Abstract
The large cytomegalovirus (CMV) US22 gene family, found in all betaherpesviruses, comprises 12 members in both human cytomegalovirus (HCMV) and murine cytomegalovirus (MCMV). Conserved sequence motifs suggested a common ancestry and related functions for these gene products. Two members of this family, m140 and m141, were recently shown to affect MCMV replication on macrophages. To test the role of all US22 members in cell tropism, we analyzed the growth properties in different cell types of MCMV mutants carrying transposon insertions in all 12 US22 gene family members. When necessary, additional targeted mutants with gene deletions, ATG deletions, and ectopic gene revertants were constructed. Mutants with disruption of genes M23, M24, m25.1, m25.2, and m128 (ie2) showed no obvious growth phenotype, whereas growth of M43 mutants was reduced in a number of cell lines. Genes m142 and m143 were shown to be essential for virus replication. Growth of mutants with insertions into genes M36, m139, m140, and m141 in macrophages was severely affected. The common phenotype of the m139, m140, and m141 mutants was explained by an interaction at the protein level. The M36-dependent macrophage growth phenotype could be explained by the antiapoptotic function of the gene that was required for growth on macrophages but not for growth on other cell types. Together, the comprehensive set of mutants of the US22 gene family suggests that individual family members have diverged through evolution to serve a variety of functions for the virus.
Herpesviruses are large and complex DNA viruses, widely found in nature. Human cytomegalovirus (HCMV), an important human pathogen, defines the betaherpesvirus family. Mouse CMV (MCMV) and rat CMV serve as biological model systems for HCMV. HCMV, MCMV, and rat CMV display the largest genomes among the herpesviruses (13, 34, 43). These genomes are essentially colinear over the central 180 kb of the 230-kb genomes. Betaherpesviruses, which include the CMVs as well as human herpesviruses 6 and 7, differ from alpha- and gammaherpesviruses by the presence of additional gene families such as the US22 gene family, which are mainly clustered at the ends of the genome (29, 30).
The US22 family was first described in HCMV (13). This gene family comprises 12 members in both HCMV and MCMV and 11 in rat CMV. Members of the US22 gene family are characterized by stretches of hydrophobic and charged residues as well as up to four conserved sequence motifs which are specific for betaherpesviruses. Motif I differs between the HCMV US and UL family members (30). In MCMV, m128 and m139 to m143 share the HCMV US-like motif I, while M23, M24, m25.1, m25.2, M36, and M43 share UL-like motif I. Motifs I and II have consensus sequences, while motifs III and IV are less well defined but have stretches of nonpolar residues (18, 24). The m139 to m141 genes contain all four of these motifs, whereas m142 and m143 (and IRS1/TRS1 of HCMV) lack motif II. In addition, m139, m140, m142, and m143 each have an acidic domain, common to herpesvirus transcriptional activators and specifically to MCMV immediate-early proteins 1 and 2 (11, 28). Several US22 gene products represent viral tegument components (1, 33, 36).
The functions of most of the US22 genes are unknown. The US22 genes TRS1/IRS1 and UL36 (HCMV) and m128, m142, and m143 (MCMV) and positional homologs to UL36 to UL38 and UL43 of human herpesvirus 6 (31) are transcribed with immediate-early kinetics. For all of these except m142 and m143, a transcriptional transactivation function was described (11, 14, 15, 22, 31, 37, 40). Besides studies on the ie2 gene product of m128, which was shown to be dispensable for growth of MCMV in vitro and in vivo (11), biological properties for some early US22 homolog genes in MCMV have been defined (12, 19, 20). A deletion mutant encompassing genes m137 through m143 could not be wild-type MCMV, indicating that the m142 or m143 gene or both genes might be essential for virus replication (12). Deletion of m140 and m141 had no effect on replication of MCMV in fibroblasts but impaired the ability of the virus to replicate in macrophages in vitro and in the spleens of mice.
The report on the combined failure of macrophage growth in vitro and altered tissue type distribution in vivo of mutants RV7 (encompassing genes m137 through m141) and RV10 (encompassing US22 family genes m139 to m141) (12, 20) prompted us to study all US22 gene family members with respect to this phenotype. It was reported that the macrophage growth phenotype was a property not of m139 but of m140 and m141, whose products acted in a cooperative and interdependent manner (19).
Furthermore, Liu and colleagues reported on an M43 mutant with a transposon insertion at codon 313 of the M43 open reading frame (ORF) (48). This mutant grew like the wild type in fibroblasts and was attenuated in salivary glands but not in other organs in vivo. Mutants of a number of US22 genes were shown to grow like wild-type virus in fibroblast cells but, except for the m139, m140, and m141 ORFs, were not tested for growth properties in macrophages (11, 33, 42, 48).
A systematic approach to an entire herpesvirus gene family has only been a theoretical option due to the time-consuming labor involved in constructing, isolating, confirming, and testing individual mutants. We recently pioneered the construction of herpesvirus genomes as infectious bacterial artificial chromosomes (BAC) in Escherichia coli and their targeted mutagenesis (2, 5, 27, 46, 47). We further described a one-step procedure for random insertional mutagenesis of herpesvirus BACs, with a Tn1721-based transposon system (9). The effectiveness of the latter method has been tested for MCMV (9), HCMV (21), and the murine gammaherpesvirus 68 (O. Fuchs, C. Menard, U. H. Koszinowski, and M. Wagner, Abstr. 26th International Herpesvirus Workshop, abstr. 3.26, 2001). The transposon insertion site can be determined by direct sequencing, and infectious virus can be recovered after transfection of permissive cells with characterized mutant genomes.
We decided to start a comparative analysis of the complete US22 gene family. For this purpose we had to limit the number of constructs carried out for each gene of all the 12 US22 gene family members, and we restricted testing to fibroblast, macrophage, and endothelial cells. We characterized about 40 mutant genomes in more detail. For two genes of the US22 family, m142 and m143, essentiality was proven. For the other 10 genes, M23, M24, m25.1, m25.2, M36, M43, m128, m139, m140, and m141, viable transposon insertion mutants were isolated. Our results show that five members of the US22 gene family, M36, M43, and m139 to m141, affect macrophage tropism. The gene products of m139, m140, and m141 apparently interact at the protein level, which explains their common phenotype. The effect of M36 on viral growth in macrophages was shown to be due to the antiapoptotic function of the gene product.
MATERIALS AND METHODS
Cells and viruses.
Murine embryo fibroblasts (MEFs) from BALB/c mice were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). NIH 3T3 fibroblasts (ATCC CRL1658) and J774-A1 macrophages (ATCC TIB67) were propagated in DMEM supplemented with 5% newborn calf serum. SVEC4-10 endothelial cells (EC) (ATCC CRL2181), a simian virus 40-transformed cell line derived from lymph node vessels, were propagated in DMEM supplemented with 10% FCS. IC-21 macrophages (ATCC TIB186), a simian virus 40-transformed peritoneal macrophage cell line, and the M2-10B4 bone marrow stromal cell line (ATCC CRL1972) were propagated in RPMI medium supplemented with 10% FCS. Fresh peritoneal exudate cells (PEC) were obtained from 8-week-old C57BL/6 mice by washing the peritoneal cavity with medium 5 days after intraperitoneal injection of 3 ml of 3% thioglycolate. The exudate cells were enriched to 90% of peritoneal macrophage cells by the thioglycolate treatment.
All MCMV mutants were generated from the parental MCMV BAC pSM3fr, which contains the complete MCMV Smith strain genome. Virus MW97.01, reconstituted from pSM3fr, was shown to have wild-type properties in vitro and in vivo (46). In all experiments, we used the wild-type virus MW97.01 except for growth on J774-A1 macrophages, PEC, and SVEC4-10 endothelial cells, where the m152 transposon mutant was used as the control. All transposon mutants expressed green fluorescent protein (GFP). Strong GFP expression by MCMV may affect virus growth. m152 does not belong to the US22 gene family, and its deletion does not affect growth in vitro (25, 44). Therefore, we used this mutant with a transposon insertion 300 bp downstream to the native ATG (nucleotide 211077) of gene m152, which expresses the GFP, as a control. For all experiments, two transposon insertion mutants for each US22 gene were tested whenever more than one transposon insertion mutant was found in the library of MCMV transposon insertion mutants.
Wild-type and mutant viruses were reconstituted by transfection of BAC DNA into NIH 3T3 cells with the Superfect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Briefly, 2 to 3 μg of BAC DNA was incubated with 100 μl of medium without serum and 10 μl of Superfect transfection reagent. The mixture was added to approximately 3 × 105 NIH 3T3 cells. Cells were washed with phosphate-buffered saline 4 h later, cultured with fresh medium, and passaged when necessary. Plaques usually appeared 4 to 6 days after transfection. All virus stocks were prepared on M2-10B4, and virus titers were determined on MEFs by plaque assay or on NIH 3T3 cells with the TCID50 (median tissue culture infectious dose) method (4, 35).
Plasmids, transposon mutagenesis, and screening for transposon insertions into US22 family genes.
The pCR3 vector (Invitrogen, Karlsruhe, Germany) was used to clone the M36 or hemagglutinin (HA)-tagged (N-terminal) M36 ORF (M36HA). The M36 and M36HA ORFs were amplified by PCR with primers 3′-M36 (5′-GAG TCT AGA CTA TCG ATA TCC CCG TGT CA-3′) and 5′-M36 (5′-CAG GAA TTC ATG TAT GAG CAA GAG GAA CA-3′) or 5′-M36HA (5′-CAG GAA TTC ATG TAC CCA TAC GAT GTT CCA GAT TAC GCG TAT GAG CAA GAG GAA CA-3′), respectively, and inserted between the XbaI and EcoRI sites of the multiple cloning site in pCR3, generating the pCR3-M36 and pCR3-M36HA vectors, respectively.
