Abstract
The Lyme disease agent Borrelia burgdorferi maintains both linear and circular plasmids that appear to be essential for mammalian infection. Recent studies have characterized the circular plasmid regions that confer autonomous replication, but the genetic elements necessary for linear plasmid maintenance have not been experimentally identified. Two vectors derived from linear plasmids lp25 and lp28-1 were constructed and shown to replicate autonomously in B. burgdorferi. These vectors identify internal regions of linear plasmids necessary for autonomous replication in B. burgdorferi. Although derived from linear plasmids, the vectors are maintained in circular form in B. burgdorferi, indicating that plasmid maintenance functions are conserved, regardless of DNA form. Finally, derivatives of these vectors indicate that paralogous gene family 49 is apparently not required for either circular or linear plasmid replication.
Lyme disease is the leading arthropod-borne disease in the United States. The causative agent of Lyme disease, Borrelia burgdorferi, has an unusual genomic structure composed of a linear chromosome and the largest plasmid complement of all characterized bacteria (3, 11). The advantages conferred by a segmented genome and linear replicons are not clearly understood. However, this type of genomic structure appears to have occurred early and successfully in the evolution of the genus Borrelia, since relapsing fever spirochetes, such as B. hermsii and B. turicatae, also maintain linear and circular replicons (10, 18, 29, 42).
Abundant evidence indicates that the extrachromosomal elements of Borrelia species are essential to their life cycles. The relapsing fever spirochetes have elaborate genetic systems for antigenic variation encoded on linear plasmids, as does B. burgdorferi, albeit on a reduced scale (32, 47). The more extensively characterized plasmids of B. burgdorferi also have loci coding for sugar transporters, nucleotide synthesis, outer surface protein (Osp) A, B, C, and EF-related proteins (which have been implicated in evasion of the mammalian complement system), and collagen fiber adhesins (11, 14, 15, 17, 21, 24, 25, 35, 41). In addition, a correlation between the loss of certain plasmids and loss of infectivity has been observed, further supporting the requirement for some plasmids in vivo (22, 30, 37, 46).
The genome sequence of B. burgdorferi strain B31 includes 21 plasmids (3, 11). Due to the large number and various forms of plasmids, a systematic nomenclature has been developed. Circular plasmid names begin with cp, and linear plasmids begin with lp; plasmid designations end with a number denoting the approximate size in kilobases (e.g., cp9, lp25). Different plasmids that exist in the same size and form are further delineated by a hyphen followed by consecutive numbers (e.g., lp28-1, lp28-2).
Despite the large number of plasmids and their significance to the life cycle of B. burgdorferi, relatively little is known concerning plasmid maintenance in the borreliae. Hinnebusch and Barbour demonstrated that the plasmids of B. burgdorferi are maintained at a 1:1 ratio with the chromosome (16). Subsequently, Picardeau et al. used CG skew analysis to predict a bidirectional mode of replication from an internal origin for the plasmids, and they experimentally demonstrated this for the linear chromosome (27, 28). Although these initial studies indicated that B. burgdorferi maintains strict control of plasmid copy number and replication, the details of these mechanisms are unknown. Indeed, it was uncertain whether linear and circular plasmids utilize the same maintenance functions, although linear plasmid replication has been shown to use a telomere resolution step (4, 20).
The first experimental evidence identifying genetic elements involved in plasmid maintenance was the development of a B. burgdorferi-Escherichia coli shuttle vector designated pBSV2 (43). This shuttle vector utilizes a 3.3-kb region of the endogenous 9-kb circular plasmid (cp9) to produce a stable replicon in B. burgdorferi. Subsequently, Eggers et al. constructed another shuttle vector from the corresponding region of a cp32 and identified a locus that conferred incompatibility in B. burgdorferi (8). These two shuttle vectors contain members of large paralogous gene families (PF) widely distributed across the B. burgdorferi plasmids (3). Due to their ubiquity among the plasmids, these PF have been proposed to function in plasmid maintenance (7, 11, 39, 48). The two shuttle vectors experimentally confirmed that several members of these PF contribute to circular plasmid maintenance.
