Abstract
IL-17 is a T cell-derived cytokine that may play an important role in the initiation or maintenance of the proinflammatory response. Whereas expression of IL-17 is restricted to activated T cells, the IL-17 receptor is found to be widely expressed, a finding consistent with the pleiotropic activities of IL-17. We have cloned and expressed two novel human cytokines, IL-17B and IL-17C, that are related to IL-17 (≈27% amino acid identity). IL-17B mRNA is expressed in adult pancreas, small intestine, and stomach, whereas IL-17C mRNA is not detected by RNA blot hybridization of several adult tissues. No expression of IL-17B or IL-17C mRNA is found in activated T cells. In a survey of cytokine induction, IL-17B and IL-17C stimulate the release of tumor necrosis factor α and IL-1β from the monocytic cell line, THP-1, whereas IL-17 has only a weak effect in this system. No induction of IL-1α, IL-6, IFN-γ, or granulocyte colony-stimulating factor is found in THP-1 cells. Fluorescence-activated cell sorter analysis shows that IL-17B and IL-17C bind to THP-1 cells. Conversely, IL-17B and IL-17C are not active in an IL-17 assay or the stimulation of IL-6 release from human fibroblasts and do not bind to the human IL-17 receptor extracellular domain. These data show that there is a family of IL-17-related cytokines differing in patterns of expression and proinflammatory responses that may be transduced through a cognate set of cell surface receptors.
IL-17 is a recently described, T cell-derived cytokine whose biological functions are only beginning to be elucidated (1, 2). When the cytokine initially was identified as a cDNA clone from a rodent T cell lymphoma, it was recognized that its sequence is similar to an ORF from a primate herpesvirus, Herpesvirus saimiri (3–5). Previous findings of the viral protein have indicated that it has many if not all of the immunostimulatory activities found for the host IL-17 (6, 7).
Human IL-17 is a 20- to 30-kDa, disulfide-linked, homodimeric protein with variable glycosylation (8, 9). It is encoded by a 155-aa ORF that includes an N-terminal secretion signal sequence of 19–23 residues. The amino acid sequence of IL-17 is similar only to the Herpesvirus protein described above and is not similar to that of other cytokines or known proteins. Of the large number of tissues and cell lines evaluated, the mRNA transcript encoding IL-17 has been detected only in activated T cells and phorbol 12-myristate 13-acetate/ionomycin-stimulated peripheral blood mononuclear cells (8, 9).
Despite its restricted tissue distribution, IL-17 exhibits pleiotropic biological activities on various types of cells, such as fibroblasts, endothelial cells, and epithelial cells (1, 2). IL-17 has been found to stimulate the production of many cytokines: tumor necrosis factor α (TNF-α) and IL-1β from macrophages (10); IL-6, IL-8, and the intracellular adhesion molecule-1 (ICAM-1) from human fibroblasts (8, 9); and granulocyte colony-stimulating factor (G-CSF) and prostaglandin 2 (PGE2) from synoviocytes (9). Through the induction of a number of cytokines, IL-17 is able to mediate a wide range of responses, mostly proinflammatory and hematopoietic.
Results from several clinical studies indicate that IL-17 may be involved in many inflammatory diseases. IL-17 is secreted by synovial T cells from rheumatoid arthritis patients and stimulates the production of inflammatory mediators (11, 12). Elevated IL-17 mRNA expression has been found in mononuclear cells from patients with multiple sclerosis, a myelin-directed autoimmune disease (13). In addition, the expression level of IL-17 mRNA is increased in T cells isolated from lesional psoriatic skin (14). The above findings have led to the suggestion that IL-17 may play a pivotal role in the initiation or maintenance of an inflammatory response (10).
Consistent with IL-17's wide range of effects, the cell surface receptor for IL-17 has been found to be widely expressed in many tissues and cell types (15). Although the amino acid sequence of the hIL-17 receptor (866 aa) predicts a protein with a single transmembrane domain and a long, 525-aa intracellular domain, the receptor sequence is unique and is not similar to that of any of the receptors from the cytokine/growth factor receptor family. This coupled with the lack of similarity of IL-17 itself to other known proteins indicates that IL-17 and its receptor may be part of a novel family of signaling protein and receptors.
