Abstract
YveA of Bacillus subtilis, a putative transporter of the amino acid/polyamine/organocation (APC) superfamily, is shown to mediate uptake of both l-aspartate and l-glutamate as well as having sensitivity to l-aspartate hydroxamate. This 14 TMS protein is the primary aspartate uptake system in B. subtilis and serves as the prototype for a new family within the APC superfamily.
Many families of transport proteins mediate the uptake of amino acids and their derivatives (6, 7). The largest such family is the amino acid/polyamine/organocation (APC) superfamily (4). This ubiquitous superfamily includes 10 well-defined families with numerous paralogues in a single organism. For example, Escherichia coli and Bacillus subtilis encode within their genomes 24 and 21 recognized paralogues of this superfamily in five and four assigned APC families, respectively (4).
Very few of the proteins of the APC superfamily are functionally characterized, and 2 of the 10 previously defined families within the APC superfamily do not include even one functionally characterized member. We have (i) cloned, (ii) overexpressed, and (iii) knocked out the gene encoding a member of a novel family within the APC superfamily from B. subtilis, yveA. We show that this protein (i) catalyzes uptake of l-aspartate, (ii) mediates sensitivity to aspartate hydroxamate, (iii) probably exhibits specificity for l-aspartate, l-glutamate, l-aspartic hydroxamate, and possibly l-asparagine and l-glutamine, (iv) allows enhanced growth with l-aspartate as the sole nitrogen source, and (v) exhibits maximal activity after growth in the presence of l-aspartate. The results show that YveA is the principal aspartate transporter in B. subtilis.
The B. subtilis yveA gene was amplified by PCR using the Pfx platinum polymerase and B. subtilis chromosomal DNA as the template. To overexpress yveA in B. subtilis, the XbaI-PstI-amplified fragment was fused to the B. subtilis promoter Pspac present on plasmid pAG58 (3). The yveA gene combined with Pspac was further subcloned (as an EcoRI-PstI DNA fragment) into the E. coli-B. subtilis shuttle vector pMK4, which encodes a chloramphenicol resistance marker (9). The Pspac promoter is known to be a weak isopropyl-β-d-thiogalactopyranoside-inducible promoter, and pMK4 is a low-copy-number plasmid (3, 9). These facts are in agreement with the results reported below (see Fig. 2).
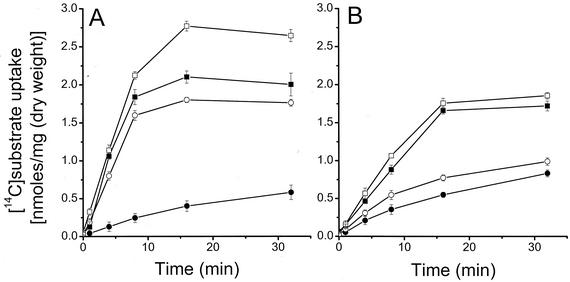
FIG. 2.
Time courses for the uptake of l-[14C]aspartate (A) and l-[14C]glutamate (B) into B. subtilis cells. M168 (wild type) (▪), knockout mutant (M168 yveA) (•), M168 cells expressing the plasmid-encoded yveA gene (M168 with Pspac-yveA) (□), and the chromosomal yveA knockout mutant expressing the yveA-bearing plasmid (○). Cells were grown for 24 h at 37°C in minimal SM medium with 0.1% d-glucose and 10 mM aspartate. Cells were harvested in the exponential growth phase, washed twice, and resuspended in 50 mM Tris-maleate-5 mM MgCl2 (TM buffer) (pH 7.0) to an optical density of 0.1. The energy source (8 mM glucose) and either l-aspartate or l-glutamate (20 μM; 5 μCi/μmol) were added to a temperature-equilibrated 1-ml cell suspension at 0 min. Transport assays were conducted at 37°C. Samples (0.1 ml) were removed at appropriate times as shown, filtered (25-mm membrane filters; 45-μm pore size), washed three times with cold TM buffer, dried, and then transferred to vials containing 10 ml of scintillation fluid for determination of radioactivity. Values reported represent the averages of two independent assays.
