Abstract
Although a wealth of knowledge exists about the molecular and biochemical mechanisms governing the swimming motility of Salmonella enterica serovar Typhimurium, its surface swarming behavior has not been extensively characterized. When inoculated onto a semisolid agar medium supplemented with appropriate nutrients, serovar Typhimurium undergoes a morphological differentiation whereby single cells hyperflagellate and elongate into nonseptate, multinucleate swarm cells. Swarm migration is a collective behavior of groups of cells. We have isolated a MudJ insertion mutant of serovar Typhimurium 14028 that failed to swarm under any conditions. The site of the MudJ insertion was determined to be in the pmrK locus within the pmrHFIJKLM operon, which was previously demonstrated to confer resistance to cationic antimicrobial peptides. β-Galactosidase assays, using the pmrK::lacZ transcriptional fusion, showed increased expression of the pmr operon in swarm cells compared to that in vegetative cells. In concurrence with the expression data, swarm cells exhibited greater tolerance to polymyxin. To compare the profiles of vegetative and swarm-cell resistance to other antibiotics, E-test strips representing a wide range of antibiotic classes were used. Swarm cells exhibited elevated resistance to a variety of antibiotics, including those that target the cell envelope, protein translation, DNA replication, and transcription. These observations, in addition to the dramatic morphological changes associated with the swarming phenotype, provide an intriguing model for examining global differences between the physiological states of vegetative and swarm cells of serovar Typhimurium.
Swarming motility is a coordinated multicellular behavior promoting rapid colony migration and expansion on semisolid surfaces (10). Swarming is preceded by differentiation of short, motile, vegetative (i.e., swimmer) cells into elongated, hyperflagellated, and multinucleate swarm cells. The mechanisms that drive chemotaxis and swimming motility in Salmonella enterica serovar Typhimurium have been extensively studied (1, 2, 7, 9, 27, 28). However, its surface swarming behavior is largely uncharacterized. In addition to inoculum density, specific medium composition triggers swarm-cell differentiation in serovar Typhimurium (15). However, the specific intra- and extracellular signals and the mechanisms that drive swarm-cell differentiation remain to be elucidated at both the physiological and molecular levels.
Although an intact chemotaxis phosphorelay system is essential, chemotaxis is dispensable for swarm-cell differentiation and migration. Swarming is abolished in cheA, cheW, cheR, and cheY mutants of serovar Typhimurium (15), but fliM and fliG flagellar motor switch mutants maintain their ability to swarm (5). It is believed that the flagellum senses an increase in the viscosity of the growth medium, which subsequently triggers swarm-cell differentiation in Vibrio parahaemolyticus (16). However, changes in the viscosity of the growth medium do not seem to trigger swarm-cell differentiation in serovar Typhimurium (31). Lipopolysaccharide (LPS) plays a pivotal role in the surface migration of serovar Typhimurium, as transposon mutations at a variety of loci associated with LPS biosynthesis or modification result in a defective swarming phenotype (31). LPS and other extracellular components may improve the wettability of the swarm medium, decreasing surface friction, thus allowing differentiated swarm cells to migrate outward.
Activation of the two-component regulatory system PmrAB results in a variety of modifications of LPS and cell envelope components of serovar Typhimurium (34). PmrA is activated either by PmrB, under conditions of moderate acidity or high Fe3+ (6, 24), or by PmrD, under Mg2+-limiting conditions via PhoPQ (17). PmrA regulates the transcription of the pmrHFIJKLM operon, which in turn adds 4-aminoarabinose (l-Ara4N) residues to the lipid A portion of LPS (14, 32, 33). This modification adds an extra positive charge to the LPS, reducing the binding potential of cationic antimicrobial peptides like polymyxin and azurocidin. Although indistinguishable by intraperitoneal challenge, the virulence of serovar Typhimurium with mutations in the pmrHFIJKLM operon was reduced compared to that of the wild type when mice were orally challenged (14). These results suggest that the pmr operon products may be important for resistance to antimicrobial peptides that are present within the intestinal environment.
In this paper, we report that the pmrK mutation in serovar Typhimurium results in loss of swarming motility. We demonstrate that pmrK expression is up-regulated under conditions that promote swarm-cell differentiation and, accordingly, swarm cells of serovar Typhimurium exhibit increased resistance to polymyxin in comparison to cells in the vegetative state. Intriguingly, swarm cells also displayed various degrees of increased resistance to a wide variety of antibiotics. The increased resistance of swarm cells to antibiotics arises because of an altered phenotypic state of the cells, not through genetic selection. This adaptive antibiotic resistance response may play a role in the survival and spread of Salmonella spp. in both the environment and hosts.