As yet, only limited information is available about the intracellular signaling induced by IL-17. It has been shown to stimulate a transient Ca2+ influx and a reduction in [cAMP]i in human macrophages (10). Treatment of fibroblasts and macrophages with IL-17 results in the activation of NF-κB (4, 10). In chondrocytes, IL-17 has been shown to activate NF-κB as well as mitogen-activated protein kinases (16).
As a part of a larger effort to clone, express, and assay a number of secreted and membrane-bound proteins, we describe in this paper the cloning and initial characterization of two novel proteins, IL-17B and IL17C, that are similar in amino acid sequence to IL-17.
Materials and Methods
Clones Encoding IL-17, IL-17B, and IL-17C.
blast (17) was used to screen expressed sequence tag (EST) sequences from publicly available databases and from the LifeSeq database from Incyte Pharmaceuticals (Palo Alto, CA) for encoded amino acid sequences similar to IL-17. For IL-17B, two EST sequences with similarity to IL-17 were identified, GenBank loci AA443286 and W74664. These were extended to include three additional, publicly available EST sequences, and one of these clones (W74558 from a human fetal heart cDNA library) was obtained from the IMAGE consortium (18) and designated DNA59294. For IL-17C, a single EST sequence in the Incyte database was identified from a human prostate cDNA library. A full-length cDNA clone (DNA62377) was isolated from a human fetal kidney library. For IL-17, the full-length hIL-17 DNA was constructed by PCR of 12 overlapping oligonucleotides based on the known sequence (8). The construct was confirmed by DNA sequencing.
RNA Expression Analysis and Reverse Transcription–PCR Analysis.
Multitissue blots containing poly(A)+ RNA (2 μg per lane) from various human tissues were purchased from CLONTECH. The entire coding regions of human IL-17B (700 bp) and IL-17C (1.1 kb) were used as hybridization probes. DNA probes were labeled with [α-32P]dCTP by random-priming DNA labeling beads (Amersham Pharmacia). Hybridization was performed with Expresshyb (CLONTECH) at 68°C for 1 hr. The blots then were washed with 2× SSC/0.05% SDS solution at room temperature for 40 min, followed by washes in 0.1× SSC/0.1% SDS solution at 55°C for 40 min with one change of fresh solution. The blots were exposed in a PhosphorImager.
Peripheral blood lymphocytes (PBL) were isolated from the blood of healthy donors by Ficoll centrifugation over the Lymphocyte Separation Medium (ICN/Cappel). The CD4+ T cell-enriched population was prepared from PBL by positive immunomagnetic selection using a MACS (Miltenyi Biotec). CD4+ T cells were activated for 40 hr with a mixture of 10 ng/ml phorbol 12-myristate 13-acetate and 500 ng/ml ionomycin. Total RNA was extracted by using RNeasy Midi kit (Qiagen) and reverse-transcribed by using an oligo(dT) primer (Perkin–Elmer) and Superscript II (GIBCO). Each cDNA sample was PCR-amplified (30 cycles of 1 min at 95°C, 1 min at 60°C, and 1 min at 72°C) by using pairs of 22-mer that hybridize to the 5′ and 3′ ends of human IL-17-, IL-17B-, or IL-17C-coding regions. The expected size for IL-17 PCR product is 469 bp, for IL-17B is 543 bp, and for IL-17C is 594 bp.
Chromosome Localization.
The human chromosomal localization of IL-17B and IL-17C was determined by using Taqman primers and probes designed in the 3′ untranslated region of the genes of IL-17B and IL-17C. PCR was performed with the Stanford Radiation Hybrid Panel G3 of human/Chinese hamster ovary clones (19, 20).
Recombinant Fc/His Fusion Proteins.
The coding sequences of IL-17, IL-17B, and IL-17C were amplified by PCR and subcloned into the EcoRI and StuI sites of pBPH.IgG to generate a C-terminal fusion with the Fc region of human IgG1 or into the EcoRI and SmaI sites of pBPH.His.c to generate a C-terminal GHHHHHHHH tag. Vectors pBPH.IgG and pBPH.His.c are derivatives of the baculovirus expression vector pVL1393 (PharMingen).