Both shuttle vectors were derived from circular plasmids, members of the cp32 plasmid family. Although most of the cp32 family members are approximately 32 kb in size and homologous to each other, two derivatives differ in size. In addition to the seven members of 32 kb size, another full copy of a cp32 has integrated into lp56, and cp9 is a smaller deletion derivative (3, 11). Hence, both shuttle vectors were derived from related circular plasmids that may be capable of sharing some functions in trans. However, elements necessary for linear plasmid maintenance have not been previously characterized.
To address specific issues of linear plasmid replication and to avoid the potential complications of studying plasmid maintenance genes in the highly similar cp32 family, we focused on linear plasmids lp25 and lp28-1 to construct circular vectors capable of autonomous replication in B. burgdorferi. Our data support an internal origin of replication for linear plasmids of B. burgdorferi. These results suggest that B. burgdorferi evolved a single system for linear and circular plasmid replication and segregation, and the main difference between them may lie in how replicated plasmid forms are resolved. Finally, these studies indicate that the proteins encoded by PF 49 do not appear to be required for either circular or linear plasmid replication.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. burgdorferi strains were grown in liquid BSK-II at 35°C or in solid BSK medium incubated at 35°C under 1% CO2 (33). Strain B31 (ATCC 35210) was originally isolated from a tick collected on Shelter Island, N.Y. (1). The genomic sequence of B. burgdorferi B31 culture MI has been determined (3, 11). Strain B31-AchbC72 is a culture-attenuated, noninfectious derivative of clone B31-A, and it lacks lp56 and lp25 as well as other plasmids (45). B. burgdorferi strain A3 is an infectious clone derived from B31 MI (9). TOP10 (Invitrogen, Carlsbad, Calif.) was the strain used in E. coli plasmid manipulations.
Construction of vectors.
The strategy for vector construction is shown in Fig. 1 and is essentially the same as previously described (43). Briefly, regions putatively involved in plasmid maintenance were PCR amplified using the Expand Long Template PCR system (Roche Molecular Biochemicals, Indianapolis, Ind.). Templates for the PCR amplifications were either genomic DNA of B. burgdorferi or E. coli plasmid DNA. Oligonucleotide primers used in this study are presented in Table 1 and below in Fig. 4. Amplified fragments were cloned into pCR-XL-TOPO (Invitrogen). DNA fragments of interest were then isolated by digestion with the relevant restriction enzymes (Table 1), gel purified by electroelution, and subcloned into compatible sites of the vector pOZK (43). Large-scale plasmid DNA isolations were performed with the QIAfilter Plasmid Maxi kit (QIAGEN, Valencia, Calif.). Vector pE18::gnt was constructed by first deleting the multiple cloning site of pBSV25 (to remove the EcoRI site) and subcloning the gentamicin resistance cassette into the remaining EcoRI site present in bbe18 (9). The bbe18 gene is 579 bases long, and the EcoRI cleavage site is located at base 357.
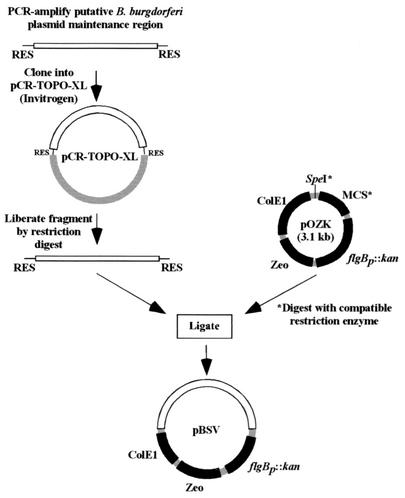
FIG. 1.
Strategy for construction of vectors used in this study. B. burgdorferi plasmid regions putatively involved in plasmid maintenance were PCR amplified from genomic DNA or from previously constructed plasmids. Primers used for the PCR amplification were designed with restriction enzyme sites (RES) for subsequent cloning steps. Fragments were cloned into pCR-TOPO-XL, liberated by specific restriction enzyme digestion, and subcloned into compatible restriction enzyme sites of pOZK, either in the multiple cloning site (MCS) or into a unique SpeI site upstream of the MCS (43). Vector pOZK contains a kanamycin resistance cassette that functions in both E. coli and B. burgdorferi. Resulting vectors were tested for their ability to autonomously replicate in B. burgdorferi. ColE1, E. coli origin of replication; Zeo, zeocin resistance marker; flgBp::kan, B. burgdorferi flgB promoter fused to the kanamycin resistance gene.