MATERIALS AND METHODS
Bacterial strains.
S. enterica serovar Typhimurium 14028 is a wild-type virulent strain (ATCC 14028). Strain 7953s is a phoPQ::Tn10 mutant of serovar Typhimurium LT2 obtained from the Salmonella Genetic Stock Centre (University of Calgary). Strain CS031 is a pmrK::MudJ mutant, and strain CS004 is an STM2532::MudJ mutant, both of which were constructed in this study from parent strain 14028 (see Fig. 1A).
FIG. 1.
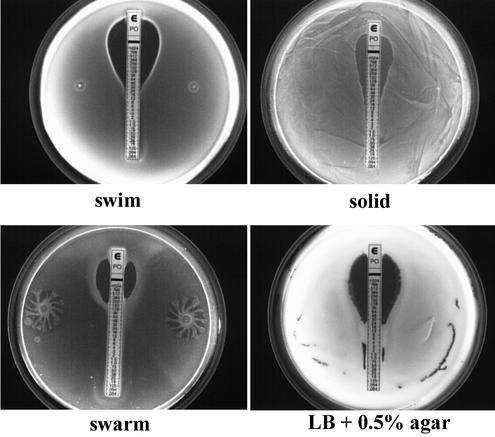
Characterization of swarm mutants of serovar Typhimurium 14028. (A) Schematic representation of MudJ insertion sites in swarm mutants CS031 and CS004. Δ indicates an insertion site, and the arrow represents the orientation of the lacZ fusion. (B) Comparison of swarming and swimming motility between wild-type and mutant strains. Strain 7953s is a phoPQ::Tn10 mutant of serovar Typhimurium LT2. All strains were inoculated onto swarm and swim plates and incubated at 37°C for 5 h.
Swarm and swim motility assays.
One microliter of an overnight culture was spotted in the middle of a swarm plate (Difco Nutrient Broth [NB], 0.5% [wt/vol] glucose, 0.5% agar [Difco]) or a swim plate (NB, 0.5% glucose, 0.25% agar) and allowed to dry for 1 h at room temperature. All plates were incubated at 37°C for 10 h or as noted otherwise. For phase-contrast microscopic analysis, swarm or swim cells were scraped from their respective fronts and then visualized in NB or saline on a glass slide.
MudJ mutagenesis and isolation of mutants defective in swarming motility.
Random MudJ insertion mutants of serovar Typhimurium LT2 were generated by using the phage P22 delivery system as previously described (19). MudJ recombinants were spotted onto swarm and swim plates, and only those that exhibited the swarm-negative and swim-positive phenotypes were subjected to further analyses. Mutants with obvious defects in the O-antigen component of LPS were first removed from the screen on the basis of the absence of P22 sensitivity, and the MudJ insertions in the remaining mutants were transferred to serovar Typhimurium 14028 by using the phage P22 delivery system. The mutants were spotted onto swarm plates containing various basal media (Luria-Bertani [LB; Difco], brain heart infusion, M9, and Mueller-Hinton II) and carbon sources (glucose, arabinose, fructose, and gluconate [0.5%, wt/vol]). Only those mutants that failed to swarm under all growth conditions were selected, and the sites of MudJ insertion were identified by arbitrary primed PCR as previously described (29).
pmrK expression analysis.
The pmrK::lacZ transcriptional fusion generated by the MudJ insertion was used to monitor the expression of the pmr operon. Four microliters of CS031, grown overnight in NB (0.5% glucose), was spread onto swarm plates containing 0, 0.3, 0.45, 0.7, 1.0, or 1.5% agar. Following 6 h of incubation at 37°C, the plate cultures were harvested by gentle scraping with a plastic coverslip and the optical densities at 600 nm of all samples were equilibrated in 0.5 to 1.5 ml of Z buffer. The β-galactosidase activity in the cell preparations was assayed as described by Miller (21). All assays were repeated three times to ensure reproducibility.
Antibiotic resistance profiling.