The bacterial strains and plasmids used in this study are described in Table 1. Recombinant plasmid was recovered in E. coli DH5α cells and transformed in B. subtilis M168 (wild-type) cells by natural competence. To construct the knockout mutant, the yveA gene was amplified by PCR and inserted into the cloning vector pPCR-Script Amp in the SrfI site. An internal fragment (500 bp) of the cloned gene was deleted by cutting with MunI. The 4-kb fragment of the cut plasmid was recovered from the gel, purified, and blunted with the Klenow enzyme. The fragment was further dephosphorylated and ligated to the kanamycin gene from pER82 (kindly provided by K. Pogliano, University of California at San Diego). The presence of the inserted gene was confirmed by PCR.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| E. coli strain | ||
| DH5α | Wild type, donor strain F−φ 80d lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) gal-phoA supE44λ−thi-1 gyrA96 relA1 | |
| B. subtilis strains | ||
| M168 Marburg | trpC2 | |
| M168 yveA− | trpC2 yveA | This study |
| M168 pMK4-yveA | trpC2 carrying the yveA gene plus upstream Pspac promoter from pAG58 | This study |
| Plasmids | ||
| pPCR-Script | Ampr, PCR cloning vector | Stratagene |
| pER82 | pDG364-Km, Kmr Ampr | 8 |
| pMK4 | Ampr Cmr Lac+, shuttle vector for B. subtilis and E. coli | 9 |
| pMK4-yveA | Ampr Cmr Lac+, carrying the yveA gene plus upstream Pspac promoter from pAG58 | This study |
| pAG58 | Ampr Cmr, expression vector containing Pspac | 4 |
| pJF751 | Ampr Cmr, isotopic integrative vector in Bacillus for translational lacZ fusions | 11 |
| pJF751-yveA | pJF751 containing a 344-bp fragment of yveA (−330 to +14) | This study |
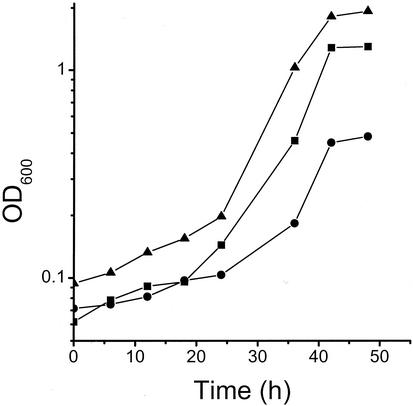
Figure 1 shows growth curves for (i) wild-type B. subtilis, (ii) a yveA knockout mutant, and (iii) the wild-type strain expressing the yveA gene from a plasmid. All cells were grown under conditions where l-aspartate served as the sole source of nitrogen. The extent of growth of the yveA knockout mutant was poor, and the estimated doubling time (dt) was ∼21 h. Growth enhancement relative to that of the wild-type strain (both the extent of growth and apparent growth rate) (dt = ∼10.4 h) was observed for the yveA plasmid expression strain (dt = ∼7.3 h). Similar behavior was observed when 0.1% Casamino Acids was included in the growth medium, although the differences were less pronounced (data not shown). These results suggested that YveA mediates uptake of l-aspartate but that it is not the sole transporter providing this function.
FIG. 1.
Growth curves for B. subtilis strain M168 (wild type) (▪), a yveA knockout mutant (M168 yveA) (•), and M168 cells overexpressing the yveA gene (M168 + Pspac-yveA) (▴). Cells were grown for 48 h at 37°C in minimal SM medium (80 mM K2HPO4, 44 mM KH2PO4, 3.4 mM trisodium citrate, 2 mM MgSO4, 6.7 mM KCl, 0.5 mM CaCl2, 5 μM MnCl2, 0.5 μM FeSO4) with 0.1% d-glucose and 10 mM aspartate. The optical density was measured at 600 nm.
Sensitivities of the same three strains used in the growth experiment shown in Fig. 1 to the toxic aspartate-asparagine analogue d,l-aspartic acid β-hydroxamate were measured. In these experiments, the bacteria in soft agar (0.7% agar in Luria-Bertani [LB] medium) were plated over hard agar (1.5% agar in LB), and a disk containing 50, 100, or 150 μg of d,l-aspartic acid β-hydroxamate was placed on the agar plates prior to incubation at 37°C. In all cases, the knockout mutant was less sensitive and the overexpressing strain was more sensitive than the wild-type strain to the aspartate analogue. For example, after a growth period of 24 h, the radius of the zone of killing on the plate with 150 μg of d,l-aspartic acid β-hydroxamate was 15 mm for the wild type, 19 mm for the overexpressing strain, and 0.8 mm for the knockout mutant. These results suggest that YveA is the major uptake system for this analogue.