TABLE 1.
Oligonucleotides used in this study
| Namea | Sequenceb | Purpose |
|---|---|---|
| 3′.ORF3.XbaI (A) | TCTAGAGCCCTATGGATTTAAGAACTG | Construction of pBSV2 |
| ORF3.RC.HindIII (A1) | AAGCTTCCTAATCACTAAATTTCTTAC | Construction of pOZK-1-3 |
| ORF3.XhoI.stop (B) | CTCGAGGCATCATTTAACTAGTAAATTTGG | Construction of pOZK-3Δ |
| BBC02.HindIII (C) | AAGCTTTGCAACATTTTCCTTAATTCAT | Construction of pOZK-1-2 |
| BBC01.XbaI (D) | TCTAGATTACGATCCAATATCAAGTAGC | Construction of pOZK-1-3 and pOZK-3Δ |
| 9026.XbaI (E) | GTCTAGACTTGACTGCTTATTCCGGGTAATTTC | Construction of pOZK-1-2 |
| 1p25.15702.XbaI (F) | TCTAGAGTTGTATCAAGGGATATTGCC | Construction of pBSV25 |
| 1p25.9932.XbaI (G) | TCTAGACATCTGCACGATAACCTGTCG | Construction of pBSV25 |
| 1p28-1.15748.XbaI | CCTCTAGAGAGTCCTCTAGTGAGTTGTGC | Construction of pBSV28-1.Aut4 |
| 1p28-1.12165.XbaI | CGTCTAGAGCAAGGGTAAAATAAATTCAAG | Construction of pBSV28-1.Aut4 |
| 1p28-1.8292.XbaI | CCTCTAGAGCGAATTTCTTTTGATGAA | Construction of pBSV28-1.Aut3 |
| 1p28-1.5598.XbaI | GCTCTAGAGAATCGGAGAAAATGTTTACC | Construction of pBSV28-1.Aut3 |
Letters in parentheses refer to oligonucleotides indicated in Fig. 4.
Relevant restriction sites are underlined. Bolded letters indicate nucleotides that were altered to produce a stop codon.
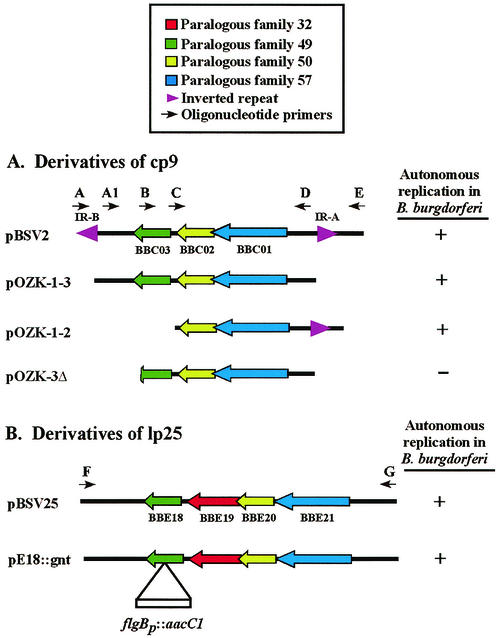
FIG. 4.
Derivatives of B. burgdorferi plasmids and their ability to autonomously replicate. Regions shown were subcloned into pOZK and tested for their ability to confer autonomous replication functions in B. burgdorferi (shown on the right). Arrowheads with letters designate oligonucleotide primers used in the PCR amplification (see Table 1). (A) Regions derived from cp9; (B) regions derived from lp25. Inactivation of bbe18 was produced by inserting the flgBp::aacC1 cassette, conferring gentamicin resistance (9).
DNA sequencing and analysis.
Nucleotide sequences were determined with the ABI Big Dye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, Calif.), using an ABI 3700 DNA sequencer. Nucleotide sequences were analyzed with DNAstar software (DNAstar, Madison, Wis.).
Electroporation of B. burgdorferi and identification of transformants.
B. burgdorferi strains were transformed by electroporation as previously described (36, 43). B. burgdorferi colonies grown on selective medium were stabbed with a sterile toothpick and inoculated into 20-μl PCR mixtures to amplify the kanamycin resistance cassette as previously described (43). Colonies positive by PCR were aspirated from the agar plate, transferred to liquid BSK, and incubated at 35°C for ∼5 days.
