Abstract
The human papillomavirus oncoproteins E6 and E7 promote cell proliferation and contribute to carcinogenesis by interfering with the activities of cellular tumor suppressors. We used a small interfering RNA molecule targeting the E7 region of the bicistronic E6 and E7 mRNA to induce RNA interference, thereby reducing expression of E6 and E7 in HeLa cells. RNA interference of E6 and E7 also inhibited cellular DNA synthesis and induced morphological and biochemical changes characteristic of cellular senescence. These results demonstrate that reducing E6 and E7 expression is sufficient to cause HeLa cells to become senescent.
Human papillomavirus (HPV) infection contributes to the development of over 90% of anogenital cancers (2). HPVs are oncogenic because they lack the DNA and RNA polymerases required for the viral life cycle and therefore must induce production of host cell replication proteins by driving the host cell into a proliferative state. The HPV oncoproteins E6 and E7 direct the progression of the cell cycle and play a major role in HPV-induced carcinogenesis by interfering with host cell regulatory proteins (14). The E7 protein inhibits the function of the retinoblastoma (Rb) family proteins, leading to cell cycle progression (13). In response to abnormal E7-driven proliferation, the host cell triggers apoptosis or senescence by activating the p53 tumor suppressor. To defeat this antiproliferative host cell response, HPVs produce the E6 protein, which targets p53 for degradation (12).
While E6 and E7 mask the activities of p53 and Rb proteins, HPV-induced malignancies generally maintain wild-type copies of the p53 and Rb genes. This suggests that reducing E6 and E7 expression in cells transformed with HPV would restore the activity of endogenous tumor suppressors and, thus, prevent proliferation of cells transformed with E6 and E7. Several laboratories have shown that the introduction of the HPV transcriptional regulator E2 into cells transformed with HPV induces either apoptosis or senescence, at least in part by inhibiting E6 and E7 expression (4, 7, 8, 9, 17). These studies raise the question of whether E2 expression is necessary to inhibit growth or whether a reduction in E6 and E7 expression alone would be sufficient. In order to determine the effects of reducing E6 and E7 levels in cells without the introduction of additional proteins, we used RNA interference (RNAi).
RNAi is a process in which double-stranded RNA (dsRNA) homologous to a specific mRNA dramatically inhibits the expression of the protein made from that mRNA (3, 16). It has been shown that small interfering dsRNA of approximately 21 bases (siRNA) potently induces RNAi in mammalian cells without provoking a nonspecific interferon response (6). Using RNAi to specifically reduce expression of E6 and E7, we found that inhibiting E6 and E7 expression alone is sufficient to induce senescence in HeLa cells.
siRNA targeting E7 reduces expression of the E6 and E7 viral oncogenes in HeLa cells.
To inhibit E6 and E7 expression in HeLa cells (a cell line transformed with HPV18), we used 19-nucleotide duplex RNAs with 2-nucleotide deoxythymidine 3′ overhangs homologous to bases 142 to 160 of the HeLa E7 coding sequence (Dharmacon Inc., or Xeragon Inc.). We hypothesized that a single siRNA species could be used to knock down the expression of both E6 and E7 because they are expressed bicistronically (18). Control cells were transfected with siRNA homologous to bases 984 to 1002 of the enhanced green fluorescent protein coding sequence (Clontech). Both siRNA sequences were screened against the human genome by using a BLAST search to avoid unintentional silencing of host cell genes. The HeLa cells were transfected with 4 μl of Oligofectamine (Invitrogen Inc.) per well of a 6-well plate. Otherwise, the procedure was as recommended by Dharmacon. The transfection efficiency under these conditions is approximately 80% (data not shown). Cultures were retransfected every 4 days to avoid overgrowth of cells that escaped transfection. To compare E6 and E7 mRNA expression levels in cells treated with E7 and those in cells treated with control siRNA, cytoplasmic RNA was prepared and analyzed by Northern blotting and hybridization to a probe complementary to the E7 coding sequence.