Figure 2 shows the uptake of l-[14C]aspartate (Fig. 2A) and l-[14C]glutamate (Fig. 2B), both at a concentration of 20 μM, as a function of time for the wild type and yveA knockout mutant with and without the overexpressing plasmid. The knockout mutant barely took up l-[14C]aspartate, while the overproducing strain took up more than the wild type. The same was observed for l-[14C]glutamate uptake, although the background activity of the yveA knockout mutant was substantially higher (over twofold higher than the wild-type aspartate uptake rate), and the increase, due to the reintroduction of the yveA gene, was less. The l-[14C]aspartate incorporation rate by the wild-type strain showed saturation at 100 μM aspartate, with a Vmax of 1.1 ± 0.1 nmol min−1 mg−1 (dry weight) and an apparent Km of 25 ± 3 μM.
Using the same conditions described in Table 2, the energetics of l-aspartate uptake were studied. Carbonyl cyanide m-chlorophenyl hydrazone and carbonyl cyanide 4-trifluoromethoxyphenylhydrazone, both at 5 μM, blocked uptake >95%. Substitution of Na+ for K+ in the uptake buffer did not result in decreased uptake (±5%). It is therefore likely that l-aspartate uptake is a proton-motive force—rather than a sodium-motive force-driven process—and that the mechanism of transport is amino acid-H+ symport.
TABLE 2.
Inhibition of the initial l-[14C]aspartate uptake rate by the l- and d-isomers of several amino acids
| Compounda | Inhibition (%) |
|---|---|
| dl-Aspartate β-hydroxamate | 95 ± 7 |
| l-Aspartate | 80 ± 3 |
| d-Aspartate | 43 ± 5 |
| l-Methyl aspartate | 61 ± 6 |
| l-Glutamate | 41 ± 2 |
| d-Glutamate | 17 ± 4 |
| l-Asparagine | 58 ± 3 |
| d-Asparagine | 46 ± 3 |
| l-Glutamine | 35 ± 6 |
| l-Threonine | 48 ± 5 |
| l-Serine | 50 ± 2 |
| l-Arginine | 21 ± 1 |
| l-Histidine | 33 ± 4 |
| l-Alanine | 28 ± 2 |
| l-Tyrosine | 23 ± 3 |
| l-Cysteine | 14 ± 6 |
The inhibitory amino acid was present at 200 μM, a 10-fold excess over the l-[14C]aspartate concentration.
Several amino acids were used to estimate substrate recognition by measuring the percent inhibition of l-aspartate uptake (Table 2). Surprisingly, d,l-aspartate β-hydroxamate was more inhibitory than l-aspartate. When a 10-fold excess of nonradioactive l-aspartate (200 μM) was added uptake of l-[14C]aspartate was reduced by 80%, while l-asparagine at 200 μM inhibited 58%. l-Glutamate and l-glutamine inhibited l-[14C]aspartate uptake 41 and 35%, respectively. Additionally, l-threonine and l-serine inhibited about 50%, although other l-amino acids tested inhibited less than 33%. d-Aspartate and d-asparagine were about equally inhibitory (∼45%), showing that substrate recognition is not strictly stereospecific. These results, together with the growth and aspartic hydroxamate inhibition results, suggest that YveA transports amino acids with relative affinities in the order of aspartate hydroxamate > l-aspartate > l-glutamate, but it recognizes a much broader range of amino acids.
Uptake of several l-amino acids was measured in the wild-type and yveA mutant strains. The cells were grown with either aspartate or proline as the sole source of nitrogen. Table 3 shows that the yveA mutant strain grown with aspartate took up less aspartate and glutamate but more asparagine, glutamine, alanine, and serine compared to the wild-type control strain. These results indicate that poor growth observed in the yveA mutant strain might have caused increased activity of other permeases. This suggestion was substantiated by the fact that proline-grown mutant cells showed decreased uptake of aspartate and glutamate but a lesser differential for most of the other amino acids when compared with the wild-type strain. The large reproducible differential observed for l-asparagine and l-glutamine is unexplained.
TABLE 3.
Uptake of14C-labeled l-amino acids in cells grown in SM medium with either aspartate or proline (10 mM) as the sole nitrogen sourcea
| Substrate | Aspartate-grown cells
|
Proline-grown cells
|
||
|---|---|---|---|---|
| WT | yveA | WT | yveA | |
| l-Aspartate | 1.6 ± 0.5 | 0.2 ± 0.2 | 0.5 ± 0.1 | 0.1 ± 0.05 |
| l-Glutamate | 0.8 ± 0.2 | 0.5 ± 0.1 | 0.6 ± 0.1 | 0.2 ± 0.1 |
| l-Asparagine | 0.7 ± 0.1 | 1.0 ± 0.1 | 0.3 ± 0.1 | 0.7 ± 0.1 |
| l-Glutamine | 0.6 ± 0.2 | 1.5 ± 0.2 | 0.4 ± 0.2 | 0.7 ± 0.1 |
| l-Alanine | 0.3 ± 0.1 | 0.6 ± 0.1 | 0.4 ± 0.2 | 0.4 ± 0.1 |
| l-Serine | 0.4 ± 0.1 | 1.1 ± 0.2 | 0.5 ± 0.1 | 0.6 ± 0.1 |
Uptake is expressed in nanomoles per milligram (dry weight) after a 16-min time interval. Values are reported ± standard deviations where at least three independent determinations were made for each value presented. Wild-type cells grown with l-proline and l-aspartate were the same ±10%, and the yveA mutation that depressed the growth rate on l-aspartate did not depress the growth rate on l-proline.