Transposon mutagenesis was performed as described elsewhere (7, 9) with the TnMax8 donor plasmid pTsTM8 (9) and the TnMax16 donor plasmid pTsTM16 (8), which is a pTsTM8 derivative containing the GFP gene under the control of the human CMV immediate-early promoter-enhancer from plasmid pEGFP-C1 (Clontech, Palo Alto, Calif.) described in detail elsewhere (7, 9). pTsTM8 or pTsTM16 and the MCMV BAC were maintained in E. coli DH10B (Invitrogen, Karlsruhe, Germany). About 2,000 and 2,600 bacterial clones were established in 96-well microtiter plates as libraries of uncharacterized MCMV::TnMax8 and MCMV::TnMax16 mutant genomes, respectively.
PCR screening of the MCMV::TnMax16 mutant library for mutants of US22 gene family members with transposon insertions at the N-terminal end of the gene was performed essentially as described previously (21). Briefly, to detect mutant genomes with transposon insertions at positions of interest, three rounds of PCR were performed with two transposon-specific primers in the same reaction, M13 forward (M13-for) (5′-GCC GCT GTA AAA CGA CGG CCA GT-3′) and reverse (M13-rev) (5′-GGC CGC AGG AAA CAG CTA TGA CC-3′) primers, which bind within the end of the transposon, and one gene-specific search primer designed to bind about 200 bp upstream of the start of the gene. The gene-specific search primers used in this study are listed in Table 1. The exact transposon insertion site in identified clones was determined by direct sequencing of the mutant BAC DNA with the M13-for and M13-rev primers as described previously (9).
TABLE 1.
Search primers used for identification of transposon insertions into pSM3fr
| Primer | Sequence |
|---|---|
| M23-for | 5′-TCT TCA GCG ACT TTG CCA C-3′ |
| M23-rev | 5′-CGA GCG GAT CGA TGA TGA AG-3′ |
| M24-for | 5′-GAG AAA GGA GGA AAA GTG GG-3′ |
| M24-rev | 5′-GAT CGA CGG TTC ATC GTT CG-3′ |
| m25-for | 5′-GTA ATT CTT GGG CCA GGT CAG C-3′ |
| m25-1-rev | 5′-GCA TTG TTC TCG TCA ATC CG-3′ |
| m25-2-rev | 5′-ACA ACC TCA CGC AGC TCA TCT C-3′ |
| M36-for | 5′-CTC CCG AGG AAG AGA TTG TG-3′ |
| M36-rev | 5′-CTG CTT GCG TCA AAA TGC TC-3′ |
| M43-for | 5′-CTC GTC CGT GCA GAG AAT CAT C-3′ |
| M43-rev | 5′-CTG ACA GCA TCG AGA ATC C-3′ |
| m128-for | 5′-GCT CGC TCG ATC CAT TCT TC-3′ |
| m128-rev | 5′-GGC GTG AAC ACG ATG TTC TTG-3′ |
| m139-for | 5′-CAG ATA CTT GGA CAG GTC CAC G-3′ |
| m139-rev | 5′-CTT GGA GAG GCA AGA GAG TCA C-3′ |
| m140-for | 5′-CAG GTG GTG GAT GCT GAA G-3′ |
| m140-rev | 5′-CTC CAC CTA GCT CAC TAA CAG C-3′ |
| m141-for | 5′-CTC GAA GCT CGA AGA GAA CAC C-3′ |
| m141-rev | 5′-CAT CAC ATA AAC CCC TCC CC-3′ |
| m142-for | 5′-ACA CAC ACA CCA ACC ACC CTT C-3′ |
| m142-rev | 5′-GGC GGG TGA CAT GAG AGA T-3′ |
| m143-for | 5′-CGA TAG ATA GAA GAT GCT GCC C-3′ |
| m143-rev | 5′-CAC CCG GAT GGT GTA AAA GAG-3′ |
| m152-for | 5′-TCC ACC TTC AAT CGT CCA CC-3′ |
| m152-rev | 5′-GGA TAC GGA GGA GAA TGT GGT G-3′ |
Site-directed mutagenesis of MCMV BAC.
For construction of the ΔM36, Δm139, ΔATG-m139, Δm143, and ΔATG-m142 mutant genomes, we used homologous recombination of linear PCR fragments with the MCMV BAC plasmid pSM3fr in E. coli as described in principle elsewhere (45). The linear fragments were generated by PCR with plasmid pACYC177 (NEB, Beverly, Mass.) (for the Δm139 and Δm143 mutant genomes) and plasmid pSLFRTKn (3) (for ΔM36 and the ΔATG mutant genomes), both of which contain the kanamycin resistance gene as the DNA template.
The following contiguous primers were used: 5′-ΔM36 (5′-TTT CTC CC CTC AC CCT CTC CGT CCC TTT CTT ATC CGT TTT CCC TCT ATC GTC GTG GAA TGC CTT CGA ATT C-3′) and 3′-ΔM36 (5′-GCT CAT TCT TTC GGG AAA GGG GTG GAG GAG GGT CGT TTG ACA GTG AAA GGA CAA GGA CGA CGA CGA CAA GTA A-3′) for ΔM36-MCMV, 5′-Δm139 (5′-CCT TGC CGC CGT CGA ACA TGT CCA TGG CGC GGA CGT AAC GAC GAT AGA AGT CGC GCG ATT TAT TCA ACA AAG CCA CG-3′) and 3′-Δm139 (5′-CAG ACC GTG AGT TGA CGG CGC CGG CGC CAG ACG GAG CAG ACA GAG AGA GAG AAG GGC CAG TGT TAC AAC CAA TTA ACC-3′) for Δm139-MCMV, 5′-Δm143 (5′-CGG TCG TGT AGC GGT ACT GCC GCT CTC GGA GGC ATT CGT GAC AAT CTC CCT CCG CCG ATT TAT TCA ACA AAG CCA CG-3′), and 3′-Δm143 (5′-AGC AGA GAG GTG GTT GCC TCG GCT CCG CTC CGC TTC GTC CGC CCG TCT CGT GCG CGG CCA GTG TTA CAA CCA ATT AAC C-3′) for the Δm143 mutant genome; 5′-ΔATG-m139 (5′-GGG GGA AGG CTC CTC TCG TCC ACG CCG CCG TAT TCT CCG AAC TTC TGG TCC GTC GTG GAA TGC CTT CGA ATT-3′) and 3′-ΔATG-m139 (5′-CGT GAG TTG ACG GCG CCG GCG CCA GAC GGA GCA GAC AGA GAG AGA GAA GGA CAA GGA CGA CGA CGA CAA GTA A-3′) for ΔATG-m139-MCMV, and 5′-ΔATG-m142 (5′-CTG GTC TCT GAA GTG ATC CGA TCG GAT CGC CGC GCA CAG GGC GTC CGT CGT GGA ATG CCT TCG AAT TC-3′) and 3′-ΔATG-m142 (5′-CCA CCC TTC TCC ACC CGT GTT CCC GCT GCC GCC CGT CGC CCT CGC CAC AAG GAC GAC GAC GAC AAG TAA-3′) for ΔATG-m142 mutant genome.
The PCR fragments were inserted by homologous recombination via the flanking 40- to 60-nucleotide homologies to the viral target sequences. Kanamycin-resistant clones were analyzed for correct insertion. BAC DNA was isolated from E. coli cultures with an alkaline lysis procedure (38) and purified with NucleoBond AX100 columns (Macherey-Nagel, Düren, Germany). For generation of ΔATG-m139 and ΔATG-m142, the kanamycin resistance cassette flanked by the minimal FLP recombinase recognition target (FRT) sites was excised by FLP recombinase. Here, the first two ATGs of ORF m139 (84 bp apart) were replaced with an 86-bp extraneous sequence, and the native ATG of ORF m142 was deleted. Correct mutagenesis was confirmed by restriction pattern analysis and sequencing.
For construction of the ΔATG-m142/m142E mutant genome to ectopically express wild-type m142, the pSM3fr BAC containing an FRT site and the GFP gene in the position of the nonessential gene m152 (pSM3fr/GFP) was first subjected to site-directed mutagenesis to delete the native ATG of m142 with the same PCR fragment used for generation of the ΔATG-m142 mutant genome in principle as described previously elsewhere (6). In a second mutagenesis step, a plasmid with a conditional origin of replication, a zeocin resistance gene, a 34-bp FRT site (to be published elsewhere), and additionally containing the wild-type m142 ORF under control of the human CMV immediate-early promoter-enhancer from plasmid pEGFP-C1 (Clontech, Palo Alto, Calif.), was inserted into the FRT site of ΔATG-m142/FRT, generating ΔATG-m142/m142E. The M36-HA virus, which contains an HA tag at the C terminus of the M36 ORF, was also constructed by homologous recombination of linear PCR fragments with the MCMV BAC plasmid pSM3fr in E. coli by methods described previously (10). The following primers were used for the PCR: 5′-M36-HA (5′-CTC CCC TCA CCC TCTC CGT CCC TTT CTT ATC CGT TTT CCC TCG TCG TGG AAT GCC TTC GAA TTC-3′) and 3′-M36-HA (5′-ATC GAG AGG AGG AGG GTC AAG CTC TTT AAG ATG ACA CGG GGA TAT CGA TAC CCA TAC GAT GTT CCA GAT TAC GCG TAG ACA AGG ACG ACG ACG ACA AGT AA-3′).
Determination of MCMV replication in fibroblasts, macrophages, and endothelial cells.
Replication of viruses in fibroblasts, macrophages, and endothelial cell lines was determined as follows. Approximately 1 × 105 to 5 × 105 cells in six-well plates were infected at a multiplicity of infection (MOI) of 0.1 for endpoint titration in NIH 3T3, IC-21, and SVEC4-10 cells or with an MOI of 5 in J774-A1 macrophages and PEC. After a 2-h incubation, the virus inoculum was removed, the cells were washed with phosphate-buffered saline, and fresh cell growth medium was added. At 6 days postinfection, supernatants were harvested and virus titers were determined on MEFs by plaque assay (35) or on NIH 3T3 cells with the TCID50 method (4). Growth of each virus was quantified at least twice and in triplicate.
Northern blot analysis.