Total genomic DNA was isolated from 20-ml cultures of B. burgdorferi with the QIAGEN Genomic-tip 20/G kit (QIAGEN) or from 5-ml cultures by using the Wizard genomic DNA purification kit (Promega, Madison, Wis.). Genomic DNA was separated on a 0.3% agarose gel and visualized by ethidium bromide staining. Total genomic DNA from transformants was used to transform E. coli and recover vectors.
Vector incompatibility with endogenous B. burgdorferi plasmids.
B. burgdorferi transformants were further characterized for plasmid content by PCR screening with plasmid-specific primers. Primers used were designed by Purser and Norris (30) or by D. Akins as described by Elias et al. (9). PCR cycling parameters were an initial denaturation at 94°C (5 min), followed by 30 cycles of 94°C (30 s), 50 or 55°C (30 s), and 72°C (1 min), with a final extension at 72°C (7 min).
Stability of vectors in B. burgdorferi.
Stability of shuttle vectors was determined as previously described (43). Randomly chosen transformants of B31-A were cultured in 5 ml of liquid BSK at 34°C in the presence or absence of 200 μg of kanamycin/ml. Cultures were inoculated to a starting concentration of ∼4 × 105 spirochetes/ml and grown to >108 spirochetes/ml. Cells were counted by dark-field microscopy with a Petroff-Hauser counting chamber. After 11 such serial passages (∼90 generations), cultures were plated in the absence of selection and 20 colonies from each condition were PCR screened for the presence of the kanamycin resistance cassette. The stability of lp25 was assessed by selecting colonies PCR positive for lp25 and inoculating them into 5 ml of liquid BSK. Cultures were treated as described above, except no antibiotic selection was imposed. After 90 generations, cultures were plated and PCR screened for the presence of lp25.
RESULTS
Construction and transformation of vectors.
Members of five PF are present in various combinations on all B. burgdorferi plasmids and have been proposed to be involved in plasmid maintenance (7, 11, 39, 48). However, only genetic elements involved in circular plasmid maintenance have been experimentally characterized (8, 43). To identify regions responsible for linear plasmid maintenance in B. burgdorferi, we focused on lp25 and lp28-1, both of which are associated with mammalian infectivity but not required for in vitro cultivation (22, 30, 37, 46).
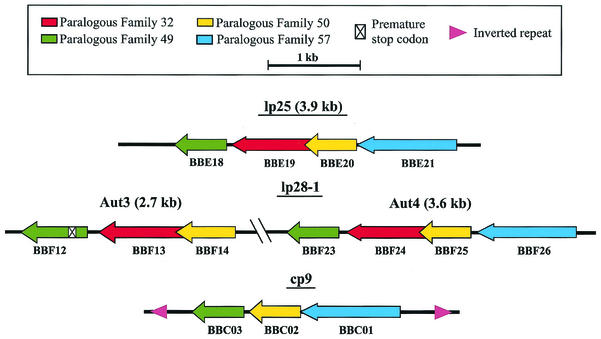
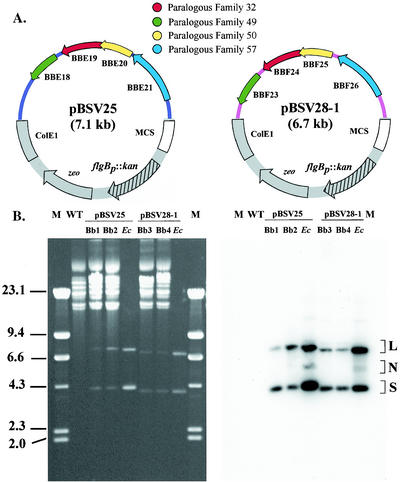
The PF members putatively involved in plasmid maintenance are located in a single, contiguous region on lp25, whereas lp28-1 has two subsets of these gene families (Fig. 2). One region of lp28-1 consists of four open reading frames (ORFs; designated the Aut 4 region), and the other region consists of three ORFs (Aut 3 region). The Aut 3 region contains a member of PF 49, bbf12, that has a premature stop codon 40 amino acids from the amino terminus of the protein (Fig. 2), as reported by The Institute for Genomic Research (3, 11). A second start codon is present 46 nucleotides downstream from the premature stop codon that could result in a truncated protein of 140 amino acids. Vectors were constructed from these three regions potentially involved in plasmid maintenance (lp25, lp28-1 Aut 3, and lp28-1 Aut 4) (Fig. 2). Transformation