The fusion proteins were expressed in H5 cells by using recommended procedures (Invitrogen). In brief, the DNA constructs were cotransfected with BaculoGold Baculovirus DNA (PharMingen) in a 7:1 ratio into adherent Sf9 cells. Cells were incubated at 28°C for 4 days, and the supernatant was harvested. The transfection supernatant was amplified and subjected to affinity purification by either protein A-Sepharose beads (Pharmacia) for Fc fusion proteins or Ni-NTA agarose beads (Qiagen) for His-tagged proteins. The control Fc fusion protein, Flt4.Fc, was prepared as described (21).
To examine the protein expression, SDS/PAGE analysis was performed on the affinity-purified, recombinant proteins under nonreducing and reducing conditions, followed by silver staining.
Biological Activities.
Human foreskin fibroblast cells (American Type Culture Collection) were cultured in MEM (10% FBS) with the test cytokine. After incubation for 18 hr at 37°C and 5% CO2, conditioned media were assayed for IL-6 by using an ELISA kit (R&D Systems). Human leukemia monocytic THP-1 cells were cultured in RPMI 1640 medium (10% FBS) with the test cytokine. After incubation for 18 hr at 37°C and 5% CO2, conditioned media were quantitated for release of IL-1α, IL-1β, IL-6, IFN-γ, granulocyte colony-stimulating factor, and TNF-α by using ELISA assay kits (R&D Systems).
Cloning of the Extracellular Domain (ECD) of hIL-17 Receptor.
Two oligonucleotide primers that hybridize to the 5′ and 3′ ends of the hIL-17R ECD were used in PCRs to amplify the ECD sequence from a human testis cDNA library with Pfu Turbo DNA polymerase (Promega). A His8-tag also was introduced at the C terminus by PCR. The final PCR product then was subcloned into an expression plasmid vector pRK5B. Sequence analysis confirmed that the insert contains a DNA fragment encoding the extracellular domain (1–320 aa) of the published hIL-17 receptor (15).
Immunoprecipitation of the IL-17R ECD.
The expression plasmid containing the IL-17R ECD (C-terminal His-tagged) was transfected into 293 cells by using SuperFect transfection reagent (Qiagen). Metabolic labeling of 293 cells was performed 16 hr after transfection by using 50 μCi/ml [35S]Cys/Met mixture for 6 hr. Conditioned medium was collected and concentrated (Centricon-10; Amicon). To examine the expression of the IL-17R ECD, Ni-NTA beads (Qiagen) were used to affinity-precipitate the His-tagged IL-17R ECD from the condition medium.
The conditioned medium diluted in RIPA buffer (1% NP-40/0.5% sodium deoxycholate/0.1% SDS in PBS) was incubated with IL-17 and the Fc fusion proteins overnight at 4°C. Protein A-agarose beads (Pierce) were added to precipitate the Fc fusion proteins. The precipitates were washed extensively in RIPA buffer, denatured in SDS sample buffer, and electrophoresed on NuPAGE 4–12% Bis-Tris gels (Novex). For IL-17 immunoprecipitation, anti-IL-17 antibody (R&D Systems) was added. In a competitive binding experiment, immunoprecipitation of IL-17R ECD by IL-17 was performed in the presence of a 5-fold molar excess of IL-17B.His or control His-tagged protein.
Fluorescence-Activated Cell Sorter Analysis.
THP-1 cells (5 × 105) were preincubated in PBS containing 5% horse serum at 4°C for 30 min to block nonspecific binding. IL-17B.Fc, IL-17C.Fc, or Flt4.Fc (1 μg each) was added and incubated with the THP-1 cells in a volume of 0.25 ml on ice for 1 hr. The cells were washed extensively and stained with FITC-conjugated goat anti-human IgG (Fc-specific, 1:100 dilution; Jackson ImmunoResearch) for 1 hr at 4°C. After thorough washes, a minimum of 5,000 cells were analyzed by using a FACScan (Becton Dickinson).
Results
cDNA Clones Encoding IL-17B and IL-17C.
As a part of large-scale screen of EST databases for novel sequences encoding secreted or membrane-bound proteins, we found two sets of sequences with similarity to IL-17. The first set contained five overlapping EST sequences. One of these clones was obtained from the IMAGE consortium and encoded full-length human IL-17B. The second set of sequences contained only a single partial-length EST; a full-length clone encoding IL-17C was isolated from a human fetal kidney library based on the sequence of this partial EST.