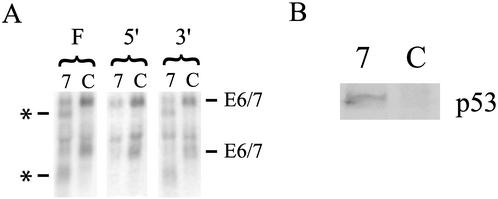
We observed a fourfold reduction in the levels of full-length cytoplasmic E6 and E7 mRNA in cells transfected with E7 siRNA compared with those in controls, when the levels were normalized to β-actin expression (Fig. 1A). This result, combined with the efficiency of transfection, suggests that more than 90% of E6 and E7 mRNA is eliminated in successfully transfected cells. Northern blot analysis of RNA from E7 siRNA-treated cells also revealed two new bands, each about 600 to 700 bases smaller than the full-length E6 and E7 mRNA species. After probing the blot with sequences corresponding to the E7 coding sequence on either side of the expected RNAi cleavage site, we found that these bands represent the 3′ cleavage products of the RNAi process (Fig. 1A). Given that most current models for the mechanism of RNAi suggest that the target mRNA should be rapidly and completely degraded (11), we were surprised to find that the 3′ products of RNAi-induced cleavage persisted in the cytoplasm. Upon review of the literature, we did not find other reports of this phenomenon. It is possible that the production of stable RNAi degradation products occurs only with a small subset of RNA targets. However, in the vast majority of studies of RNAi in mammalian cells, reverse transcriptase PCR is used to determine the level of RNA. Depending on the positions of the primers used, this method could easily miss the presence of mRNA fragments. Thus, the persistence of RNAi cleavage products may be more common than previously thought, which could have implications for the interpretation of future RNAi-based work.
FIG. 1.
Effects of E7-targeted siRNA on E6, E7, and p53 expression. (A) Northern analysis of cytoplasmic RNA isolated from HeLa cells transfected with either E7 (7) or control (C) siRNA and hybridized with a probe complementary to the full E7 coding sequence (F), the 5′ end of the E7 coding sequence (5′), or the 3′ end of the E7 coding sequence (3′). The positions of the major bands of the full-length E6 and E7 mRNA at 3.5 and 1.6 kb are labeled (E6/7). The positions of bands representing presumed 3′ RNAi degradation products are labeled with an asterisk. (B) Western analysis of p53 protein expression using extracts from HeLa cells transfected with either E7 (7) or control (C) siRNA and probed with an antibody to p53. The position of this band corresponded to a molecular mass of approximately 53 kDa.
Although large fragments of E6 and E7 mRNA remain stable in the cytoplasm, expression of E6 and E7 proteins should still be reduced. siRNA-induced cleavage of E6 and E7 mRNA in the region of homology would disrupt the E7 coding sequence and separate the E6 coding sequence from the 3′ poly(A) tail, which is generally required for efficient translation. To confirm that the level of E6 protein was reduced by E7 siRNA, despite the potentially intact E6 coding sequence, we assessed E6 protein expression. Because E6 is expressed in HeLa cells at levels too low to be detected by Western blotting, we chose to examine the expression of the p53 protein. p53 is targeted for degradation by E6 and thus can serve as a marker of E6 expression. Expression of p53 was considerably higher in cells transfected with E7 siRNA than in controls (Fig. 1B). The increase in p53 protein levels cannot be explained by the inhibition of E7 expression because a reduction in E7 alone would result in lower levels of p53 protein (13). Therefore, the presence of more p53 protein in cells treated with E7 siRNA is likely to reflect reduced E6 protein levels.
E7 siRNA inhibits DNA replication.
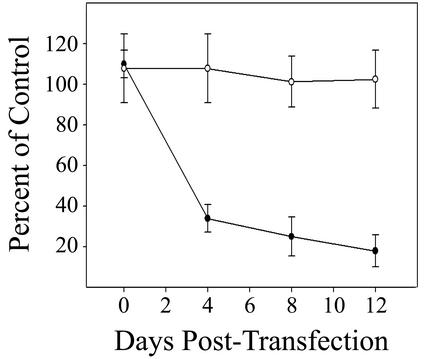
To determine rates of DNA synthesis for E7 siRNA-treated cells and controls, sparsely plated cells were incubated with 5 μCi of [3H]thymidine/ml for 4 h prior to harvesting, washed with cold 10% trichloroacetic acid, and lysed with cold 0.2 N NaOH. The amount of radioactivity incorporated into cellular DNA was measured in a scintillation counter. The cells were assayed prior to transfection and at various time points after the initial transfection. Introduction of E7 siRNA significantly reduced cellular DNA replication. There was a notable decrease in DNA synthesis in E7 siRNA-treated cells by 4 days after the first transfection (Fig. 2). The rate of replication in treated cells continued to decline for 12 days after the first transfection to 18% of the control replication rate. We found that E7 siRNA had no effect on the growth of an HPV-negative cervical cancer cell line (C33A), confirming that E7 siRNA inhibited growth specifically by interfering with HPV gene expression (Fig. 2). The reduction in HeLa cell DNA synthesis induced by E7 siRNA was not as complete as that induced by E2 expression (9). This is most likely because transfection with siRNA is less efficient than infection with E2-expressing viruses. However, it is possible that E2 provides some additional growth-suppressive activity that is independent of a reduction in E6 and E7 expression (4, 17).