To compare the polymyxin sensitivities of strains 14028 and CS031 in broth, overnight LB broth cultures were diluted in fresh LB medium and 104 to 105 CFU were inoculated into 96-well plates containing 100 μl of LB medium with a series of twofold dilutions of polymyxin in concentrations ranging from 500 to 0.000125 μg ml−1. The minimum concentration of polymyxin that inhibited visible growth was declared the MIC. E-test strips (AB Biodisk, Solna, Sweden) were used to compare the relative resistances of swarm and vegetative cells of serovar Typhimurium 14028 to a wide variety of antibiotics. Vegetative cells were analyzed on either solid (NB, 0.5% glucose, 1.5% agar) or swim plates. On solid plates, 100 μl containing 104 to 105 cells of 14028 was evenly spread and incubated at room temperature for 20 min and the E-test strip was placed in the middle of each plate. On swim plates, 1 μl of the seed culture was inoculated at two spots and the E-test strip was placed in the middle of the plate between the two inocula. Swarm plates were inoculated in the same manner as the swim plates. All plates were incubated overnight at 37°C, and the E-test results were interpreted in accordance with the manufacturer's instructions. All assays were repeated at least two times to ensure reproducibility.
RESULTS AND DISCUSSION
pmrK and STM2532 mutations result in loss of swarming motility.
Strain CS031, harboring a pmrK::MudJ insertion mutation, failed to exhibit swarming motility when inoculated onto a swarm plate (Fig. 1). Toguchi et al. (31) previously reported that a variety of LPS biosynthesis or modification mutants of serovar Typhimurium failed to exhibit swarming motility. All LPS mutants, however, retained the ability to swim and became hyperflagellated when grown on swarm plates. Similarly, growth in the swim plates at 37°C revealed that CS031 was still motile (Fig. 1B), and under growth conditions that promote swarm-cell differentiation, a heterogeneous mixture of short and elongated (i.e., 2 to 3 cell lengths) cells were observed by phase-contrast microscopic analysis (data not shown). Thus, CS031 maintains its ability to differentiate into swarm cells and the pmrK mutation exerts a nonflagellar physical defect that prevents the differentiated swarm cells from migrating outward. No swarming or swimming defect was observed in phoPQ mutant strain 7953s (Fig. 1B), indicating that PhoPQ-dependent expression of pmrK is not required under our experimental conditions.
The site of MudJ insertion in strain CS004, another unconditional swarm-negative, swim-positive mutant of 14028, was determined to be in STM2532, an uncharacterized open reading frame that exists in an operon upstream of pbpC encoding an Escherichia coli homolog of penicillin-binding protein 1C (Fig. 1A). Albeit with a reduced colony diameter, CS004 maintained its ability to swim (Fig. 1B). When propagated on swarm media, CS004 also differentiated into swarm cells, whose average length (n = 50) was determined to be slightly greater than that of 14028 and CS031 cells (data not shown). All of the strains exhibited cell elongation on swarm plates, regardless of the basal media, given that they were supplemented with glucose, fructose, or arabinose (see Materials and Methods). Gluconate did not support swarm-cell differentiation of strains 14028, CS004, and CS031 (data not shown).
Up-regulation of pmrHFIJKLM operon expression confers increased polymyxin resistance to swarm cells.
The pmrK gene was recently shown to encode an inner membrane enzyme that transfers l-Ara4N residues to lipid A (32), a modification that confers increased resistance to polymyxin and other cationic antimicrobial peptides (14). To determine whether pmrK expression is differentially regulated under swarming conditions, the chromosomal pmrK::lacZ fusion, generated by the MudJ insertion in CS031, was used to compare expression under swim, swarm, and nonmotile conditions. Although pmrK expression did not differ dramatically among the different agar concentrations, pmrK expression was consistently higher under conditions that promote swarming motility in wild-type 14028 (Fig. 2). The lack of dramatic differences in pmrK expression between the different growth conditions may be due largely to the fact that CS031 was spread plated onto swarm plates. Since CS031 fails to migrate on swarm plates, the entire plate was harvested for the expression assay, which contains a heterogeneous population of short and elongated cell types. Nonetheless, the expression results suggested that serovar Typhimurium could exhibit increased resistance to polymyxin in the swarm state compared to the cells in the vegetative state. Previously, McCoy et al. (20) demonstrated that an LPS mutant of Proteus mirabilis that lacked the l-Ara4N modification was severely defective in swarming motility and exhibited increased sensitivity to polymyxin and other antimicrobial peptides.
FIG. 2.
Transcriptional analysis of pmrK in serovar Typhimurium CS031 grown in various agar concentrations. β-Galactosidase assays of cells in the vegetative state (swim and solid) and cells grown under conditions that promote swarm-cell differentiation were performed. All cells were grown in NB with 0.5% glucose supplemented with agar at the concentrations indicated.