An alignment of the predicted amino acid sequence of IL-17B and IL-17C with the known sequence of IL-17 (8, 9) shows that this is a family of related polypeptides with 26–28% amino acid identity between the three members (Fig. 1). All three contain a hydrophobic motif at the N terminus that is expected to function as a secretion signal sequence of 18–20 aa. The predicted size range for the members of this family is from 155 to 197 aa (mature monomer, ≈17–20 kDa). The alignment shows several conserved amino acids including a tryptophan residue and five cysteines in the C-terminal half of the proteins.
Figure 1.
Alignment of the protein sequences of IL-17, IL-17B, and IL-17C. The putative signal sequences are underlined, potential N-linked glycosylation sites are double-underlined, and conserved tryptophan and cysteine residues are marked with asterisks. IL-17, IL-17B, and C share 26–28% amino acid identity with each other.
mRNA Expression.
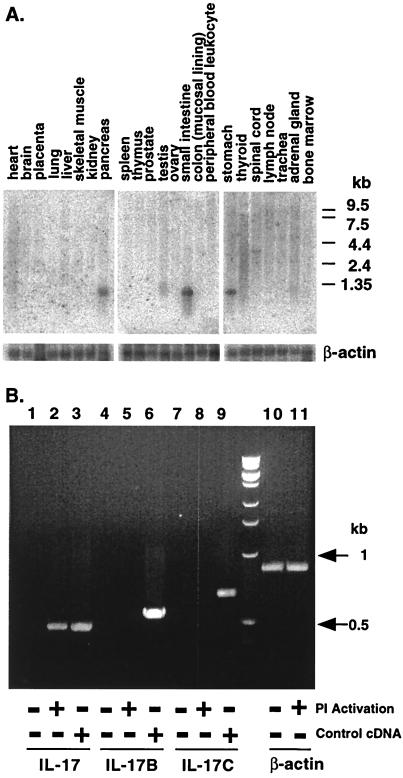
The expression of IL-17B and IL-17C in various human tissues was examined by RNA blot analysis by using probes derived from the coding sequences. For IL-17B, an 800-bp mRNA transcript was found in pancreas, small intestine, and stomach of adult human tissues; a weaker band was detected in testis (Fig. 2A). IL-17C expression was examined in the same set of adult human tissues, but no detectable signals were observed (data not shown). This result is consistent with the suggestion of low abundance inferred from the finding of only a single EST in the database searches. Whereas IL-17 mRNA is induced upon activation of CD4+ T cells (8, 9), no such induction of IL-17B or IL-17C mRNA was detected by reverse transcription–PCR amplification (Fig. 2B).
Figure 2.
RNA blot analysis of IL-17B and reverse transcription–PCR (RT-PCR) analysis. (A) Northern blot of mRNA from human tissues hybridized to a human IL-17B-specific radiolabeled probe. RNA size markers are shown on the right. A rehybridization of the same blot with a human β-actin cDNA probe is shown at the bottom. (B) Total RNA from unactivated (lanes 1, 3, 4, 6, 7, 9, and 10) and activated (lanes 2, 5, 8, and 11) CD4+ T cells was RT-PCR-amplified with primers to the 5′ and 3′ ends of human IL-17 (lanes 1–3), IL-17B (lanes 4–6), or IL-17C (lanes 7–9) coding regions. As a positive control, DNA fragments containing either IL-17 (lane 3) or IL-17B (lane 6) or IL-17C (lane 9) coding regions were diluted (1:10,000) into the cDNAs made from untreated total RNA. cDNA samples also were amplified by using human β-actin primers (lanes 10 and 11).
Chromosome Mapping.
The chromosome localization of IL-17B and IL-17C was obtained by radiation hybrid analysis of the Stanford G3 panel (19, 20). The IL-17B gene was mapped to human chromosome 5q32–34, whereas IL-17C was localized to chromosome 16q24. hIL-17 is found on chromosome 2q31 (3). Thus, this gene family is dispersed in the genome.
Biological Activity of Recombinant IL-17B and IL-17C.