FIG. 2.
Effect of E7 siRNA on cellular DNA synthesis. DNA synthesis was measured by [3H]thymidine incorporation in HeLa cells (closed circles) and C33A cells (open circles) before and after transfection. The counts per minute in cells transfected with E7 siRNA is expressed as a percentage of the counts per minute in cells transfected with control siRNA. Error bars represent 2 standard deviations.
E6 and E7 siRNA induces senescence in HeLa cells.
Based on previous work, we hypothesized that the observed reduction in replication was most likely to be due to either senescence or apoptosis (4, 9). To determine whether the decrease in proliferation was due to apoptosis or any other type of cell death, transfected cells were assayed for viability by staining with trypan blue. In both cells transfected with E7 siRNA and controls transfected with green fluorescent protein siRNA, more than 97% of cells were viable through at least 14 days posttransfection, the last time point tested (data not shown).
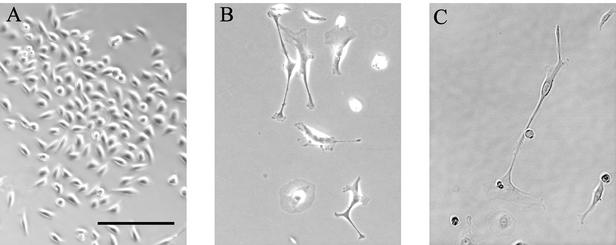
The absence of cell death suggested that the cells might have become senescent. This hypothesis was confirmed when observation of the morphology of E7 siRNA-treated cells revealed changes characteristic of senescent keratinocytes (9, 15). Subtle morphological changes were first apparent 6 to 7 days after the original transfection. By 10 to 12 days posttransfection, there were striking morphological differences between E7 siRNA-treated cells and control cells (Fig. 3). The E7 siRNA-treated cells were larger and flatter than controls. They were frequently multinucleate and had long cytoplasmic projections. They were also more dispersed than control cells.
FIG. 3.
Effect of E7 siRNA on HeLa cell morphology. Cells transfected with control (A) or E7 (B and C) siRNA were photographed 14 days after the original transfection. The scale bar represents 200 μm for all panels.
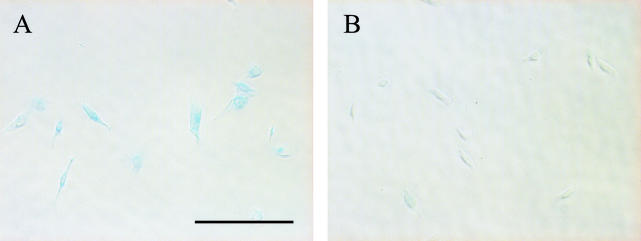
To further demonstrate that these cells had become senescent, the cells were stained as described by Dimri et al. for senescence-associated β-galactosidase (SAβ-gal) activity, a well-established marker of senescence in keratinocytes (5). The majority of E7 siRNA-treated cells showed intense blue staining, reflecting high levels of SAβ-gal activity (Fig. 4). In contrast, control cells did not show significant staining. The cells stained for SAβ-gal activity did not display the characteristic senescent morphology because they were replated 24 h prior to staining. We found that E7 siRNA induced neither morphological changes nor SAβ-gal activity in the HPV-negative C33A cells (data not shown).
FIG. 4.
E7 siRNA induces SAβ-gal expression. Cells transfected with E7 (A) or control (B) siRNA were stained for SAβ-gal activity and photographed 12 days after the original transfection. The scale bar represents 200 μm for both panels.