Since swarm cells dedifferentiate into vegetative cells when grown in liquid media, resistance to polymyxin was compared between the two states by using polymyxin E-test strips on swarm, swim, and solid plates. E-test strips (AB Biodisk) are plastic strips containing a predefined immobilized antibiotic gradient that provide a continuous antibiotic gradient when transferred to an agar surface. The efficacy of E-test strips has been successfully compared to that of the standard agar dilution MIC methods and demonstrated to be highly reproducible against a wide variety of microorganisms (4). As predicted, swarm cells of wild-type strain 14028 exhibited an eightfold increase in resistance to polymyxin compared to vegetative cells (Fig. 3; Table 1). Unlike P. mirabilis, serovar Typhimurium does not differentiate into swarm cells when grown on media containing >1% (wt/vol) agar (our unpublished observations; 15). Thus, only vegetative cells are produced when serovar Typhimurium is propagated on swim or solid plates. There was virtually no difference in the polymyxin MICs on the solid and swim plates, indicating that the relative differences in the agar concentrations did not significantly alter the efficacy of the E-test strips in our assays. This notion is further supported by the fact that when the cells were spread plated onto LB medium containing 0.5% agar, the same MIC was obtained as with the NB-based vegetative plates (Fig. 3). LB medium (0.5% agar) does not promote swarm-cell differentiation; thus, serovar Typhimurium remained in the vegetative state. The observed phenotype was a consequence of swarm-cell differentiation and not the result of an effect of agar concentration on the function of the E-test strip.
FIG. 3.
E-test strip comparison of polymyxin resistance of strain 14028 in the vegetative and swarm states. Cells were plated on NB containing either 0.25 or 1.5% agar (vegetative cells) or 0.5% agar (swarm cells). In addition, cells were also spread plated on LB plates containing 0.5% agar because this growth condition does not promote swarming.
TABLE 1.
Comparison of antibiotic resistance profiles of vegetative and swarm cells of serovar Typhimurium 14028
| Target | Group | Antibiotic(s) | MIC (μg/ml)a
|
||
|---|---|---|---|---|---|
| Vb | S | S/V ratio | |||
| Cell envelope | β-Lactam | Penicillin G | >32 | >32 | |
| Oxacillin | >256 | >256 | |||
| Ampicillin | 0.38 | 0.38 | 1 | ||
| Amoxicillin | 1 | 1 | 1 | ||
| Amoxicillin-clavulanic acid | 0.75 | 0.75 | 1 | ||
| Piperacillin | 3 | 16 | 5.3 | ||
| Meropenem | 0.047 | 0.25 | 5.3 | ||
| Imipenem | 0.125 | 0.125 | 1 | ||
| Ceftazidime | 0.25 | 2 | 8 | ||
| Phosphonate peptide | Fosfomycin | 4 | 4 | 1 | |
| Bacitracin | >256 | >256 | |||
| Colistin | 16 | 96 | 6 | ||
| Polymyxin | 24 | 192 | 8 | ||
| Protein synthesis | Macrolide | Clarithromycin | >256 | >256 | |
| Erythromycin | >256 | >256 | |||
| Lincosamide | Clindamycin | >256 | >256 | ||
| Aminoglycoside | Kanamycin | 12 | >256 | >21 | |
| Streptomycin | 12 | >256 | >21 | ||
| Tobramycin | 6 | >256 | >42 | ||
| Gentamicin | 3 | >256 | >85 | ||
| Tetracycline | Doxycycline | 1.5 | 2 | 1.3 | |
| Tetracycline | 0.5 | 0.5 | 1 | ||
| DNA gyrase | Quinolone | Nalidixic acid | 2 | >256 | >128 |
| Ciprofloxacin | 0.064 | 6 | 93 | ||
| RNA polymerase | Antimycobacterial | Rifampin | 6 | >256 | >42 |
| Metabolism | Folic acid antagonist | Trimethoprim | >32 | >32 | |
According to the instructions of the manufacturer, in situations where inhibition of growth occurred between two MICs, the higher value was declared the MIC. V, vegetative; S, swarm.
Results from solid or swim plates; the plate with the higher MIC was chosen when the results were slightly different.