NIH 3T3 cells were infected with the wild-type and mutant viruses at an MOI of 5 PFU/cell, and total RNA was harvested 24 h postinfection. Total RNA was isolated with the RNeasy kit (Qiagen, Hilden, Germany), and mRNA was harvested with the Oligotex direct mRNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The probe used to analyze m139 to m141 gene expression corresponds to a DNA fragment from nucleotide positions 196317 to 197220 of the MCMV genome. To analyze M35 and M37 gene expression, DNA fragments from nucleotide positions 46153 to 46737 and 49680 to 50385 of the MCMV genome, respectively, were used as probes. DNA probes were labeled with [α-32P]dCTP with the nick translation kit from Amersham (Amersham Biosciences Europe, Freiburg, Germany) according to the manufacturer's instructions. To verify loading of equal amounts of viral RNA, aliquots of the same RNA samples were hybridized with a probe specific to the m04 gene (nucleotide positions 3419 to 3978 of the MCMV genome).
Southern blot analysis.
Southern blot analysis was carried out to detect the presence of the transposon within the viral genome. Briefly, genomic DNA was digested with BamHI, separated on a 0.8% agarose gel, transferred to a nylon membrane (Hybond-N; Roche, Mannheim, Germany), and hybridized to a DNA probe corresponding to the NheI-PstI fragment from plasmid pEGFP-C1 (Clontech, Palo Alto, Calif.) described in detail elsewhere (7, 9). The probe was prepared with the DIG DNA labeling and detection kit from Roche (Roche, Mannheim, Germany) according to the manufacturer's instructions.
Immunoprecipitation and Western blot analysis of viral gene expression.
The MCMV m139 rabbit polyclonal antiserum has been described (19). For the m140 and m141 proteins, 15-amino-acid peptides from the C terminus (SVLTTRPDRNRDTRT, amino acid positions 431 to 446) and the N terminus (ATGGDQNARRRAIER, amino acid positions 25 to 40), respectively, were used to generate rabbit polyclonal antibodies (Eurogentec, Seraing, Belgium). The m04-3 antiserum, specific for m04/gp34 (23), was used as a control for viral infection. The anti-HA antibody (clone 3F10) directly coupled to peroxidase was purchased from Roche (Roche, Mannheim, Germany), and the anti-caspase-3 and anti-poly(ADP-ribose) polymerase (PARP) antibodies were purchased from Cell Signaling (Beverly, Mass.) and Transduction Laboratories (Lexington, Ky.), respectively. The anti-caspase-8 (clone 5F7) antibody was purchased from Upstate Biotechnology (Biozol, Eching, Germany).
Immunoprecipitation was performed as described previously (16). In brief, subconfluent layers of cells were mock infected or infected at an MOI of 1 with the wild-type control or the indicated MCMV mutant. At 24 h postinfection, the cells were labeled with [35S]methionine and [35S]cysteine (Amersham Biosciences, Braunschweig, Germany) for 40 min at a concentration of 350 μCi/ml. Cells were lysed in buffer (140 mM NaCl, 5 mM MgCl2, 20 mM Tris [pH 7.6], 1 mM phenylmethylsulfonyl fluoride) containing 1% (vol/vol) Nonidet P-40 (Sigma, St. Louis, Mo.) in the presence of a protease inhibitor mixture (Biomol, Plymouth Meeting, Mass.). After removal of nuclei by centrifugation, lysates were cleared with the appropriate preimmune serum and immunoprecipitated with the anti-m141 purified antiserum and protein A-Sepharose (Amersham Biosciences, Uppsala, Sweden).
For Western blot detection of proteins encoded from genes m139, m140, and m141, cells were infected at an MOI of 1 (NIH 3T3) or 3 (IC-21), and lysates were harvested 24 h postinfection in lysis buffer. Different lysis buffer contents were used for isolation of the proteins: 1% Triton X-100-4 mM MgCl2-140 mM NaCl-20 mM Tris (pH 7.3) to isolate the m140, m141, and M36-HA proteins; 50 mM Tris-1% sodium dodecyl sulfate (pH 7.5) for the m139 protein; and 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-NaOH (pH 6.5)-2 mM EDTA-0.1% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate)-5 mM dithiothreitol-20 μg of leupeptin per ml-10 μg of pepstatin per ml-10 μg of aprotinin per ml-1 mM phenylmethylsulfonyl fluoride for caspase-3 and PARP. From 12 to 15 μg of total protein was loaded per lane. Proteins were separated on 12.5% polyacrylamide gels for m04, m139, and m141 proteins, caspase-3, caspase-8, HA, and PARP or on 10% polyacrylamide gels for the m140 protein under denaturing conditions. All blots were blocked with Tris-buffered saline-0.1% Tween 20 (TBST) and either 5% milk (for the m04 and m139 proteins, caspase-3, caspase-8, and PARP) or 5% bovine serum albumin (for the m140 and m141 proteins).
For immunoprecipitation and Western blot analysis, 293 cells in a 10-cm dish were transfected with 10 μg of the pCR3-M36 or the pCR3-M36-HA vector with the Superfect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. After 24 h, the cells were lysed in the same buffer used for immunoprecipitation analysis and split into two aliquots for immunoprecipitation with either anti-HA or anti-caspase-8 antibodies. Immunoprecipitates were separated on 12.5% polyacrylamide gels under denaturing conditions for Western blot analysis as described above.
Apoptosis assay.
Fas-mediated apoptosis in NIH 3T3 and IC-21 cells was induced by exposure to the Jo2 anti-mouse Fas antibody (BD PharMingen, Heidelberg, Germany). About 5 × 105 NIH 3T3 or 1 × 105 IC-21 cells were mock infected or infected with wild-type or ΔM36 MCMV at MOIs of 1 and 5, respectively. At 48 h postinfection, cells were exposed to 0.5 μg of anti-Fas antibody and 12.5 μg of cycloheximide per ml or medium alone as controls. Cells were trypsinized, and cell viability was determined by visual inspection of cells under a phase contrast microscope with the trypan blue dye exclusion assay (41). The results are expressed as percentage of viable cells compared to that in nontreated controls. The caspase-8 activity in infected cells was determined with the ApoAlert caspase colorimetric assay kit (Clontech, Palo Alto, Calif.). About 106 IC-21 macrophages were mock treated or infected at an MOI of 7. At 36 h postinfection, cell lysates were prepared, and the assays were carried out according to the manufacturer's instructions.
RESULTS
Concept of US22 gene family mutant analysis.
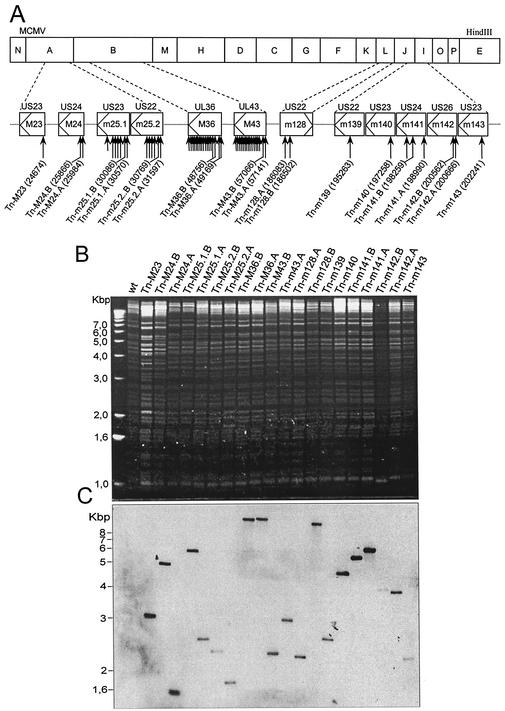
As an extension to previous work on MCMV cell type tropism (8, 12, 19, 20), in this study we tested US22 gene mutants for host cell range effects in vitro and focused on macrophage growth. Considering that cytopathic effects in macrophages can be difficult to observe upon virus infection, we constructed a library of MCMV transposon insertion mutants with the pTsTM16 transposon donor plasmid (8). As the TnMax16 transposon contains the GFP gene, all mutant viruses expressed GFP from the transposon. Altogether, 2,600 clones were screened, and 31 different clones, each with a transposon insertion within genes of the US22 gene family, were selected (Fig. 1).
FIG. 1.
Transposon insertions and analysis of US22 gene family in MCMV. (A) Locations of the transposon integration sites in US22 genes. The top line of boxes represent HindIII fragments of the 230-kb MCMV genome. The second line of boxes represent the ORFs of the US22 genes in MCMV. Arrows within the boxes indicate the direction of gene transcription. Designations M and m indicate ORFs with and without direct sequence homology to individual HCMV genes, respectively (34). The HCMV genes with the greatest homology are listed above each box. The long arrows indicate the transposon insertion sites of isolated mutants. The shorter arrows indicate additional clones that were detected by PCR screening but not analyzed further. The exact nucleotide positions of the transposon insertions are indicated in parentheses after the names of the mutants. (B) Structural analysis of the wild-type (wt) and mutant BAC plasmids. The ethidium bromide-stained agarose gel shows BamHI-digested DNA from the BAC plasmids. The majority of bands remain conserved, and specific alterations occur as a result of transposon insertion. (C) Southern blot analysis of BamHI-digested BAC plasmids hybridized with a GFP-specific probe shows a single transposon insertion for every mutant.
The observation of a large number of insertions within genes M36 and M43 and fewer insertions in other genes demonstrated that, in this new library, the distribution of transposon insertions was not completely random and that it differed from that of the library generated previously (17). All the mutant genomes used in this study were analyzed for genome integrity by restriction enzyme digestion (Fig. 1B). There were no unanticipated alterations in the number or sizes of the restriction fragments. All mutants were sequenced at the transposon insertion site. Multiple transposon insertions would have become apparent from the inability to read a clear sequence. In addition, Southern blot analysis with a GFP-specific probe was performed (Fig. 1C). Since the restriction enzyme also cleaves the transposon, multiple transposon insertions, even at the same site, would have resulted in multiple bands. Collectively, these controls indicated that only single insertions had occurred, and there were no apparent alterations elsewhere in the genome.