A yveA-lacZ fusion was constructed using the pJF751 plasmid (11). This vector allows the construction of a translational fusion with codon 8 of the promoterless β-galactosidase gene and inserts into the chromosome by homologous recombination. The N-terminal fragment of the yveA gene (positions −229 to +14) was amplified by PCR, cut with EcoRI and BamHI, and ligated into pJF751 which had been cleaved with the same enzymes. The recombinant plasmid was transferred to B. subtilis M168 by natural competence, and the effects of different compounds included in the growth medium on β-galactosidase production were measured. Cells were grown in minimal salts medium with the added amino acid present at 10 mM. The presence of l-aspartate resulted in a threefold increase in β-galactosidase activity compared with cells grown with (NH4)2SO4 or any one of most other amino acids (Table 4). Proline, asparagine, and glutamine induced activity to lesser extents (about twofold), while all other amino acids induced minimally.
TABLE 4.
β-Galactosidase activity of a yveA-lacZ translational fusion after growth with different amino acids as the sole nitrogen sourcea
| SM medium plusb | β-Galactosidase |
|---|---|
| (NH4)2SO4 | 1.0 ± 0.1 |
| (NH4)2SO4 + Asp | 3.1 ± 0.3 |
| Asp | 3.1 ± 0.8 |
| Arg | 0.9 ± 0.2 |
| Glu | 1.5 ± 0.1 |
| Gln | 2.1 ± 0.4 |
| Asn | 2.2 ± 0.4 |
| Ala | 1.5 ± 0.7 |
| Leu | 1.4 ± 0.2 |
| Pro | 2.0 ± 0.3 |
β-Galactosidase activity is expressed in Miller units per milligram of protein.
Individual amino acids were added at a concentration of 10 mM and served as the sole source of nitrogen for growth. Serine was not tested because it did not support growth of the cells as the sole nitrogen source.
Immediately downstream of the yveA gene and transcribed in the same direction is the yvdT gene encoding a transcriptional regulator of the TctR/AcrR family. YvdT is the most likely mediator of yveA induction by l-aspartate. Downstream of yvdT are two small genes, yvdS and yvdR, probably sequence-divergent members of the small multidrug resistance family within the drug/metabolite transporter superfamily (TC 2.A.7) (5). Preliminary evidence suggests that YvdR may mediate ammonium efflux (reference 1 and Y. J. Chung and M. H. Saier, Jr., unpublished observations).
In this communication we have demonstrated that YveA is the primary l-aspartate transporter of B. subtilis following growth under standard laboratory conditions. Based on the inhibition studies with aspartate hydroxamate, it must also transport this aspartate analogue. The system also transports l-glutamate but with lower affinity and efficiency. The main glutamate transporter, GltP (P39817), has been described previously (2, 10).
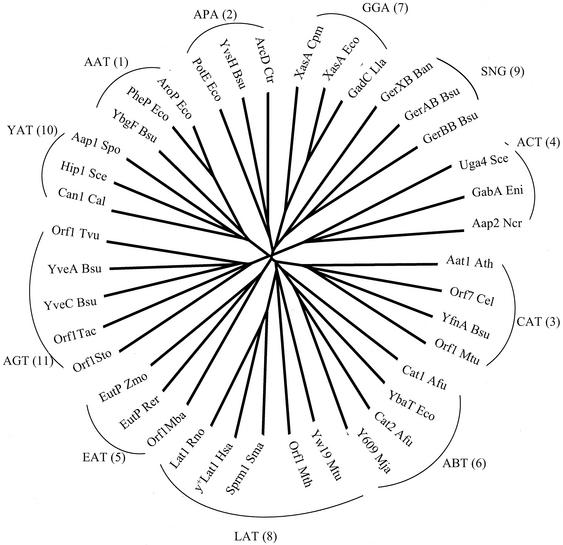
YveA is the first member of a new family within the APC superfamily to be characterized functionally. In an earlier communication, 10 families were described for the APC superfamily (4). Figure 3 shows a phylogenetic tree for this superfamily, including representative members of the 10 previously defined families (families 1 to 10 in Fig. 3). As can be seen in the figure, members of these 10 families cluster in accordance with the expectation based on the results of Jack et al. (4). However, the tree shown in Fig. 3 defines a new family that we have called the aspartate/glutamate transporter (AGT) family (family 11; TC 2.A.3.11). YveA of B. subtilis is the only characterized member of the AGT family, and it is therefore the prototype for this family. In contrast to all other prokaryotic members of the APC superfamily (4), all of the members of this family exhibit 14 rather than 12 putative transmembrane segments (TMSs). The two extra TMSs are found C-terminal to the 12 TMSs that are common to other members of the superfamily. It should be noted that YveA was included as a highly divergent member of the ABT family by Jack et al. (4). The expansion of this family due to the increased sequence data now available allows segregation of the previously specified ABT family into two families of different topological protein types (Fig. 3).