CS031 was more susceptible to polymyxin (MIC, 0.5 μg ml−1) than was 14028 (MIC, 8 μg ml−1) in broth, consistent with previous reports of pmrK mutants (14). However, compared with that obtained with polymyxin E-test strips on solid media, the MIC was higher, at 24 μg ml−1, for both 14028 and CS031 (Table 2). In addition to the l-Ara4N modification of lipid A, the mig-14 gene product was recently shown to confer resistance to polymyxin by an unknown mechanism that is independent of PhoPQ or PmrAB (3), and four additional genes have been implicated in polymyxin resistance that are independent of the l-Ara4N modification (30). Therefore, other factors may have also contributed to the observed increase in resistance in both CS031 and 14028 under the growth conditions associated with the E-test assay. There was virtually no differences in STM2532::lacZ activity in CS004 under growth conditions that promote vegetative growth or swarm-cell differentiation (data not shown). It is unclear whether the loss of swarming motility in CS004 is the result of the inactivation of the STM2532 gene or due to polar effects on the downstream pbpC gene. PbpC is a bifunctional enzyme with both transpeptidase and transglycosylase activities in E. coli (22). The potential loss of pbpC regulation may have altered some aspect of cell division and other associated events, leading to a swarm-negative phenotype.
TABLE 2.
Comparison of antibiotic resistance profiles of swarm mutant strains CS031 and CS004 on solid plates
| Antibiotic(s) | MIC (μg/ml)
|
|
|---|---|---|
| CS031 | CS004 | |
| Ampicillin | 0.38 | 0.38 |
| Amoxicillin-clavulanic acid | 0.75 | —a |
| Meropenem | 0.023 | — |
| Imipenem | 0.094 | 0.094 |
| Ceftazidime | 0.25 | 0.25 |
| Colistin | 12 | 12 |
| Polymyxin | 24 | — |
| Streptomycin | 4 | — |
| Tobramycin | 3 | 2 |
| Nalidixic acid | 2 | 3 |
| Rifampin | 6 | 3 |
—, antibiotic not tested.
Swarm-cell differentiation is linked to elevated resistance to a wide variety of antibiotics.
On the basis of the observation that mutations in genes that had been previously implicated in antibiotic resistance result in loss of swarming motility, we questioned whether swarm-cell differentiation in serovar Typhimurium is coupled with increased resistance to other antibiotics. E-test strips representing a wide variety of antibiotics were used in the same manner as in the polymyxin resistance assay to compare vegetative and swarm cells of strain 14028. As summarized in Table 1, swarm cells exhibited an antibiotic resistance profile different from that of cells in the vegetative state. Swarm cells were more resistant to three of the nine β-lactam antibiotics tested. The dramatic morphological changes associated with swarm-cell differentiation may be indicative of an altered cell wall structure with increased resistance to disruption, consequently decreasing the effectiveness of these cell wall-targeting antibiotics. The mechanism(s) underlying the increased resistance of swarm cells to colistin is most likely the same as those described for polymyxin (i.e., LPS modification), since it too is a cationic polypeptide.
The elevated antibiotic resistance phenotype of swarm cells extended beyond antibiotics that target the cell envelope to antibiotics that target the components within the cytoplasm, including protein translation, DNA replication, and transcription (Table 1). Dramatic differences were observed with all aminoglycosides, as the swarm fronts migrated beyond the strips' maximum MIC of 256 μg/ml (Fig. 4). The general mechanism of bacterial resistance to aminoglycosides is based on enzymatic modifications of the drug (8) that reduce both the influx rate and activity within the cytoplasm. These specific enzymes are not present in strain 14028; thus, swarm cells may exhibit elevated resistance to aminoglycosides either by increasing the activity of other preexisting enzymes or up-regulating the expression of novel proteins. The influx rate of aminoglycosides can be significantly increased by using cell wall-targeting antibiotics that act synergistically, such as penicillin (8). In an inverse fashion, the cell envelope (and LPS) of swarm cells may already be physically distinct from that of the vegetative cells, which may indirectly, yet effectively, block the entry of the aminoglycosides and other antibiotics by a nonenzymatic mechanism. In support of this view, Macfarlane et al. (18) demonstrated that mutations in Pseudomonas aeruginosa that may increase outer membrane stability decrease the permeability of the outer membrane to aminoglycosides. Furthermore, membrane-associated LPS and other released exopolysaccharides that are essential for swarming in serovar Typhimurium (31) may initially bind and restrict the penetration of aminoglycosides, as demonstrated with tobramycin in P. aeruginosa (13, 23).