For comparison, fibroblasts and macrophage cells were infected in parallel with individual reconstituted mutants to screen for loss or impairment of ability to grow on macrophage cells. Mutants with a selective macrophage phenotype were also tested on endothelial cells. The phenotype of a single insertion mutant cannot define a function of the targeted ORF. Either a revertant with a wild-type phenotype or an identical phenotype obtained with independent mutants of the same ORF would indicate that adventitious mutations elsewhere in the genome are probably not responsible for the observed phenotype. To limit the work on confirmatory targeted gene mutations but to draw conclusions on the function of specific genes nevertheless, we took the following strategy. If a mutant had no selective macrophage phenotype or if the phenotype merely confirmed already published work, we limited the analysis to two independent mutants with insertions located in the first half of the ORF whenever possible. Macrophage growth phenotypes described for the first time or mutant phenotypes different from published ones were retested by targeted mutations. These targeted mutations were either complete deletions of the ORF, mutations of ATG, or reinsertions of the complete gene into an ectopic position of the viral genome.
Genes m142 and m143 are essential for MCMV replication.
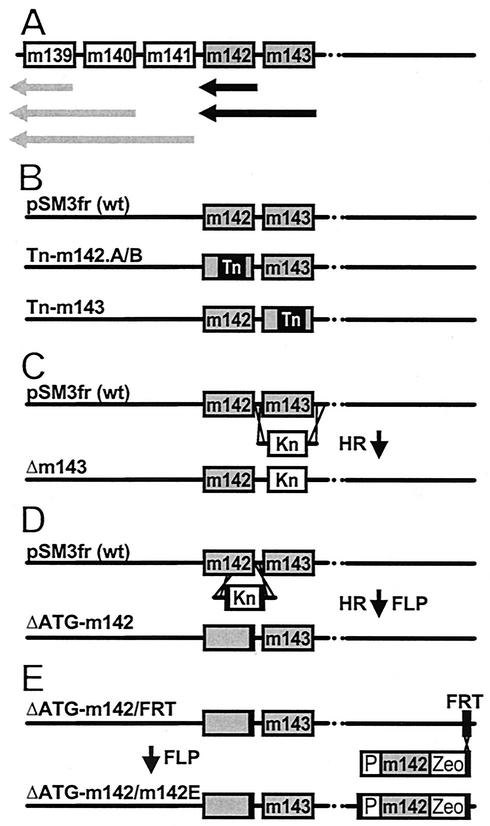
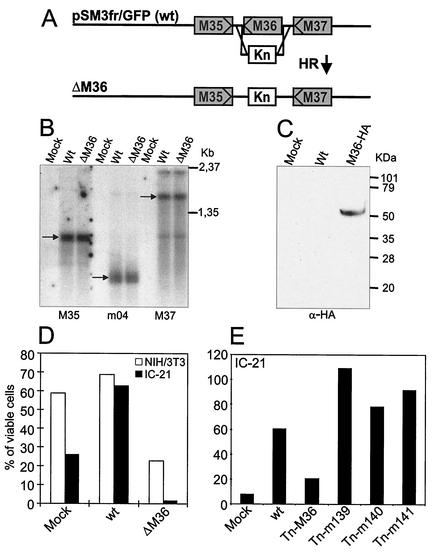
Three mutant genomes with transposon insertions in m142 and m143 (Tn-m142.A, Tn-m142.B, and Tn-m143) failed to generate infectious progeny despite numerous attempts, whereas all other MCMV mutants in the US22 family of genes could be reconstituted as viruses by transfection of the genomes into NIH 3T3 cells. Genes m139 to m143 belong to a complex transcriptional region (18). The transcripts of m142 and m143 coterminate downstream of m142 (Fig. 2A). The transposon library contained only one m143 transposon insertion mutant (Fig. 2B). Therefore, we constructed a targeted deletion of m143 and confirmed the lethal phenotype of the m143 gene (Fig. 2C). The additional 3-kb transposon insertion into m142 might destabilize the essential m143 transcript (Fig. 2A and B).
FIG. 2.
Targeted mutations of m142 and m143 genes. (A) Transcript map of genes m139 to m143. Black and grey arrows denote the major transcripts of genes m142 and m143 and genes m139 to m141, respectively, according to Hanson et al. (18). (B) transposon insertion into the wild-type (wt) MCMV BAC pSM3fr generated the mutant genomes Tn-m142.A, Tn-m142.B, and Tn-m143. (C) Deletion of the entire m143 ORF and (D) deletions of the first ATG of gene m142 were achieved by homologous recombination (HR) between pSM3fr and a linear PCR fragment containing a kanamycin resistance gene (Kn). By flanking the kanamycin cassette with minimal FRT sites (black boxes), the kanamycin cassette was excised by FLP recombinase (FLP). (E) Complementation of the ΔATG-m142 genome by insertion of the m142 gene at an ectopic position. The genome of ΔATG-m142/FRT is identical to that of ΔATG-m142 but carries a 48-bp FRT site in gene m152. This FRT site was used for insertion by FLP recombinase (FLP) of the m142 gene under the control of its native promoter (P). Zeocin (Zeo) was used for selection. The resulting genome, ΔATG-m142/m142E, gave rise to virus progeny.
In order to minimize the possible polar effects of the transposon insertion within m142 on m143 mRNA expression, we restricted the mutagenesis to the m142 start codon. This was carried out in a two-step procedure (44, 45). First, the ATG codon was deleted by insertion of a kanamycin marker by PCR based mutagenesis with linear DNA fragments as described previously (45). To excise the kanamycin resistance marker from the BAC, the kanamycin resistance gene was flanked by minimal FRT sites. Expression of the FLP recombinase excised the kanamycin resistance marker, and only one FRT site remained. (Fig. 2D). Because ΔATG-m142 did not allow virus reconstitution, we concluded that m142 is also essential for virus replication. For confirmation, we reintroduced the m142 gene into ΔATG-m142 at an ectopic position (Fig. 2E). This genome, ΔATG-m142/m142E, gave rise to progeny and definitely proved the essentiality of m142. Thus, both of the US22 family genes m142 and m143 are essential for virus replication.
Majority of US22 gene mutants have no selective macrophage phenotype.
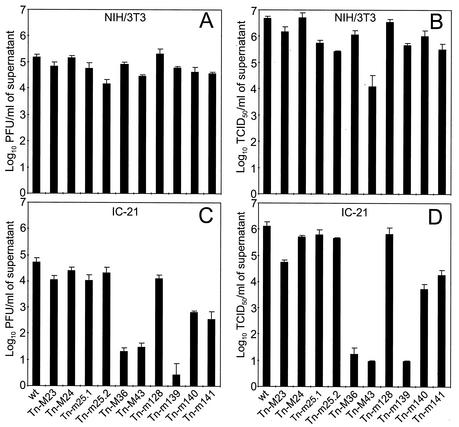
Transposon insertions into the other 10 US22 genes (M23, M24, m25.1, m25.2, m128, M36, M43, m139, m140, and m141) allowed virus growth. Viable mutants were first tested with respect to growth on NIH 3T3 fibroblasts and on IC-21 macrophages in comparison with wild-type virus. Figure 3 shows one representative experiment. Virus progeny in tissue culture supernatants was quantified by plaque assay on MEFs (Fig. 3A and C) and by the TCID50 method on NIH 3T3 fibroblasts (Fig. 3B and D). There were no significant differences between wild-type MCMV and the transposon mutants of ORFs M23, M24, m25.1, m25.2, and m128 with respect to growth on NIH 3T3 fibroblasts and on IC-21 macrophages.
FIG. 3.
Growth of US22 gene family mutants in NIH 3T3 fibroblasts and IC-21 macrophages. NIH 3T3 fibroblasts (A and B) and IC-21 macrophages (C and D) were infected in triplicate at an MOI of 0.1 with wild-type (wt) or mutant MCMV. At day 6 postinfection, supernatant was harvested from individual wells, and infectious virus was quantified on MEF cells by plaque assay (A and C) or on NIH 3T3 fibroblasts by the TCID50 endpoint titration method (B and D). Error bars indicate standard deviations between triplicates.
The two mutants of the M43 gene with transposon insertions 4 bp (shown) and 78 bp downstream of the ATG, showed a growth deficit when titrated on NIH 3T3 cells (Fig. 3B) but not when titrated on MEFs (Fig. 3A). The difference was in the range of about 2 log units compared to wild-type MCMV. This discrepancy between titration on NIH 3T3 and MEFs for the Tn-M43 mutants was consistently observed in repeated experiments. A growth reduction was also apparent when the Tn-M43 mutants were tested in IC-21 macrophages (Fig. 3C and D). There was no growth deficit of the Tn-M43 mutants on J774-A1 macrophages or SVEC4-10 endothelial cells and a slight reduction in growth on PEC (Table 2). Because the M43 mutation also affected viral growth in another cell type, the mutation does not define a macrophage-specific host range effect.
TABLE 2.
In vitro growth differences between wild-type MCMV and the US22 gene mutants at day 6 postinfection in fibroblasts, macrophages, and endothelial cells
| MCMV mutant | In vitro growtha
|
||||
|---|---|---|---|---|---|
| NIH 3T3 fibroblasts | IC-21 macrophages | J774-A1 macrophages | PEC | SVEC4-10 | |
| Tn-M36 | − | +++ | ++/+++ | ++ | − |
| Tn-M43 | ++b | +++ | − | + | − |
| Tn-m139 | − | +++ | − | − | − |
| Tn-m140 | −/+ | ++/+++ | − | − | −/+ |
| Tn-m141 | −/+ | ++/+++ | − | − | − |
−, no difference; +, ++, and +++, 1, 2, and 3 log units of difference, respectively; −/+, <1 log unit of difference.
The titers were at least 2 log units lower when titrated on NIH 3T3 cells.
Mutation of genes M36, m139, m140, and m141 affects growth on IC-21 macrophages.