of B. burgdorferi with these vectors demonstrated that the lp28-1 Aut 4 and lp25 regions conferred autonomous replication in B. burgdorferi, whereas the lp28-1 Aut 3 region did not (Fig. 3 and data not shown). Hence, all subsequent experiments were performed with the vectors derived from lp28-1 Aut 4 and lp25. These Borrelia shuttle vectors were designated pBSV25 and pBSV28-1, denoting the plasmid from which they were derived. The vectors, shown in Fig. 3A, are capable of autonomous replication in E. coli and B. burgdorferi (Fig. 3B and Table 2). Vectors could be recovered by transforming E. coli with total genomic DNA from B. burgdorferi transformants (data not shown). Both pBSV25 and pBSV28-1 had transformation frequencies and efficiencies similar to those of pBSV2 in noninfectious clone B31-AchbC72 and infectious clone B31 A3 (Table 2). Sequence analysis of pBSV25 identified a single nucleotide substitution in bbe21, relative to the published sequence, that resulted in the amino acid change I143T (11).
FIG. 2.
PF from B. burgdorferi plasmids putatively involved in plasmid maintenance. These regions were tested for their ability to autonomously replicate in B. burgdorferi. Two sets of PF members are present on lp28-1. The region of cp9 conferring autonomous replication was previously characterized (43).
FIG. 3.
Vectors derived from linear plasmids autonomously replicate in B. burgdorferi. (A) Plasmid maps of pBSV25 and pBSV28-1. ColE1, E. coli plasmid origin of replication; MCS, multiple cloning sites; flgBp::kan, B. burgdorferi flgB promoter fused to the kanamycin resistance gene. (B) B. burgdorferi plasmid DNA separated on a 0.3% agarose gel and stained with ethidium bromide (left panel) and the corresponding Southern blot hybridized with the radiolabeled kanamycin gene (right panel). Molecular size standards (in kilobases) of HindIII-digested lambda DNA are given on the left; brackets on the right indicate the autonomously replicating vectors pBSV25 and pBSV28-1 and are consistent with linear (L), nicked (N), and supercoiled (S) plasmid forms. The parental A3 (WT), two transformants of each vector, and the corresponding plasmid DNA isolated from E. coli are shown.
TABLE 2.
Plasmid stabilities and transformation frequencies and efficiencies in B. burgdorferi infectious clone A3 (9) and noninfectious clone B31-AchbC72 (45)
| Plasmid | Transformation frequencya
|
Transformation efficiencyb
|
Stabilityc (%)
|
|||
|---|---|---|---|---|---|---|
| A3 | B31-AchbC72 | A3 | B31-AchbC72 | + Kan | − Kan | |
| pBSV2 | 5.7 × 10−7 | 2.8 × 10−4 | 7 | 2,207 | 100 | 100 |
| pBSV25 | 3.6 × 10−7 | 9.7 × 10−5 | 2 | 2,041 | 80 | 19 |
| pBSV28-1 | 5.3 × 10−7 | 3.0 × 10−4 | 4 | 6,538 | 100 | 100 |
| 1p25 | NAd | NA | NA | NA | NA | 85 |
Transformation frequency was calculated as the ratio of transformants relative to the total number of CFU on medium without kanamycin.
Transformation efficiency was calculated as the number of transformants per microgram of DNA.
Stability was measured as the percentage of total colonies that retain the vector after ∼90 generations compared to the total number of colonies arising (40 colonies per culture condition were PCR screened for the presence of each vector).
NA, not applicable.
Plasmid incompatibility in B. burgdorferi.