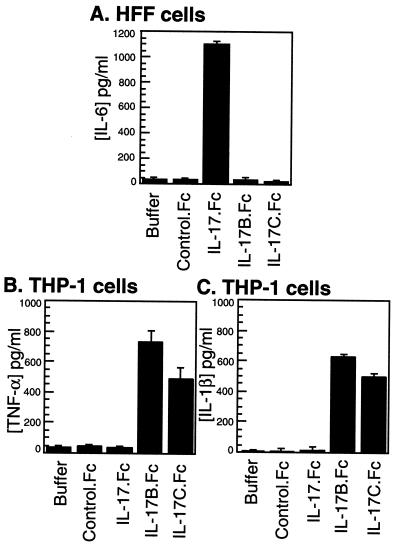
One of the biological activities of IL-17 is the induction of IL-6 release in fibroblast cells (8). To assess the biological functions of IL-17B and IL-17C, IgG Fc-tagged proteins were expressed in baculovirus-transfected Sf9 insect cells and affinity-purified. Human foreskin fibroblasts were treated with the Fc fusion proteins and assayed for IL-6 production. In contrast to the high level of IL-6 induced by IL-17, both IL-17B and IL-17C failed to stimulate IL-6 secretion in fibroblast cells (Fig. 3A). A human leukemic monocytic cell line, THP-1, reported to express IL-17 receptor (15) then was used to assay for the release of TNF-α by IL-17 or the two homologs. Although IL-17 induced only a low level of TNF-α in THP-1 cells, both IL-17B and IL-17C (as Fc fusion proteins) increased TNF-α production in THP-1 cells (Fig. 3B). The control Fc fusion protein, Flt4.Fc, had no effect. Similar results also were observed in the treatment of THP-1 cells with His-tagged IL-17B (data not shown). In addition, we surveyed the induction of cytokines that are known to be secreted by monocytes and macrophage cells. In THP-1 cells, both IL-17B and IL-17C induced high levels of IL-1β, whereas IL-17 had no effect (Fig. 3C). No stimulation by IL-17, IL-17B, or IL-17C was observed in THP-1 cells for IL-1α, IL-6, IFN-γ, or granulocyte colony-stimulating factor (data not shown).
Figure 3.
Biological activities of IL-17, IL-17B, and IL-17C. (A) Human foreskin fibroblast (HFF) cells were cultured with IL-17.Fc, IL-17B.Fc, IL-17C.Fc, or the control Fc fusion protein, Flt4.Fc (50 nM each), for 18 hr, and the conditioned media were assayed for IL-6 as described in Materials and Methods. Human leukemic cell line THP-1 was treated with the same cytokines (50 nM each) as above under the same conditions, and the supernatants were assayed for the level of TNF-α (B) or IL-1β (C). Results are expressed as the mean ± SE of triplicate determinations from one representative experiment.
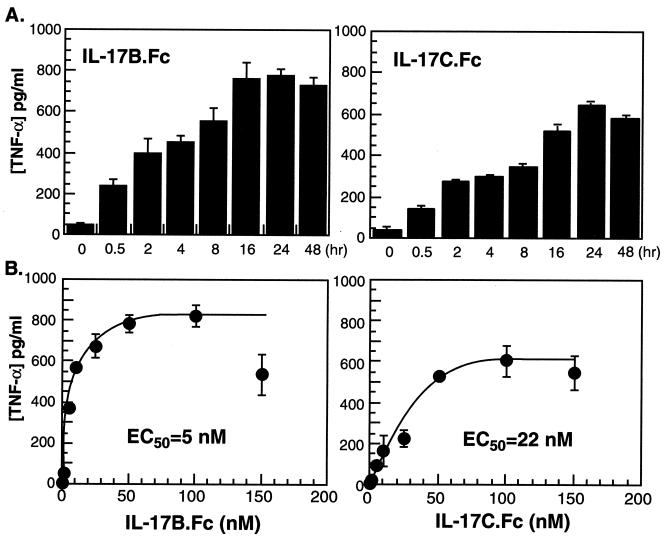
To characterize further the stimulation of TNF-α release by IL-17B and IL-17C, the time course and concentration dependence of the response were assayed in THP-1 cells. The results show that IL-17B and IL-17C stimulate the release of TNF-α in a time- and concentration-dependent manner (Fig. 4). The EC50 of TNF-α release for IL-17B stimulation is 5 nM; the EC50 for IL-17C is 22 nM.
Figure 4.