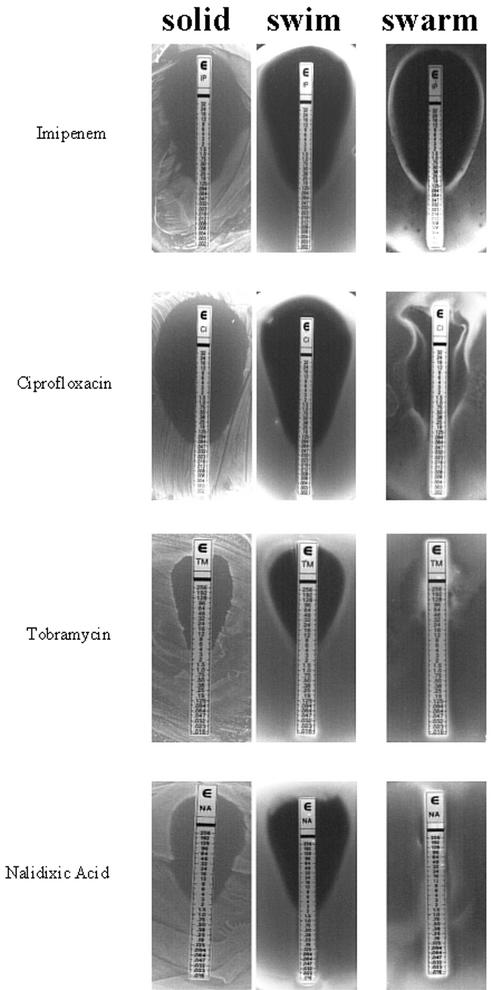
FIG. 4.
Representative E-test strip analyses of vegetative and swarm cells of strain 14028. Swarm cells exhibit either no difference (imipenem) or elevated resistance (ciprofloxacin, tobramycin, and nalidixic acid) compared to vegetative cells (solid and swim). Note the formation of a secondary swarm front in response to ciprofloxacin. All of the E-test strip results obtained are summarized in Table 1.
Distinct waves of swarm fronts were associated with heightened resistance to piperacillin, ceftazidime, ciprofloxacin, and rifampin. As shown in Fig. 4, an initial swarm front formed in the presence of ciprofloxacin at 0.25 μg/ml and a secondary swarm front formed at 6 μg/ml. These distinct fronts are similar in appearance to the concentric rings generated by Proteus spp., representing phases of consolidation, where swarm cells dedifferentiate into vegetative cells and subsequently redifferentiate into swarm cells (10). Although Salmonella spp. do not normally exhibit phases of consolidation (15), the observed fronts may represent phases of adaptation and/or selection in response to increasing levels of antibiotics.
The observed increased resistance of swarm cells to antibiotics was the result of a transient physiological state. Dedifferentiated swarm cells (i.e., swarm cells growing at the elevated antibiotic concentrations inoculated and incubated overnight in fresh liquid NB [0.5% glucose]) exhibited the same sensitivity to the respective antibiotics as the primary vegetative cells (Table 3), confirming that the E-test swarm plate assay did not simply select for antibiotic-resistant mutants. The altered antibiotic resistance of swarm cells therefore represents an adaptive change in the cells. When swarm cells of 14028 were inoculated into LB broth, the cells were completely killed within 150 min of incubation with lethal doses of polymyxin and kanamycin (data not shown). The reversion to more sensitive states upon dedifferentiation suggests that changes in gene expression associated with the swarm-cell differentiation and/or the dilution of the modified LPS upon dedifferentiation are responsible for the observed phenotypic changes. Interestingly, it was recently shown that dramatic global transcriptome changes occur in serovar Typhimurium when it is exposed to sublethal concentrations of a variety of antibiotics (12). Since the swarm cells physically migrate from a region that lacks antibiotics toward an increasing antibiotic gradient in our assay, they would be preexposed to subinhibitory concentrations of antibiotics. However, if such preexposure alone were responsible for the observed elevated resistance, the vegetative swim cells should have exhibited a similar phenotype. The antibiotic resistance profiles of mutant strains CS031 and CS004 were much like that of wild-type strain 14028 in the vegetative state (Table 2). The resistance profiles of the strains in the swarm state could not be compared conclusively because CS031 and CS004 cannot migrate on swarm plates. When the mutant strains were spread plated onto swarm plates, the resistance profiles were virtually the same as those observed on solid plates (data not shown). This was most likely due to the fact that the initial inocula were rapidly destroyed in the kill zone before the cells could properly differentiate into swarm cells.