Although viruses with mutations in M36, m139, m140, and m141 grew comparably to the wild-type control virus on fibroblasts, growth on IC-21 macrophages was reduced by two to four log units (Fig. 3C and D). This was seen with both M36 transposon mutants with transposon insertions 98 bp downstream of the ATG and 363 bp downstream of the start of exon2 (shown); for the m141 mutants with insertions 241 (shown) and 972 bp downstream of the ATG; and for the transposon insertion mutants of m139 and m140 analyzed in this study. Compared with wild-type MCMV, the transposon mutants of genes M36 and m139 were severely impaired for growth on IC-21 macrophages, whereas the transposon mutants of genes m140 and m141 were less compromised with respect to growth on this macrophage cell line. However, in repeated experiments, the growth differences between the M36, m139, m140, and m141 mutants were less pronounced, and the attenuation of these mutants was in the range of two to three log units compared to wild-type MCMV. Since we only found one transposon mutant for m139 in the library, we constructed a deletion mutant for m139 (Δm139-MCMV) lacking almost the complete m139 ORF except for the last 200 bp at the C terminus. This mutant also showed an attenuation of three log units for growth on IC-21 macrophages (data not shown).
Interaction among the gene products of m139, m140, and m141 explains the common phenotype.
The m139, m140, and m141 genes belong to a complex transcriptional unit and have 3′-coterminal transcripts (Fig. 4A) (18). It is less likely that a transposon insertion in the m141 ORF would affect the transcription of the downstream m139 and m140 genes because they have independent transcription start sites downstream of ORF m141. Therefore, we concluded that the two independent transposon insertion mutants for m141 probably define the phenotype of m141 gene inactivation, which confirms similar conclusions drawn before (19). More likely, the transposon insertion in the m140 gene could also affect the expression of the m141 gene and thereby cause the observed phenotype in macrophages. A comparable m140 mutant phenotype has been described previously with a targeted deletion mutant (19), and this deletion reduces the stability of the m141 protein. Finally, the transposon insertion in the m139 ORF could affect the transcription rate or the transcript stability of the upstream m140 and m141 genes.
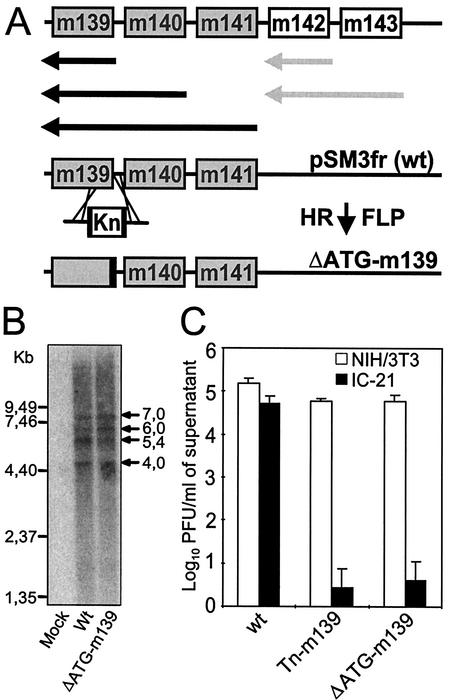
FIG. 4.
Construction, analysis, and growth of mutant ΔATG-m139. (A) Transcript map of genes m139 to m143 and construction of the ATG deletion mutant of m139. Black and grey arrows denote the major transcripts of genes m139 to m141 and genes m142 and m143, respectively, according to Hanson et al. (18). Deletion of the first two ATGs of m139 was achieved by site-directed mutagenesis with a linear PCR fragment containing a kanamycin resistance gene (Kn) flanked by FRT sites (black boxes) and by 50 nucleotides homologous to the up- and downstream sequences. After insertion of the kanamycin resistance gene by homologous recombination (HR), the kanamycin cassette was excised by FLP recombinase, generating the ΔATG-m139 mutant genome. (B) Northern blot analysis of the m139 to m141 transcripts. NIH 3T3 cells were infected at 5 PFU/cell with wild-type (wt) or ΔATG-m139 MCMV, and mRNA was isolated as described in Materials and Methods. The probe used for detection of these transcripts binds to the m140 ORF. (C) Growth of the corresponding ΔATG-m139 MCMV mutant in NIH 3T3 fibroblasts and IC-21 macrophages. Monolayers of NIH 3T3 fibroblasts and IC-21 macrophages were infected at an MOI of 0.1. The titers of cell-free viruses were determined on day 6 postinfection.
To determine whether the transposon insertion in m139 had an effect on expression of the neighboring genes, a targeted mutation of the m139 ORF (ΔATG-m139) was constructed by site-directed mutagenesis of the wild-type MCMV BAC plasmid (see Fig. 4A). Since an alternative ATG in frame is located 86 bp downstream of the native ATG, we decided to delete the first 86 bp of m139, thereby deleting both ATGs. After excision of the kanamycin resistance marker by FLP recombination, 86 bp of noncoding sequence was left behind in the BAC, corresponding exactly to the length of the deleted viral sequence. The first two start ATGs of m139 were thus deleted, but the lengths of the m139 to m141 mRNA transcripts were not altered.
We first analyzed transcription of the neighboring m140 and m141 genes in this mutant by Northern blot analysis. The transcripts characteristic of this transcription unit seen in the wild-type virus, as already shown by Hanson et al. (18), were also present in cells infected with the ΔATG-m139 mutant (Fig. 4B). Nevertheless, ΔATG-m139 MCMV showed the same growth defect in IC-21 macrophages as the Tn-m139 mutant (Fig. 4C). These results show for the first time that inactivation of the m139 gene of the US22 gene family also affects MCMV growth on IC-21 macrophages.
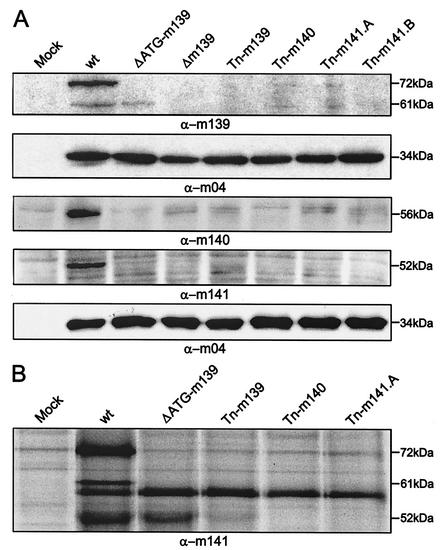
Next, we tested whether the proteins encoded by m139, m140, and m141 interact posttranslationally at the protein level. The transcripts and proteins are expressed at early times within the MCMV replication cycle (18, 42). Western blot analysis of the ΔATG-m139, Δm139, Tn-m139, Tn-m140, Tn-m141.A, and Tn-m141.B MCMV mutants was performed with m139-, m140-, and m141-specific polyclonal antisera (Fig. 5A and 5B). Detection of the 34-kDa m04/gp34 gene product served as a control for comparable viral protein loading. Due to different biochemical properties, the protein samples were prepared in different buffers and applied on different gel systems. In wild-type MCMV-infected cells, two proteins of 72 and 61 kDa were detected with the m139-specific antiserum (Fig. 5A), consistent with previous data (19). In cells infected with the Δm139 and the Tn-m139 MCMVs, no m139-specific signal was seen, whereas with the ΔATG-m139 MCMV, a smaller protein of 61 kDa was still present, suggesting that this protein is encoded by the m139 transcript, probably originating at the third alternative start ATG of the m139 ORF at nucleotide position 195767. Remarkably, none or very little of the two proteins was detectable in the Tn-m140, Tn-m141.A, and Tn-m141.B MCMV mutants (Fig. 5A). Thus, the m140- and m141-encoded proteins either directly or indirectly affected the stability of the m139 proteins.
FIG. 5.
Interaction of m139, m140, and m141 proteins. (A) Western blot analysis. NIH 3T3 cells were infected at an MOI of 1 with wild-type MCMV (wt) or MCMV mutants. Cell lysates were harvested 24 h postinfection and separated by polyacrylamide gel electrophoresis. Western blot analysis with rabbit polyclonal antisera against m139, m140, and m141 was performed. An anti-m04 antiserum which recognizes the gp34 gene product of m04 was used to control viral protein expression. (B) Coimmunoprecipitation analysis. NIH 3T3 cells were infected at an MOI of 1 with wild-type or mutant MCMVs. After 24 h, cell lysates were subjected to immunoprecipitation with an anti-m141 antiserum. The anti-m141 rabbit polyclonal antiserum precipitated the 52-kDa gene product of m141 and the 72- and 61-kDa proteins of m139 in wild-type MCMV.
The peptide antisera to the m140 and m141 products detected single viral proteins with the expected sizes of 56 kDa and 52 kDa, respectively, in wild-type MCMV-infected fibroblasts (Fig. 5A), consistent with previous data (19). Mutation of m139, m140, or m141 strongly reduced or abolished the presence of the m140 and m141 proteins.
Immunoprecipitation from wild-type MCMV-infected cells with the peptide antiserum to m141 showed a 72- and a 61-kDa protein coprecipitating with the 52-kDa m141 product (Fig. 5B). The 72-kDa protein most probably represents the large m139 protein, since this protein was absent in cells infected with ΔATG-m139. The 61-kDa protein most probably represents the truncated m139 product. In cells infected with the ΔATG-m139 mutant, the m141 product was still detectable, but not the 61-kDa protein of m139. Perhaps the 61-kDa protein requires the presence of the large m139 product in order to be coprecipitated by the m141 peptide antiserum. However, in cells infected with the Tn-m139 mutant, which does not express either of these two m139 proteins, only a trace of the m141 product was detected. This indicates that the presence of the 61-kDa protein of m139 expressed by the ΔATG-m139 mutant was required for detectable levels of the m141 product. In cells infected with Tn-m140 MCMV and, as expected, in cells infected with Tn-m141 MCMV, the 52-kDa m141 protein was missing. The m141 peptide antiserum cross-reacted with another unknown protein of about 59 kDa in infected cells (Fig. 5B).