Time course and concentration dependence of IL-17B- and IL-17C-activated TNF-α release from THP-1 cells. (A) THP-1 cells were incubated with 50 nM IL-17B.Fc or IL-17C.Fc for 0.5–48 hr, the conditioned media were harvested, and the TNF-α concentration was quantitated as described in Materials and Methods. (B) THP-1 cells were treated with IL-17B.Fc and IL-17C.Fc at a concentration range from 0 to 150 nM for 18 hr, and the TNF-α release was determined.
Although the IL-17B and IL-17C preparations used in these experiments contained an undetectable level of endotoxin (less than 1 endotoxin unit/ml), we performed additional control experiments to confirm that the TNF-α release from THP-1 cells was not artifactual. The IL-17B and IL-17C activities could be abolished by heat treatment, yet they were unaffected by the addition of polymyxin B, an endotoxin-binding protein (data not shown). These observations indicate further that the proteins themselves were responsible for the activities, not any contaminating endotoxin.
IL-17 Receptor Binding.
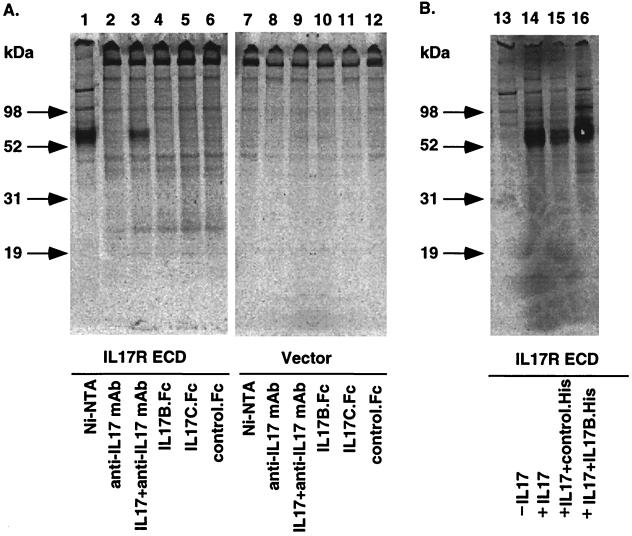
The differential activity of IL-17 when compared with IL-17B and IL-17C suggested that they might bind and activate different cell surface receptors. To directly test whether IL-17B or IL-17C binds to the IL-17 receptor, a His-tagged form of the receptor ECD was expressed in 293 cells and metabolically labeled. The IL-17R ECD migrated as a 60-kDa band when purified via its histidine tag (Fig. 5A, lane 1). As expected, this IL-17 receptor ECD could be precipitated in a complex with IL-17 and an IL-17 antibody (lane 3). However, both IL-17B and IL-17C failed to precipitate the labeled receptor band (lanes 4 and 5). In addition, IL-17B failed to compete for the binding of IL-17 to the labeled IL-17 receptor ECD (Fig. 5B, lane 15). These results suggest that IL-17B and IL-17C do not bind to the IL-17 receptor.
Figure 5.
Immunoprecipitation of IL-17R ECD with IL-17, IL-17B, and IL-17C. His-tagged IL-17 receptor ECD was expressed in 293 cells and metabolically labeled with 35S as described in Materials and Methods. The supernatant was collected. To examine the expression of the IL-17R ECD, Ni-NTA beads were used to affinity-precipitate the His-tagged IL-17R ECD in the supernatant (lane 1). (A) IL-17, IL-17B.Fc, IL-17C.Fc, or the control Fc fusion protein was incubated with the supernatant, and protein A-agarose beads were added to precipitate the Fc fusion proteins. For IL-17 immunoprecipitation reaction, anti-IL-17 antibodies were included. (B) For the competitive binding experiment, immunoprecipitation of IL-17R ECD by IL-17 was performed in the presence of a 5-fold excess of IL-17B.His and control His-tagged proteins. Precipitates in both A and B were analyzed by electrophoresis on NuPAGE (4–12% Bis-Tris) gels. Molecular mass markers are indicated at the left of A and B.
Binding to THP-1 Cells.