TABLE 3.
Comparison of antibiotic resistance profiles of vegetative cells of serovar Typhimurium 14028 prior to swarm-cell differentiation and following dedifferentiation from swarm cells that had been exposed to an antibiotic
| Antibiotic | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Initial inoculuma
|
Swarm front inoculumb
|
|||
| Solid | Swim | Solid | Swim | |
| Piperacillin | 1.5 | 1.5 | 1.5 | 1.5 |
| Ceftazidime | 0.19 | 0.19 | 0.19 | 0.125 |
| Polymyxin | 24 | 24 | 24 | 24 |
| Kanamycin | 6 | 12 | 6 | 6 |
| Streptomycin | 8 | 12 | 8 | 12 |
| Ciprofloxacin | 0.064 | 0.064 | 0.064 | 0.064 |
| Rifampin | 4 | 4 | 4 | 6 |
Single colonies incubated overnight in NB (0.5% glucose) in the absence of antibiotics.
Cells that were harvested from the swarm front that formed in the presence of an antibiotic and were than incubated overnight in NB (0.5% glucose).
Discussion of global experimental approaches by which to elucidate the regulatory mechanisms underlying swarming motility and associated phenotypes.
Elevated resistance to a broad spectrum of antibiotics would provide distinct advantages to Salmonella spp. in a clinical setting. Although swarming behavior has been demonstrated to play a significant role in the pathogenesis of numerous organisms (10), the biological significance of the swarm phenomenon in Salmonella spp. remains elusive. Construction of isogenic constitutive swarm-positive and phenotypically nonpleiotropic swarm-negative mutants may shed light on this issue. All of the clinical isolates of Salmonella spp. and many of the E. coli isolates that we have examined to date possess the ability to swarm, and interestingly, E. coli O157:H7 has exhibited the most prolific swarming phenotype (V. Sood and M. Surette, unpublished observations).
Initiation of swarm-cell differentiation is intimately associated with the nutrient composition of the growth medium and the population density (10, 15). Thus, multiple sensory and regulatory pathways are likely integrated to coordinate the appropriate expression of genes and/or modification of proteins to drive swarm-cell differentiation and colony migration in serovar Typhimurium. Extensive transposon mutagenesis of serovar Typhimurium generated a large list of candidate genes that may be involved in swarm-cell differentiation and migration (31). However, it is difficult to elucidate the roles of these genetic elements in swarm-cell differentiation, since all of the reported mutations resulting in the swarm-negative phenotype were rescued by simply changing the source of the agar (31). A reductive approach, such as random transposon mutagenesis, can identify key factors required for a cell to go from one state to another. Antithetically, systemic global approaches can provide information on the changes that occur as wild-type cells are exposed to various growth conditions, resulting in distinct physiological states. In combination with the utility of readily available genomic information, a global approach may generate more comprehensible data to aid in the elucidation of mechanisms underlying complex phenomena such as the swarming behavior of serovar Typhimurium. The phenotypic increase in resistance to multiple antibiotics observed in swarm cells is not likely coupled to the mechanisms of swarm-cell differentiation but rather reflects the unique physiological state of the swarm cells.
Transcriptomic and proteomic profiling may provide insight into other physiological phenomena that may be linked to swarming, including elevated antibiotic resistance. Intriguingly, expression of numerous virulence factors has been shown to be coregulated with swarm-cell differentiation in P. mirabilis (11). Proteomic comparison of serovar Typhimurium in the swarm and vegetative states revealed numerous readily detectable differences (W. Kim and M. G. Surette, unpublished results). Experiments are currently in progress to identify these differentially expressed and modified proteins. This should provide insight not only into the molecular and biochemical mechanisms that govern swarming but also into the physiological basis underlying the coregulation of other linked phenotypes.