Altogether, these results indicated that the products of genes m139, m140, and m141 interact at the protein level and that lack of any of these genes affects the steady-state level of the other proteins. These data are consistent with previous reports that at least the m140 and m141 proteins form a stable complex within infected cells (Z. Karabekian, L. K. Hanson, J. S. Slater, and A. E. Campbell, Abstr. 25th International Herpesvirus Workshop, abstr. 9.26, 2000; A. E. Campbell, Z. Karabekian, L. K. Hanson, and J. S. Slater, Abstr. 8th International Cytomegalovirus Conference, p. 26, 2001). Therefore, we concluded that the common host range phenotype detected by deletion of each of these three genes reflects their interaction at the protein level.
M36 mutants have a general macrophage phenotype.
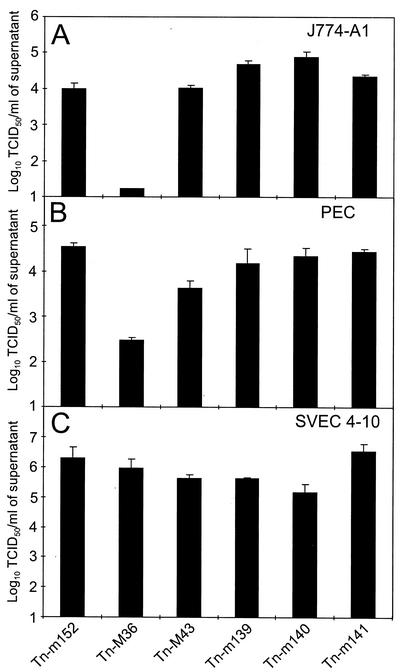
M36, M43, m139, m140, and m141 mutants were further analyzed for viral replication on J774-A1 macrophages and on PEC in order to discriminate between cell type- and cell line-specific effects (Fig. 6). On J774-A1 macrophages, which are semipermissive for MCMV replication (26), growth of only the Tn-M36 MCMV mutants with transposon insertions 98 bp downstream of the ATG (shown) and 363 bp downstream of the start of exon 2, was strongly reduced by three log units compared to Tn-m152 MCMV (Fig. 6A). M43, m139, m140, and m141 mutants showed no growth deficits. On PEC, the growth of the Tn-M36 MCMV mutants (transposon insertion 363 bp downstream of the start of exon 2 is shown) was reduced by two log units (Fig. 6B). The Tn-m139, Tn-m140, and Tn-m141 MCMV mutants were not growth deficient in PEC. These data are in contrast to a previous report that mutant MCMV deleted of m139, m140, and m141 is defective for replication in lipopolysaccharide-activated PEC (20). The growth of the Tn-M43 MCMV mutants in PEC (transposon insertion 4 bp downstream of the ATG is shown) was reduced by only one log unit (Fig. 6B). Therefore, we concluded that only the mutation of the M36 gene caused a robust and constant macrophage phenotype independently of whether primary macrophages, either stimulated or activated, or established cell lines were used.
FIG. 6.
Growth of M36, M43, m139, m140, and m141 mutants on the J774-A1 macrophage cell line, on peritoneal exudate cells, and on the SVEC4-10 endothelial cell line. J774-A1 macrophage cells (A) and peritoneal exudate cells (B) were infected at an MOI of 5 and the SVEC4-10 endothelial cells (C) were infected at an MOI of 1 with either wild-type or mutant MCMV. Six days postinfection, virus titers in the supernatant were determined on NIH 3T3 fibroblasts. Error bars indicate standard deviations between triplicates.
MCMV mutants Tn-M36, Tn-M43, Tn-m139, Tn-m140, and Tn-m141 were further analyzed for viral replication on the SVEC4-10 endothelial cell line (Fig. 6C). All mutants showed no significant reduction in growth compared to Tn-m152 MCMV. This indicates that the function of genes m139, m140, and m141 may be influenced by the state of macrophage differentiation, while gene M36 has a more fundamental role in MCMV replication in macrophage cells but not in endothelial cells. The collective data for the host cell range effects of the M36, M43, m139, m140, and m141 mutants are shown in Table 2.
M36-encoded protein has antiapoptotic function.
To prove the phenotype-gene connection of Tn-M36 mutants, we generated a targeted deletion of the M36 ORF in which the M36 ORF was replaced by a kanamycin resistance gene (see Fig. 7A and Materials and Methods). First, we examined whether deletion of the M36 ORF interfered with transcription of the neighboring M35 and M37 genes. Northern blot analysis were conducted with two probes specific for the first part of the M35 ORF and for the first two thirds of the M37 ORF. Transcripts for the M35 and M37 ORFs were detected in NIH 3T3 cells infected with wild-type MCMV or the ΔM36 MCMV mutant (Fig. 7B). Identical transcripts for M35 and M37 could be detected in wild-type and ΔM36 MCMV-infected cells, confirming that deletion of M36 did not affect transcription of the neighboring genes. To detect the M36 gene product, an MCMV virus containing a C-terminally HA-tagged M36 ORF was constructed. By Western blot analysis of lysates from infected NIH 3T3 cells, the anti-HA antibody detected a protein of about 50 kDa in cells infected with the M36-HA virus but not in cells infected with wild-type MCMV (see Fig. 7C).
FIG. 7.
ΔM36 mutant and antiapoptotic activity of the M36 protein. (A) Construction of the ΔM36 mutant genome. Deletion of the M36 gene was achieved by homologous recombination (HR) in E. coli with a linear PCR fragment containing a kanamycin resistance gene (Kn). (B) Northern blot analysis of M35 and M37 transcripts. NIH 3T3 cells were infected with 5 PFU of wild-type (wt) or ΔM36 MCMV per cell, and total RNA was isolated 24 h postinfection. Probes used for each of the neighboring genes are indicated under the blot. An m04 probe was used to control loading of equal amounts of viral RNA. (C) Expression of the M36-HA protein in NIH 3T3 fibroblasts infected with wild-type or M36-HA MCMV. NIH 3T3 cells were infected with 3 PFU/cell. Cell lysates were harvested 24 h postinfection, and equal volumes of lysates were separated on a 12.5% acrylamide gel for Western blot analysis with an anti-HA monoclonal antibody. (D and E) ΔM36 and Tn-M36 MCMV-infected cells are susceptible to anti-Fas-induced apoptosis. Cells were mock infected or infected with 1 or 3 PFU of virus per cell for NIH 3T3 (D and E) and IC-21 (D) cells, respectively. At 48 h postinfection, cells were exposed for 16 h to 0.5 μg of anti-Fas plus 12.5 μg of cycloheximide per ml. Surviving cells were counted with the trypan blue dye exclusion assay.
After infection of cells with ΔM36-MCMV, small cell fragments reminiscent of apoptotic bodies could be observed (data not shown). Therefore, the antiapoptotic function of M36 was examined by studying the viability of wild-type and ΔM36 MCMV-infected NIH 3T3 fibroblasts and IC-21 macrophages and of mock-infected cells after proapoptotic anti-Fas treatment (Fig. 7D). In NIH 3T3 fibroblasts, there was a 4.5-fold difference in the number of surviving cells after infection with wild-type and ΔM36 MCMV, whereas the difference was 65-fold in IC-21 macrophages. In fact, wild-type MCMV infection in comparison to mock infection protected IC-21 macrophages against the proapoptotic challenge (Fig. 7D). The m139, m140, and m141 mutants were also tested for an antiapoptotic phenotype, but no loss of protection against Fas-mediated apoptosis could be detected (Fig. 7E). After infection with some mutants, there were more surviving cells than after infection with wild-type MCMV. This may be due to the fact that the m139, m140, and m141 mutants grow poorly on macrophages and therefore had less of a lytic effect than wild-type MCMV.
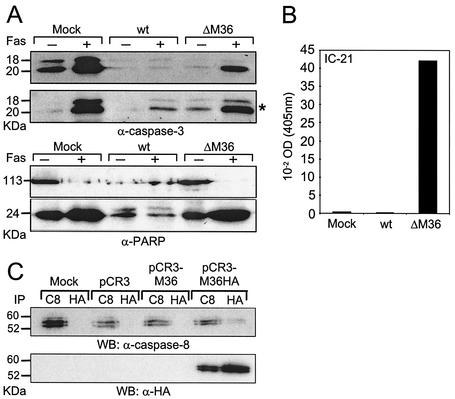
To further analyze at which step of the receptor-mediated apoptosis pathway the M36 protein acts, we examined a number of steps of the apoptotic cascade (Fig. 8). As expected, the proteolytic processing and activation of the executioner caspase-3 and the cleavage of PARP was inhibited in NIH 3T3 cells infected with the wild-type control but not in fibroblasts infected with ΔM36 MCMV and not in mock-infected controls (Fig. 8A). The total amount of cleaved and uncleaved PARP in wild-type virus-infected cells appeared smaller, probably due to the effect of 64 h of virus infection. However, unlike in wild-type-infected cells, no uncleaved PARP was detectable in ΔM36 MCMV-infected fibroblasts after induction of apoptosis with anti-Fas. Next, we determined the caspase-8 activity in IC-21 macrophages without proapoptotic stimulation. After infection with ΔM36 MCMV, the caspase-8 protease activity was 160 times higher than that after infection with wild-type MCMV, and there was a 90-fold difference between ΔM36 MCMV-infected cells and uninfected cells (Fig. 8B). These results confirmed that the M36 protein acts in the apoptosis pathway at or above the caspase-8 level.
FIG. 8.