Incompatibility of B. burgdorferi plasmids carrying identical maintenance sequences was previously demonstrated for circular plasmids (8, 43). However, incompatibility functions on lp25 and lp28-1 are more difficult to assess due to the presence of a putative restriction-modification system encoded on lp25. Lawrenz and colleagues recently reported that all pBSV2 transformants examined lacked lp25 (23). Their results suggest that the associated loss of lp25 in pBSV2 transformants does not relate to incompatibility but to the presence of a putative restriction-modification system encoded on lp25. The proposed model speculates that shuttle vector transformation occurs in a preexisting subpopulation of cells lacking lp25. Therefore, cells retaining lp25 pose a barrier to shuttle vector transformation and make it difficult to demonstrate incompatibility functions from linear plasmids of B. burgdorferi.
Nevertheless, we assessed the presence of the parental plasmids in the shuttle vector transformants. Ten pBSV25 transformants were PCR screened, and all lacked lp25. Four of the 10 pBSV25 transformants had also lost lp28-1, most likely explained by the frequent loss of lp28-1 observed during in vitro cultivation of B. burgdorferi (22, 30, 46). Six B31-A3 transformants of pBSV28-1 were examined, and all lacked lp28-1. However, all six pBSV28-1 transformants also lacked lp25, reinforcing the observations of Lawrenz and colleagues (23).
Stability of vectors.
To determine if the plasmid maintenance region of a linear plasmid was stable in a circular form, pBSV25 and pBSV28-1 transformants were continuously passaged with and without antibiotic selection. After 90 generations, 100% of the colonies tested retained pBSV28-1, in both the presence and absence of kanamycin (Table 2). In contrast, pBSV25 was much less stable, with only 19% of the colonies retaining the shuttle vector after ∼90 generations in the absence of selection. Stability was greater in the presence of kanamycin, with 80% of the colonies tested retaining pBSV25 (Table 2). The parental plasmid, lp25, was more stable than the derivative pBSV25, with 85% of the colonies examined retaining lp25 (Table 2).
PF 49 is not required for circular or linear plasmid replication.
Eggers et al. demonstrated that a cp32-based shuttle vector, containing only PF 57 and upstream regions (including an inverted repeat), replicated autonomously (8). Previously, we reported that three cp9 ORFs (bbc01-PF 57, bbc02-PF 50, and bbc03-PF 49) were the minimal elements necessary for autonomous replication, and the inverted repeats that flank these three ORFs were unnecessary (43). Subsequently, we constructed a derivative of pBSV2 containing inverted repeat A (IR-A) and upstream sequences, plus bbc01 and bbc02. This vector, designated pOZK-1-2 (Fig. 4A), was capable of autonomous replication in B. burgdorferi. Stability assays (described above) showed pOZK-1-2 was present in 100% of the colonies examined after ∼90 generations, with or without selection (data not shown). Therefore, a cp9-based vector requires either IR-A and upstream sequences or bbc03 and downstream sequences for autonomous replication and stable maintenance.
To further delineate the genetic requirements of cp9 maintenance, a premature stop codon was engineered at a unique SpeI site, resulting in a protein lacking the carboxy-terminal 27 amino acids. All sequences downstream of this new stop codon were deleted. This vector, pOZK-3Δ, lacked both IRs (Fig. 4A). Electroporation of pOZK-3Δ into B. burgdorferi cells did not result in any transformants, despite repeated attempts. Taken together, these results suggest that sequences downstream of the SpeI site are required for cp9 replication in the absence of the IR-A region.
The requirement for a PF 49 paralog in linear plasmid replication was assessed by inactivating bbe18 by using a gentamicin resistance cassette (9) (Fig. 4B). This construct, designated pE18::gnt, was designed to determine whether the BBE18 protein was necessary (a trans requirement) or if a DNA sequence present within or downstream of bbe18 was necessary (a cis requirement). Vector pE18::gnt autonomously replicated in B. burgdorferi (Fig. 4B), indicating that the BBE18 protein does not appear to be required for linear plasmid maintenance.
DISCUSSION
Why B. burgdorferi evolved a genome segmented between a chromosome and multiple plasmids, or minichromosomes, remains unknown. Although plasmids can be lost during in vitro cultivation, it seems likely that most B. burgdorferi plasmids either confer a selective advantage or are required for survival in vivo. As Casjens observed, infectious strains isolated from nature almost always maintain the full plasmid complement, and many functions presumed essential reside on the plasmids of B. burgdorferi (2). Several extrachromosomal elements appear to be ubiquitous, including cp26 and the cp32 plasmid family (2, 40, 44). Additionally, the importance of linear plasmids to mammalian infection, specifically lp25 and lp28-1, has been observed for over a decade (22, 30, 37, 46).