Although the specific mechanisms of elevated antibiotic resistance remain to be elucidated, it is tempting to speculate that some common traits may be associated with the increased antibiotic resistance observed in biofilm cells. As reviewed by Stewart (26), the mechanisms of antibiotic resistance in biofilms are believed to be quite distinct from the conventional antibiotic resistance mechanisms. Multiple factors are believed to confer increased antibiotic resistance in biofilms, including their innate refractoriness to penetrating antibiotics, coupled with the formation of persister cells around the perimeters of a growing biofilm, which are believed to be specifically differentiated to elicit an elevated protective phenotype. Although Spoering and Lewis (25) provided convincing evidence that planktonic cells in the stationary phase of growth exhibit levels of antibiotic resistance comparable to those of biofilm cells, they also attributed the mechanism of elevated antibiotic resistance to the presence of persister cells in the stationary-phase planktonic population. Similarly, the formation of distinct swarm fronts may be indicative of adaptively differentiating swarm cells, resulting in a persister-like phenotype. Moreover, the adaptive resistant state observed in swarm cells does not appear to be confined to a subset of cells but is observed in the entire population. This will greatly facilitate the analysis of the mechanism(s) of this behavior by proteomic approaches. The adaptive resistance response described here likely plays a significant role in the successful expansion of a migrating colony, both in and outside a clinical setting.
Acknowledgments
We thank D. Woods and J. Davies for providing the E-test strips and J. Davies for reviewing the manuscript.
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR). M.G.S. is also an Alberta Heritage Foundation for Medical Research Senior Scholar and Canada Research Chair in Microbial Gene Expression.
REFERENCES
- 1.Berry, R. M., and J. P. Armitage. 1999. The bacterial flagella motor. Adv. Microb. Physiol. 41:291-337. [DOI] [PubMed] [Google Scholar]
- 2.Bouret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 3.Brodsky, I. E., R. K. Ernst, S. I. Miller, and S. Falkow. 2002. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J. Bacteriol. 184:3203-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. F., and L. Brown. 1991. Evaluation of the E test, a novel method of quantifying antimicrobial activity. J. Antimicrob. Chemother. 27:185-190. [DOI] [PubMed] [Google Scholar]
- 5.Burkart, M., A. Toguchi, and R. M. Harshey. 1998. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2568-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamnongpol, S., W. Dodson, M. J. Cromie, Z. L. Harris, and E. A. Groisman. 2002. Fe(III)-mediated cellular toxicity. Mol. Microbiol. 45:711-719. [DOI] [PubMed] [Google Scholar]
- 7.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow, J. W. 2000. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31:586-689. [DOI] [PubMed] [Google Scholar]
- 9.Dahlquist, F. W. 2002. Amplification of signaling events in bacteria. Sci. STKE 2002:PE24. [DOI] [PubMed]
- 10.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630-635. [DOI] [PubMed] [Google Scholar]
- 11.Fraser, G. M., L. Claret, R. Furness, S. Gupta, and C. Hughes. 2002. Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiology 148:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goh, E.-B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, C. A., N. A. Hodges, and C. Marriott. 1988. Antibiotic interaction and diffusion through alginate and exopolysaccharide or cystic fibrosis-derived Pseudomonas aeruginosa. J. Antimicrob. Chemother. 22:667-674. [DOI] [PubMed] [Google Scholar]
- 14.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harshey, R. M., and T. Matsuyama. 1994. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. USA 91:8631-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawagashi, I., M. Imagawa, Y. Imae, L. L. McCarter, and M. Homma. 1996. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol. Microbiol. 20:693-699. [DOI] [PubMed] [Google Scholar]
- 17.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macfarlane, E. L. A., A. Kwasnicka, and R. E. W. Hancock. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543-2554. [DOI] [PubMed] [Google Scholar]
- 19.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 20.McCoy, A. J., H. Liu, T. J. Falla, and J. S. Gunn. 2001. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichols, W. W., S. M. Dorrington, M. P. Slack, and H. L. Walmsley. 1988. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob. Agents Chemother. 32:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart, P. S. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292:107-113. [DOI] [PubMed] [Google Scholar]
- 27.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 28.Stock, J. B., M. N. Levit, and P. M. Wolanin. 2002. Information processing in bacterial chemotaxis. Sci. STKE 2002:PE25. [DOI] [PubMed]
- 29.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamayo, R., S. S. Ryan, A. J. McCoy, and J. S. Gunn. 2002. Identification and genetic characterization of PmrA-regulated genes involved in polymyxin B resistance in Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6770-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 33.Wosten, M. M., and E. A. Groisman. 1999. Molecular characterization of the PmrA regulon. J. Biol. Chem. 274:27185-27190. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, Z., A. A. Ribeiro, S. Lin, R. J. Cotter, S. I. Miller, and C. R. Raetz. 2001. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PmrA-dependent 4-amino-4-deoxy-l-arabinose and phosphoethanolamine incorporation. J. Biol. Chem. 276:43111-43121. [DOI] [PubMed] [Google Scholar]