Effect of M36 on caspase activation. (A) Caspase-3 and PARP activation in ΔM36 MCMV- but not in wild-type MCMV-infected cells. NIH 3T3 (upper panel for caspase-3 and both panels for PARP) or IC-21 cells (*, lower panel for caspase-3) were mock infected or infected with 1 PFU of virus per cell. At 48 h postinfection, cells were treated for 16 h with 0.5 μg of anti-Fas plus 12.5 μg of cycloheximide per ml or left untreated. Cell lysates were prepared, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and blotted onto a nitrocellulose membrane. The uncleaved and cleaved forms of PARP and the activated form of caspase-3 were detected by immunoblotting. (B) Caspase-8 activity in ΔM36- and wild-type MCMV-infected macrophages. IC-21 macrophages were mock infected or infected with 7 PFU/cell. At 36 h postinfection, cell lysates were prepared, and the substrate of caspase-8 was added. The pNA chromophore was quantified after cleavage by activated caspase-8. (C) The M36-HA protein can be coprecipitated with the endogenous procaspase-8. 293 cells were mock transfected or transfected with the empty pCR3 vector or the pCR3 vector expressing the M36 and the M36-HA proteins. At 48 h posttransfection, cells were lysed, and protein lysates were immunoprecipitated with either anti-procaspase-8 (C8) or anti-HA (HA) antibody. Immunoprecipitates were then used for Western blot analysis with either anti-caspase-8 or anti-HA antibody. The procaspase-8 was slightly recognized in lysates which were immunoprecipitated with the anti-HA antibody, and the M36-HA protein was strongly recognized in lysates which were immunoprecipitated with the anti-procaspase-8 antibody.
Recently, Skaletskaya et al. showed that the isolated gene UL36 of HCMV has antiapoptotic function and binds to procaspase-8 (39). To test if the M36 protein also binds to procaspase-8, we cloned the M36 gene and an HA-tagged version of the M36 gene (M36-HA) into the pCR3 expression vector. These constructs and the empty vector were transfected into 293 cells, and cell lysates were subjected to immunoprecipitation with either anti-caspase-8 or anti-HA antibodies. The immunoprecipitates were then analyzed by Western blot (Fig. 8C). The Western blot probed with anti-caspase-8 antibodies showed procaspase-8 in all lysates immunoprecipitated with anti-caspase-8. A faint specific signal for coimmunoprecipitation with the M36-HA protein was seen only in cells transfected with the pCR3-M36-HA construct, indicating that procaspase-8 can be coprecipitated with the M36-HA protein. In the Western blot probed with the anti-HA antibody, the M36-HA protein could be detected only in cells transfected with the pCR3-M36-HA vector after immunoprecipitation with anti-HA, as expected. Interestingly, the M36 protein signal was also present in the lysates of pCR3-M36-HA-transfected cells when first immunoprecipitated with the anti-caspase-8 antibody. Therefore, we concluded that M36 binds to the endogenous procaspase-8 and that the M36 antiapoptotic function determines the macrophage cell phenotype of the ΔM36 MCMV mutant.
DISCUSSION
The data presented here are the first comprehensive mutational approach to the entire US22 gene family of MCMV based on a BAC-cloned viral genome. Random transposon insertional mutagenesis was combined with targeted mutations in the search for macrophage cell-specific growth differences. We report on five genes with cell growth differences (M36, M43, m139, m140, and m141) and two essential genes (m142 and m143). In contrast, 5 of the 12 genes conferred no detectable phenotype. Thus, members of this gene family are widely divergent in function and degree of relevance to viral replication.
Random versus targeted mutagenesis of BAC-cloned herpesvirus genomes.
When we started this systematic analysis, we could only generate mutants of numerous genes by either shuttle mutagenesis or transposon insertion mutagenesis. Shuttle mutagenesis results in the desired mutant, but construction of the appropriate shuttle plasmid with its long regions of homology often requires numerous cloning steps (47). Transposon insertion is technically easier but requires a collection of at least 2,000 mutants to obtain a library that covers all CMV genes, since the distribution of insertion sites is not completely random (17). Even with a library of 2,600 clones, it was not possible to isolate at least two independent mutants for each of the genes under study. We found that in more than 95% of the mutant genomes, the transposon insertion was not associated with deletions in the genome. When deletions were found, they occurred at the insertion site and probably resulted from multiple transposon insertions followed by elimination of intervening sequences by the transposon-specific resolvase (9). High antibiotic concentrations during mutant selection favor selection of multiple transposon insertions and increase the number of deletions. Analysis of transposon mutant DNA by restriction enzyme digestion, by direct sequencing, and by Southern blotting excluded these unwanted side effects for the mutants discussed here.
Whether transposon insertion mutagenesis is a method of choice in the future depends on the number of genes to be studied. In parallel to the work on the US22 gene mutants, we adapted PCR-based targeted mutagenesis procedures to BAC-cloned herpesvirus genomes (45). Presently, the targeted mutagenesis of a dozen genes probably takes no more effort than the generation of a library. Libraries, however, will keep their value for questions of forward genetics (47).
M43 gene.
The two insertion mutants independently isolated for gene M43 showed a growth deficit that was not uniformly seen on all cells, but the growth phenotype was not restricted to macrophages. The group of Liu has described transposon mutagenesis of genomic subfragments of MCMV (49). To obtain mutant virus, each plasmid carrying a transposon needs to be reinserted into the MCMV genome by homologous recombination in cells. This led to the identification of a mutant with an insertion in codon 313 of the M43 ORF which had no phenotype in NIH 3T3 fibroblasts (48), possibly reflecting the expression of a partial but functional product. The two mutants studied by us had insertions in codons 2 and 26. Also, in our hands the fibroblast phenotype was different between NIH 3T3 and MEF cells. Thus, the M43 phenotype is complex and cannot be explained by a selective effect on macrophages.
Essentiality of genes m142 and m143.
Transposon insertions leading to lethal phenotypes in fibroblasts were controlled by targeted mutations. Here we identified the US22 gene family members m142 and m143 as being essential for virus replication. The essential nature of these genes is consistent with a previous report that mutant MCMV with a deletion spanning genes m137 through m143 (RV9) could not be purified from wild-type virus, while mutants deleted of m137 through m141 were independently replication competent (12). To date, no one has successfully constructed viruses with mutations in these two genes. Clearly, no conclusion with regard to the essentiality of individual genes can be made from this type of negative recombination experiment in cells. However, the data suggested that either m142 or m143 as individual genes, as a tandem, or in combination with other genes deleted in RV9 might be essential for growth.
Transcript mapping of the m142 to m144 region revealed that the transcripts derived from this region use a common polyadenylation signal downstream of m142 (18). Our Tn-m142 and Tn-m143 mutant genomes failed to produce progeny, as did a deletion mutant of the complete m143 ORF. Transcription of m142 is most probably not affected by transposon insertion into m143, since m142 and m143 have independent transcripts and the transposon insertion in the m143 mutant was located 1,493 bp upstream of the m142 start ATG. Although this suggested that m143 is essential for MCMV replication, the transposon insertion within m142 could have resulted in destabilization of the m143 transcript. However, a start ATG mutant of the m142 ORF, which should not affect the m143 transcript, also failed to produce progeny. Furthermore, the m142 revertant, which contained the m142 gene in an ectopic position, rescued virus growth. Therefore, we conclude that both m142 and m143 are essential for virus replication and form a new subfamily of essential US22 family genes expressed at immediate-early times after infection. It will be of interest to study their functions and to see whether this prediction can be confirmed by studying the homologous genes of HCMV.
Genes m139 to m141.
A recent publication demonstrated an interaction between m140 and m141 at the protein level, since a reduction in the m141 protein was observed after m140 deletion (19). The m141 (and m140) deletion also resulted in reduced levels of the m139 proteins within the nuclear fraction of IC-21 macrophages, whereas deletion of m139 did not lead to a detectable difference in the steady-state levels of the other two proteins (19). Thus, there was an effect of m140 and m141 on m139 but not vice versa. With regard to replication in macrophages, the deletion of either m140 or m141 caused the same phenotype, whereas deletion of m139 had no effect.
Our studies confirm the relevant aspects of that paper, namely, the cooperativity of the gene products. We also conclude that the macrophage phenotype is defined at the level of protein stability. However, there are differences. Different from the previous report, we found a comparable macrophage growth phenotype regardless of which of the three genes was inactivated. When we analyzed the interaction of the protein products, we found, in accordance with the biological properties, that mutagenesis of one of the genes affected the stability of the other two gene products. When we deleted the first two ATGs in the m139 ORF, we observed that the mutants still produced a smaller 61-kDa m139 product. Our immunoprecipitation studies revealed that the m141 protein was synthesized in ΔATG-m139-infected cells (Fig. 5B); nevertheless, the expression of the 61-kDa product did not suffice to stabilize the steady-state expression of m140 and m141 (Fig. 5A). Altogether, these data suggest that m139, m140, and m141 interact at the protein level in the Smith strain genome that we used and that this is the cause of the common phenotype. The differences are not explained by technical differences because the phenotype of the m139 mutant was confirmed in two laboratories.
The most likely explanation is that the Smith substrains kept in the two laboratories differ in this genomic region. Fresh HCMV isolates differ in biological properties and gene content and individual gene sequences from laboratory strains as a result of continuous cultivation in cell lines in vitro without the physiological selection pressure found in vivo. Resequencing of the MCMV Smith substrains in that region should reveal the differences. Whether interdependence or independence of the m139, m140, and m141 genes represents the wild-type situation will only be solved by BAC cloning and mutagenesis of a new wild-type isolate of MCMV. We are in the process of BAC cloning new virus isolates from mice. An alternative explanation for the differences is that BAC cloning requires fewer passages of progeny virus in cell culture than does the former method of cloning. It is possible that the repeated passages required to purify recombinant progeny from wild-type parental virus may select, by way of a growth advantage, viruses with a compensatory mutation elsewhere in the genome.
M36, antiapoptotic gene required for growth on macrophages.
In a previous study, we identified the transposon insertion into gene M45 with a forward genetic approach (8). The disruption of M45 prevented MCMV replication in endothelial cells, and this host cell range restriction could be explained by an antiapoptotic function of M45 that is particularly relevant in infected endothelial cells. Therefore, we checked for all host range mutants whether the growth reduction could be due to an antiapoptotic function. Among the US22 gene family mutants, only the deletion of M36 resulted in apoptosis of infected macrophages. In addition, it enhanced apoptosis following proapoptotic stimulation of infected fibroblasts (Fig. 7D). This is consistent with a lack of the Tn-M36 MCMV growth defect in fibroblasts without extrinsic proapoptotic stimulation (Fig. 3A).