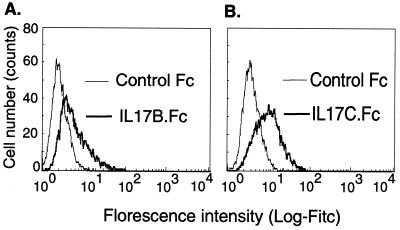
Because IL-17B and IL-17C can activate TNF-α and IL-1β release in THP-1 cells, their binding to THP-1 cells was assessed by fluorescence-activated cell sorter analysis. Both IL-17B and IL-17C (as Fc fusion proteins) displayed binding to THP-1 cells as compared with the control Fc fusion protein, Flt4.Fc (Fig. 6). In this experiment, binding to the human Fc receptors was minimized by the addition of horse serum to the binding buffer.
Figure 6.
Fluorescence-activated cell sorter analysis of the binding of IL-17B.Fc and IL-17C.Fc to THP-1 cells. THP-1 cells were incubated with IL-17B.Fc (A) or IL-17C.Fc (B) or the control Fc fusion protein, Flt4.Fc, in PBS (5% horse serum) and followed by the addition of FITC-conjugated anti-Fc secondary antibodies.
Discussion
With the isolation and initial characterization of two new relatives of IL-17, we have established and expanded the potential role this family of cytokines may play in proinflammatory immune and other responses. The three members of the family, IL-17, IL-17B, and IL-17C, are modestly related in primary structure with ≈27% overall amino acid identity including five conserved cysteine residues (Fig. 1). In addition to the primary sequence similarity, the three family members are 150–200 aa in length and are secreted from cells via a hydrophobic secretion signal sequence.
Although members of the same family based on amino acid sequence similarity, the three proteins are expressed in different tissues and are dispersed in the genome. IL-17 expression has been reported only in activated T cells (8, 9), whereas we show that IL-17B is expressed in normal human adult pancreas, small intestine, and stomach (Fig. 2A). The expression pattern of IL-17C is much more restricted, having been found only as a rare EST in an adult prostate and fetal kidney libraries. In contrast to IL-17, IL-17B and IL-17C mRNA transcripts were not detected in activated CD4+ T cells (Fig. 2B). The human genes for the family members have been localized to 2q31 for IL-17 (3), to 5q32–34 for IL-17B, and to 16q24 for IL-17C.
Our initial characterization of the biological activities of IL-17B and IL-17C shows that they differ considerably from the established activities for IL-17. IL-17B and IL-17C fail to induce the induction of IL-6 in human foreskin fibroblasts (Fig. 3A), in clear contrast to the marked induction known for IL-17 (4, 8). Conversely, IL-17B and IL-17C induce the release of TNF-α and IL-1β from the monocytic cell line, THP-1, whereas IL-17 has a very small effect (Fig. 3 B and C). None of the three family members induces IL-1α, IL-6, IFN-γ, or granulocyte colony-stimulating factor in THP-1 cells. The stimulated release of TNF-α in THP-1 cells by IL-17B and IL-17C is time- and concentration-dependent (Fig. 4), with IL-17B being about 4-fold more potent than IL-17C (EC50 = 5 nM for IL-17B vs. 22 nM for IL-17C). Thus, although still promoting the release of proinflammatory cytokines, the newly isolated IL-17B and IL-17C are likely to differ in their biological role in the immune response.
The differing biological effects of IL-17 as compared with IL-17B or IL-17C suggest that they may function via a different cell surface receptor (or some differing receptor components) than the known IL-17 receptor (15). In an initial effort to examine the question of receptor specificity directly, we have shown that IL-17B and IL-17C fail to bind to the IL-17 receptor ECD (Fig. 5A) and also that IL-17B fails to compete for the binding of IL-17 to its receptor ECD (Fig. 5B). IL-17B and IL-17C do bind to the surface of THP-1 cells, where they have activity (Fig. 6). The interaction is specific at least to the extent that a control Fc fusion protein fails to bind to these cells. These results suggest that there could be a set of receptors that bind and transduce the signal from the family of IL-17 cytokines, a receptor/ligand model that has been found for many cytokine and growth factor families. The complete lack of amino acid sequence similarity of the IL-17 family cytokines with other known proteins coupled with the lack of sequence similarity of the IL-17 receptor with any other known sequence shows that this will be a novel receptor/ligand family.
In summary, we have isolated two new cytokines related to IL-17, thus establishing the IL-17 family of cytokines. These two cytokines, IL-17B and IL-17C, differ from IL-17 in their patterns of expression and biological activities. These findings coupled with a lack of interaction with the known IL-17 receptor suggest an expanded role for the IL-17 family in the proinflammatory immune response.
