores virus production to wt levels. The mutant virus is incapable of viral DNA replication, and late viral protein expression is severely attenuated in G50 KO(BAC)-infected cells, even at a high MOI. Therefore, we conclude that gene 50 is essential for γHV68 replication in vitro.
The question still remains whether there are other genes in γHV68 that are critical for initiation of the lytic cycle. As mentioned previously, γHV68 does not appear to encode a homolog of the EBV BZLF1 gene, an immediate-early gene with a demonstrated importance for lytic cycle initiation in EBV. Recently, it was shown that, in addition to gene 50, the genes K3, M8, and ORF 73 are expressed with immediate-early kinetics (26), although γHV68 ORF 73 is likely a latency-associated gene (41). Further studies are required to identify other candidate immediate-early genes and determine whether they play a role in initiating the viral lytic cascade. Importantly, analysis of viral gene expression in the absence of functional gene 50 expression should aid in the identification of other immediate-early genes.
Currently γHV68 infection of mice is the only tractable animal model for dissecting host and viral factors involved in regulating in vivo latency and reactivation. To date, there has been no report of the in vivo characterization of a null mutant in a gammaherpesvirus immediate-early gene. The generation of a null mutation in γHV68 gene 50, a conserved immediate-early gene, will allow for the first time an investigation of the role of gammaherpesvirus immediate-early genes for the maintenance of latency in vivo, as well as reactivation from latency. In addition, it will be of interest to determine whether infection of immunocompromised mice with replication-defective γHV68 mutants leads to an enhanced ability to induce lymphomas and other tumors due to the absence of reactivation-associated disease.
Acknowledgments
S. H. Speck was supported by NIH grants CA43143, CA52004, CA58524, and CA87650. H. W. Virgin IV was supported by NIH grants CA74730, HL60090, and AI39616.
We thank members of the Speck and Virgin laboratories, as well as David Leib and members of his laboratory, for helpful comments and advice on this research.
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2001. Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. J. Virol. 75:5692-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 24:468. [PubMed] [Google Scholar]
- 4.Blaskovic, D., D. Stanekova, and J. Rajcani. 1984. Experimental pathogenesis of murine herpesvirus in newborn mice. Acta Virol. 28:225-231. [PubMed] [Google Scholar]
- 5.Cardin, R. D., J. W. Brooks, S. R. Sarawar, and P. C. Doherty. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J. Exp. Med. 184:863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 7.Clambey, E. T., H. W. Virgin, and S. H. Speck. 2000. Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency. J. Virol. 74:1973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehtisham, S., N. P. Sunil-Chandra, and A. A. Nash. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J. Virol. 67:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 12.Francis, A., T. Ragoczy, L. Gradoville, L. Heston, A. El Guindy, Y. Endo, and G. Miller. 1999. Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of early lytic cycle genes and synergy with the Epstein-Barr virus R transactivator. J. Virol. 73:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin, D. J., M. S. Walters, P. G. Smith, M. Thurau, H. Fickenscher, and A. Whitehouse. 2001. Herpesvirus saimiri open reading frame 50 (Rta) protein reactivates the lytic replication cycle in a persistently infected A549 cell line. J. Virol. 75:4008-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruffat, H., S. Portes-Sentis, A. Sergeant, and E. Manet. 1999. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) encodes a homologue of the Epstein-Barr virus bZip protein EB1. J. Gen. Virol. 80:557-561. [DOI] [PubMed] [Google Scholar]
- 16.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 17.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, S., I. V. Pavlova, H. W. Virgin, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 21.Mistrikova, J., and D. Blaskovic. 1985. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 29:312-317. [PubMed] [Google Scholar]
- 22.Paulson, E., J. Fingeroth, J. Yates, and S. Speck. 2002. Methylation of the EBV genome and establishment of restricted latency in low-passage EBV-infected 293 epithelial cells. Virology 299:109-121. [DOI] [PubMed] [Google Scholar]
- 23.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajcani, J., D. Blaskovic, J. Svobodova, F. Ciampor, D. Huckova, and D. Stanekova. 1985. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 29:51-60. [PubMed] [Google Scholar]
- 25.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 75:4955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seaman, W. T., D. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus 1. Virology 263:436-449. [DOI] [PubMed] [Google Scholar]
- 29.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 30.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 33.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 34.Svobodova, J., D. Blaskovic, and J. Mistrikova. 1982. Growth characteristics of herpesviruses isolated from free living small rodents. Acta Virol. 26:256-263. [PubMed] [Google Scholar]
- 35.Swenson, J. J., A. E. Mauser, W. K. Kaufmann, and S. C. Kenney. 1999. The Epstein-Barr virus protein BRLF1 activates S phase entry through E2F1 induction. J. Virol. 73:6540-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson, J. M., and W. A. Parrott. 1998. pMECA: a cloning plasmid with 44 unique restriction sites that allows selection of recombinants based on colony size. BioTechniques 24:922-928. [DOI] [PubMed] [Google Scholar]
- 37.Usherwood, E. J., J. P. Stewart, and A. A. Nash. 1996. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J. Virol. 70:6516-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Santen, V. L. 1991. Characterization of the bovine herpesvirus 4 major immediate-early transcript. J. Virol. 65:5211-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Santen, V. L. 1993. Characterization of a bovine herpesvirus 4 immediate-early RNA encoding a homolog of the Epstein-Barr virus R transactivator. J. Virol. 67:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virgin, H. W., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Virgin, H. W., R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weck, K. E., A. J. Dal Canto, J. D. Gould, A. K. O'Guin, K. A. Roth, J. E. Saffitz, S. H. Speck, and H. W. Virgin. 1997. Murine gamma-herpesvirus 68 causes severe large-vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus-induced vascular disease. Nat. Med. 3:1346-1353. [DOI] [PubMed] [Google Scholar]
- 43.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73:4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitehouse, A., M. Cooper, K. T. Hall, and D. M. Meredith. 1998. The open reading frame (ORF) 50a gene product regulates ORF 57 gene expression in herpesvirus saimiri. J. Virol. 72:1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zacny, V. L., J. Wilson, and J. S. Pagano. 1998. The Epstein-Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J. Virol. 72:8043-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]