While we were studying the antiapoptotic activity of M36 in more detail, Goldmacher and colleagues reported the constructing and screening of an HCMV genomic DNA library for genes with antiapoptotic function (39). They found that the isolated UL36 gene product formed a complex with procaspase-8. Our data allowed analysis of M36 function in the viral context and showed that the MCMV M36 protein also forms a complex with procaspase-8. Our data contribute the observation that murine macrophages need the function of M36 even in absence of proapoptotic stimuli for highly productive MCMV infection. Further studies in vitro and in vivo will show whether this phenotype extends to other cell types that MCMV can infect.
Altogether, we could segregate insertion mutations in all members of the US22 gene family of MCMV with regard to essentiality and growth properties in fibroblasts and macrophages. We were able to associate the macrophage growth defect of the M36 mutants with the antiapoptotic function of this gene product, whereas the function of the m139 to m141 genes was difficult to resolve due to the interaction at the protein level. Although US22 family genes share sequence motifs, perhaps indicative of a common origin, evolution and gene duplications created new functions that apparently dominate the unknown original function, delineated by the motif signature. Whether the definition of related ORFs as protein families is still valid remains a matter of debate (32). Therefore, we expect that the comparison between positional homologs of US22 family genes in HCMV and MCMV will reveal more common features than the comparison of gene families in general. This assumption has held true for the M36 and UL36 genes.
Acknowledgments
We thank L. K. Hanson for technical controls. We thank L. Cicin-Sain for providing primary cells and S. Mathys for the pSM3fr/GFP BAC. We also thank A. Bubeck for comments on the manuscript.
This work was supported by grants from the Boehringer Ingelheim Fonds (C.M.), the Deutsche Forschungsgemeinschaft through DFG SFB 455 (U.H.K.) and SFB 479 (W.B.), and PHS grant CA41451 (A.E.C.).
REFERENCES
- 1.Adair, R., E. R. Douglas, J. B. Maclean, S. Y. Graham, J. D. Aitken, F. E. Jamieson, and D. J. Dargan. 2002. The products of human cytomegalovirus genes UL23, UL24, UL43 and US22 are tegument components. J. Gen. Virol. 83:1315-1324. [DOI] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atalay, R., A. Zimmermann, M. Wagner, E. Borst, C. Benz, M. Messerle, and H. Hengel. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J. Virol. 76:8596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blacklaws, B. A., P. Bird, D. Allen, D. J. Roy, I. C. MacLennan, J. Hopkins, D. R. Sargan, and I. McConnell. 1995. Initial lentivirus-host interactions within lymph nodes: a study of maedi-visna virus infection in sheep. J. Virol. 69:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borst, E. M., S. Mathys, M. Wagner, W. Muranyi, and M. Messerle. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 75:1450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brune, W. 2002. Random transposon mutagenesis of large DNA molecules in Escherichia coli. Methods Mol. Biol. 182:165-171. [DOI] [PubMed] [Google Scholar]
- 8.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 9.Brune, W., C. Menard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:360-364. [DOI] [PubMed] [Google Scholar]
- 10.Bubeck, A., U. Reusch, M. Wagner, T. Ruppert, W. Muranyi, P. M. Kloetzel, and U. H. Koszinowski. 2002. The glycoprotein gp48 of murine cytomegalovirus L proteasome-dependent cytosolic dislocation and degradation. J. Biol. Chem. 277:2216-2224. [DOI] [PubMed] [Google Scholar]
- 11.Cardin, R. D., G. B. Abenes, C. A. Stoddart, and E. S. Mocarski. 1995. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology 209:236-241. [DOI] [PubMed] [Google Scholar]
- 12.Cavanaugh, V. J., R. M. Stenberg, T. L. Staley, H. W. Virgin, M. R. MacDonald, S. Paetzold, H. E. Farrell, W. D. Rawlinson, and A. E. Campbell. 1996. Murine cytomegalovirus with a deletion of genes spanning HindIII-J and -I displays altered cell and tissue tropism. J. Virol. 70:1365-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 14.Colberg, P. A. 1996. Functional roles of immediate early proteins encoded by the human cytomegalovirus UL36-38, UL115-119, TRS1/IRS1 and US3 loci. Intervirology 39:350-360. [DOI] [PubMed] [Google Scholar]
- 15.Colberg, P. A., L. D. Santomenna, P. P. Harlow, P. A. Benfield, and D. J. Tenney. 1992. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J. Virol. 66:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.del Val, M., H. Hengel, H. Hacker, U. Hartlaub, T. Ruppert, P. Lucin, and U. H. Koszinowski. 1992. Cytomegalovirus prevents antigen presentation by blocking the transport of peptide-loaded major histocompatibility complex class I molecules into the medial-Golgi compartment. J. Exp. Med. 176:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutermann, A., A. Bubeck, M. Wagner, U. Reusch, C. Menard, and U. Koszinowski. 2002. Strategies for the identification and analysis of viral immune evasion genes—cytomegalovirus as example. Curr. Top. Microbiol. Immunol. 269:1-22. [DOI] [PubMed]
- 18.Hanson, L. K., B. L. Dalton, Z. Karabekian, H. E. Farrell, W. D. Rawlinson, R. M. Stenberg, and A. E. Campbell. 1999. Transcriptional analysis of the murine cytomegalovirus HindIII-I region: identification of a novel immediate-early gene region. Virology 260:156-164. [DOI] [PubMed] [Google Scholar]
- 19.Hanson, L. K., J. S. Slater, Z. Karabekian, G. Ciocco-Schmitt, and A. E. Campbell. 2001. Products of US22 genes M140 and M141 confer efficient replication of murine cytomegalovirus in macrophages and spleen. J. Virol. 75:6292-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson, L. K., J. S. Slater, Z. Karabekian, H. W. Virgin, C. A. Biron, M. C. Ruzek, N. van Rooijen, R. P. Ciavarra, R. M. Stenberg, and A. E. Campbell. 1999. Replication of murine cytomegalovirus in differentiated macrophages as a determinant of viral pathogenesis. J. Virol. 73:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavanagh, D. G., M. C. Gold, M. Wagner, U. H. Koszinowski, and A. B. Hill. 2001. The multiple immune-evasion genes of murine cytomegalovirus are not redundant: m4 and m152 inhibit antigen presentation in a complementary and cooperative fashion. J. Exp. Med. 194:967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides, T., A. T. Bankier, S. C. Satchwell, E. Preddy, and B. G. Barrell. 1988. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology 165:151-164. [DOI] [PubMed] [Google Scholar]
- 25.Krmpotic, A., M. Messerle, M. I. Crnkovic, B. Polic, S. Jonjic, and U. H. Koszinowski. 1999. The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T cell control in vivo. J. Exp. Med. 190:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, M. A., J. S. Slater, L. I. Leverone, and A. E. Campbell. 1990. Enhancement of interleukin-1 activity by murine cytomegalovirus infection of a macrophage cell line. Virology 178:452-460. [DOI] [PubMed] [Google Scholar]
- 27.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munch, K., M. Messerle, B. Plachter, and U. H. Koszinowski. 1992. An acidic region of the 89K murine cytomegalovirus immediate early protein interacts with DNA. J. Gen. Virol. 73:499-506. [DOI] [PubMed] [Google Scholar]
- 29.Neipel, F., K. Ellinger, and B. Fleckenstein. 1991. The unique region of the human herpesvirus 6 genome is essentially collinear with the UL segment of human cytomegalovirus. J. Gen. Virol. 72:2293-2297. [DOI] [PubMed] [Google Scholar]
- 30.Nicholas, J. 1996. Determination and analysis of the complete nucleotide sequence of human herpesvirus. J. Virol. 70:5975-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholas, J., and M. E. Martin. 1994. Nucleotide sequence analysis of a 38.5-kilobase-pair region of the genome of human herpesvirus 6 encoding human cytomegalovirus immediate-early gene homologs and transactivating functions. J. Virol. 68:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novotny, J., I. Rigoutsos, D. Colemann, and T. S. Shenk. 2001. In silico structural and functional analysis of the human cytomegalovirus (human herpesvirus5) genome. J. Mol. Biol. 310:1151-1166. [DOI] [PubMed] [Google Scholar]
- 33.Patterson, C. E., and T. Shenk. 1999. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J. Virol. 73:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddehase, M. J., F. Weiland, K. Munch, S. Jonjic, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romanowski, M. J., G. E. Garrido, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J. T., and D. Russel.2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stasiak, P. C., and E. S. Mocarski. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzung, S. P., K. M. Kim, G. Basanez, C. D. Giedt, J. Simon, J. Zimmerberg, K. Y. Zhang, and D. M. Hockenbery. 2001. Antimycin A mimics a cell-death-inducing Bcl-2 homology domain 3. Nat. Cell Biol. 3:183-191. [DOI] [PubMed] [Google Scholar]
- 42.Vieira, J., H. E. Farrell, W. D. Rawlinson, and E. S. Mocarski. 1994. Genes in the HindIII J fragment of the murine cytomegalovirus genome are dispensable for growth in cultured cells: insertion mutagenesis with a lacZ/gpt cassette. J. Virol. 68:4837-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner, M., and U. H. Koszinowski. Mutagenesis of viral BACs with linear PCR fragments (ET recombination). Methods Mol. Biol., in press. [DOI] [PubMed]
- 45.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Syst. excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 48.Xiao, J., T. Tong, X. Zhan, E. Haghjoo, and F. Liu. 2000. In vitro and in vivo characterization of a murine cytomegalovirus with a transposon insertional mutation at open reading frame M43. J. Virol. 74:9488-9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhan, X., M. Lee, G. Abenes, R. Von, I., C. Kittinunvorakoon, P. Ross-Macdonald, M. Snyder, and F. Liu. 2000. Mutagenesis of murine cytomegalovirus with a Tn3-based transposon. Virology 266:264-274. [DOI] [PubMed] [Google Scholar]