Acknowledgments
We thank Amy Carlow, Thad Baker, Iqbal Grewal, Sarah Schilbach, Jacob Tropea, and Wendy Shillinglaw for their technical assistance. We also thank Ruey-bing Yang, Melanie Mark, Yisheng Jin, James Lee, and Alane Gray for their help and suggestions.
Abbreviations
- EST
expressed sequence tag
- ECD
extracellular domain
- TNF-α
tumor necrosis factor α
Footnotes
References
- 1.Spriggs M K. J Clin Immunol. 1997;17:366–369. doi: 10.1023/a:1027360106635. [DOI] [PubMed] [Google Scholar]
- 2.Broxmeyer H E. J Exp Med. 1996;183:2411–2415. doi: 10.1084/jem.183.6.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouvier E, Luciani M F, Mattei M G, Denizot F, Golstein P. J Immunol. 1993;150:5445–5456. [PubMed] [Google Scholar]
- 4.Yao Z, Fanslow W C, Seldin M F, Rousseau A M, Painter S L, Comeau M R, Cohen J I, Spriggs M K. Immunity. 1995;3:811–821. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy J, Rossi D L, Zurawski S M, Vega F, Jr, Kastelein R A, Wagner J L, Hannum C H, Zlotnik A. J Interferon Cytokine Res. 1996;16:611–617. doi: 10.1089/jir.1996.16.611. [DOI] [PubMed] [Google Scholar]
- 6.Fleckenstein B, Desrosiers R C. In: The Herpesviruses. Roizman I B, editor. New York: Plenum; 1982. pp. 253–332. [Google Scholar]
- 7.Biesinger B, Muller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Z, Painter S L, Fanslow W C, Ulrich D, Macduff B M, Spriggs M K, Armitage R J. J Immunol. 1995;155:5483–5486. [PubMed] [Google Scholar]
- 9.Fossiez F, Djossou O, Chomarat P, Flores-Romo L, Ait-Yahia S, Maat C, Pin J J, Garrone P, Garcia E, Saeland S, et al. J Exp Med. 1996;183:2593–2603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jovanovic D V, Di Battista J A, Martel-Pelletier J, Jolicoeur F C, He Y, Zhang M, Mineau F, Pelletier J P. J Immunol. 1998;160:3513–3521. [PubMed] [Google Scholar]
- 11.Chabaud M, Fossiez F, Taupin J L, Miossec P. J Immunol. 1998;161:409–414. [PubMed] [Google Scholar]
- 12.Chabaud M, Durand J M, Buchs N, Fossiez F, Page G, Frappart L, Miossec P. Arthritis Rheum. 1999;42:963–970. doi: 10.1002/1529-0131(199905)42:5<963::AID-ANR15>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Matusevicius D, Kivisakk P, He B, Kostulas N, Ozenci V, Fredrikson S, Link H. Mult Scler. 1999;5:101–104. doi: 10.1177/135245859900500206. [DOI] [PubMed] [Google Scholar]
- 14.Teunissen M B, Koomen C W, de Waal Malefyt R, Wierenga E A, Bos J D. J Invest Dermatol. 1998;111:645–649. doi: 10.1046/j.1523-1747.1998.00347.x. [DOI] [PubMed] [Google Scholar]
- 15.Yao Z, Spriggs M K, Derry J M, Strockbine L, Park L S, VandenBos T, Zappone J D, Painter S L, Armitage R J. Cytokine. 1997;9:794–800. doi: 10.1006/cyto.1997.0240. [DOI] [PubMed] [Google Scholar]
- 16.Shalom-Barak T, Quach J, Lotz M. J Biol Chem. 1998;273:27467–27473. doi: 10.1074/jbc.273.42.27467. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennon G, Auffray C, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 19.Boehnke M, Lange K, Cox D R. Am J Hum Genet. 1991;49:1174–1188. [PMC free article] [PubMed] [Google Scholar]
- 20.Walter M A, Spillett D J, Thomas P, Weissenbach J, Goodfellow P N. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, Gray A, Yuan J, Luoh S M, Avraham H, Wood W I. Proc Natl Acad Sci USA. 1996;93:1988–1992. doi: 10.1073/pnas.93.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]