Reducing E6 and E7 levels by RNAi causes HeLa cells to undergo dramatic changes that are characteristic of senescent cells: reduction in DNA replication, enlargement and flattening of cells, and induction of the expression of SAβ-gal. This work demonstrates that a reduction in E6 and E7 expression alone is sufficient to induce senescence in HeLa cells. Despite the many mutations HeLa cells have undergone, they have retained an intrinsic, functional pathway that results in senescence and does not require introduction of exogenous proteins such as E2. As indicated by the failure of control cells treated with siRNA targeting green fluorescent protein to show any of these changes, senescence is not simply a nonspecific reaction to dsRNA. The absence of any effect of E7 siRNA on an HPV-negative cell line demonstrates that it is acting specifically by interfering with HPV gene expression.
We found that the E7 cognate siRNA species reduced expression of both E6 and E7 as evidenced by Northern and Western blotting results. In contrast, work was recently published reporting that siRNAs could be used to specifically target either E6 or E7 in cell lines transformed with HPV16 (10). Because of alternate splicing, it is possible to target E6 expression without completely eliminating E7 expression. However, it is perplexing to us that the authors report targeting E7 without affecting E6 because, in HPV16, the E6 coding sequence is always expressed on an mRNA with the entire E7 coding sequence (1). Of note, in the experiments that demonstrated the specificity of E7 siRNA, reverse transcription-PCR was used to measure RNA levels, which could give misleading results.
Finally, the work presented here suggests the possibility of using RNAi to treat or even prevent HPV-related disease; a single molecule could be used to silence expression of the two most potent HPV oncogenes. Several features of RNAi make it attractive for use in a clinical setting. Perhaps most importantly, RNAi is exquisitely sequence specific. Thus, an RNAi-based treatment should reduce expression of targeted viral genes without disrupting expression of normal host cell genes. In addition, the potency of interfering RNA combined with the relatively accessible location of most anogenital carcinomas might be expected to minimize problems of drug delivery.
Acknowledgments
We are grateful to Daniel DiMaio, Edward Goodwin, and Yin-Xiong Li for helpful discussions. We thank Joshua Frederick for advice about thymidine incorporation assays and Rachel Lovingood for help with preliminary experiments. We also thank the Duke University Cancer Center Cell Culture Facility for excellent technical assistance.
This work was supported by a grant from the Children's Miracle Network. A.H.S.H. was supported by the Duke University Medical Scientist Training Program. K.A.A. was partially supported by R01 CA081214 from the National Institutes of Health.
Footnotes
Dedicated to the memory of Colleen Haberstroh.
REFERENCES
- 1.Baker, C., and C. Calef. 1995. Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences, p. III-3-III-19. [Online.] Los Alamos National Laboratory, Los Alamos, N.Mex. http://hpv-web.lanl.gov/stdgen/virus/hpv/compendium/htdocs/COMPENDIUM_PDF/95PDF/3/maps.pdf.
- 2.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 3.Cullen, B. R. 2002. RNA interference: antiviral defense and genetic tool. Nat. Immunol. 3:597-599. [DOI] [PubMed] [Google Scholar]
- 4.Desaintes, C., C. Demeret, S. Goyat, M. Yaniv, and F. Thierry. 1997. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 16:504-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, and O. Pereira-Smith. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 7.Francis, D. A., S. I. Schmid, and P. M. Howley. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 74:2679-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodwin, E. C., L. K. Naeger, D. E. Breiding, E. J. Androphy, and D. DiMaio. 1998. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J. Virol. 72:3925-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin, E. C., E. Yang, C. J. Lee, H. W. Lee, D. DiMaio, and E. S. Hwang. 2000. Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. USA 97:10978-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, M., and J. Milner. 2002. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 21:6041-6048. [DOI] [PubMed] [Google Scholar]
- 11.Kitabwalla, M., and R. M. Ruprecht. 2002. RNA interference—a new weapon against HIV and beyond. N. Engl. J. Med. 347:1364-1367. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani, F., and L. Banks. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874-7887. [DOI] [PubMed] [Google Scholar]
- 13.Munger, K., J. R. Basile, S. Duensing, A. Eichten, S. L. Gonzalez, M. Grace, and V. L. Zacny. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 14.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norsgaard, H., B. F. Clark, and S. I. Rattan. 1996. Distinction between differentiation and senescence and the absence of increased apoptosis in human keratinocytes undergoing cellular aging in vitro. Exp. Gerontol. 31:563-570. [DOI] [PubMed] [Google Scholar]
- 16.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 17.Webster, K., J. Parish, M. Pandya, P. L. Stern, A. R. Clarke, and K. Gaston. 2000. The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. J. Biol. Chem. 275:87-94. [DOI] [PubMed] [Google Scholar]
- 18.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]