FIG. 3.
Phylogenetic tree for representative members of the APC superfamily. The 10 previously identified families (labeled 1 to 10 in parentheses following the family abbreviation) were as described by Jack et al. (4). Abbreviations of the proteins in these families were as defined therein. The new family to which YveA belongs is the AGT family (TC 2.A.3.11). The tree was derived from a multiple alignment obtained using the ClustalX program as detailed previously (4). Members of the AGT family include YveA Bsu (B. subtilis gi1945680); YbeC Bsu (B. subtilis gi2632498); Orf1 Sto (Sulfolobus tokodaii gi15922067); Orf1 Tvo (Thermoplasma volcanium gi13542093); and Orf1 Tac (Thermoplasma acidophilum gi16081310).
Two B. subtilis proteins are found within the AGT family, the other being YbeC. Three other proteins, all from archaea, two from two different species of Thermoplasma and one from Sulfolobus tokodaii, are also included in this family. As revealed by the tree shown in Fig. 3, these five proteins form a clear cluster, although they are all distant homologues of each other. The two Bacillus paralogues cluster loosely together. Further studies will be required to determine the range of substrates transported by members of the AGT family.
Acknowledgments
We thank Y. J. Chung for helpful discussions and Mary Beth Hiller for assisting in the preparation of the manuscript.
G. Lorca and B. Winnen contributed equally to the work reported.
This work was supported by NIH grant GM64368. G.L.L. was supported by a fellowship from the Pew Latin American Program in the Biomedical Sciences.
REFERENCES
- 1.Chung, Y. J., and M. H. Saier, Jr. 2001. SMR-type multidrug resistance pumps. Curr. Opin. Drug Discov. Dev. 4:237-245. [PubMed] [Google Scholar]
- 2.de Vrij, W., R. A. Bulthuis, P. R. van Iwaarden, and W. N. Konings. 1989. Mechanism of l-glutamate transport in membrane vesicles from Bacillus stearothermophilus. J. Bacteriol. 171:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaacks, K. J., J. Healy, R. Losick, and A. D. Grossman. 1989. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 171:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jack, D. L., I. T. Paulsen, and M. H. Saier, Jr. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797-1814. [DOI] [PubMed] [Google Scholar]
- 5.Jack, D. L., N. M. Yang, and M. H. Saier, Jr. 2001. The drug/metabolite transporter superfamily. Eur. J. Biochem. 268:3620-3639. [DOI] [PubMed] [Google Scholar]
- 6.Saier, M. H., Jr. 2000. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146:1775-1795. [DOI] [PubMed] [Google Scholar]
- 7.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somkuti, G. A., D. K. Solaiman, and D. H. Steinberg. 1995. Native promoter-plasmid vector system for heterologous cholesterol oxidase synthesis in Streptococcus thermophilus. Plasmid 33:7-14. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan, M. A., R. E. Yasbin, and F. E. Young. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli, which allow rapid detection of inserted fragments. Gene 29:21-26. [DOI] [PubMed] [Google Scholar]
- 10.Tolner, B., T. Ubbink-Kok, B. Poolman, and W. N. Konings. 1995. Cation-selectivity of the l -glutamate transporters of Escherichia coli, Bacillus stearothermophilus and Bacillus caldotenax: dependence on the environment in which the proteins are expressed. Mol. Microbiol. 18:123-133. [DOI] [PubMed] [Google Scholar]
- 11.Weinrauch, Y., T. Msadek, F. Kunst, and D. Dubnau. 1991. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J. Bacteriol. 173:5685-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]