Because of their importance to mammalian infection, and to better understand linear plasmid maintenance in B. burgdorferi, derivatives of lp25 and lp28-1 were constructed and designated pBSV25 and pBSV28-1, respectively (Fig. 2 and 3). Recent evidence suggested B. burgdorferi linear plasmids use an internal origin of replication (4, 27). The data presented here confirm this prediction. The regions from linear plasmids lp25 and lp28-1 conferring autonomous replication are located internally and contain members of PF 49, PF 50, PF 32, and PF 57 (Fig. 2).
Vectors pBSV25 and pBSV28-1 were maintained in a circular form, despite being derivatives of linear replicons. This suggests that both linear and circular plasmids of B. burgdorferi can utilize the same mechanism for replication initiation, segregation, and compatibility, thereby minimizing the machinery necessary for plasmid maintenance. A major difference between linear plasmid replication and circular replication would be the ability to resolve the replicated form (a circular dimer in the case of linear plasmids) (2, 4, 19). Recently, the enzyme that carries out this function in B. burgdorferi, ResT, was identified, purified, and characterized (20).
Conservation of replication initiation functions for linear and circular plasmids is further supported by the conversion of a B. burgdorferi circular plasmid to a linear form by introduction of synthetic telomeres (4). The report by Ferdows et al. of the spontaneous conversion of a linear plasmid of B. hermsii to a stable circular form suggests a common replication initiation mechanism for both linear and circular plasmids (10). Finally, CG skew analysis predicted a common mode of replication for both linear and circular replicons in B. burgdorferi (27, 28). Together, these results reinforce the theory that plasmid maintenance in B. burgdorferi is largely conserved, regardless of DNA form. Similarly, linear plasmids with internal origins and bidirectional modes of replication have been observed in various actinomycetes and are capable of driving replication in a circular form (5, 6, 26, 31, 38). A general principal of bacterial linear plasmid replication appears to be the adaptation of preexisting replication and segregation functions.
Although both pBSV25 and pBSV28-1 were capable of autonomous replication, these vectors exhibited striking differences in stability after 90 generations in B. burgdorferi (Table 2). The pBSV25 shuttle vector was unstable, even in the presence of selection, whereas pBSV28-1 was completely stable in the presence or absence of selection (Table 2). Both lp25 and lp28-1, as well as cp9, are frequently lost during in vitro growth (22, 30), yet vectors derived from lp28-1 and cp9 are extremely stable (43). Since pBSV28-1 and pBSV2 are stable even in the absence of selection, the frequently noted loss of lp28-1 and cp9 with in vitro propagation does not appear to be due to a defect in their ability to autonomously replicate and segregate. Rather, lp28-1 and cp9 may not encode essential gene products for in vitro growth and may be lost from some cells at a low frequency and become fixed in the population. However, the instability of pBSV25 does not appear to relate to the stability of the parental plasmid lp25, which was lost in only 15% of the population over the same time period (Table 2). Possibly, lp25 may have dispersed genetic elements necessary for stable plasmid maintenance that pBSV25 does not include. The phage N15 is maintained as a linear plasmid within E. coli cells and has telomeric ends structurally similar to the linear plasmids of B. burgdorferi (34). Interestingly, N15 contains four separate centromeres dispersed across its length, with each centromere adding to N15 stability (13). Likewise, lp25 may have more than one region necessary for stable inheritance, and not all regions may have been incorporated into pBSV25. Alternatively, sequence analysis of pBSV25 identified a single nucleotide change resulting in an amino acid substitution in BBE21 (PF 57). The amino acid change, an isoleucine-to-threonine substitution, replaces a hydrophobic residue with a hydrophilic one. Eggers et al. demonstrated that the corresponding PF 57 member from a cp32 plasmid was the only coding region required for replication of shuttle vector pCE316 (8). This single amino acid change may be responsible for the instability of pBSV25.
Surprisingly, conversion of the circular shuttle vector pBSV2 to a linear form by addition of 35-bp telomeres to the multiple cloning site, as reported by Chaconas et al. (4), resulted in a highly unstable plasmid and reduced the copy number by fivefold (data not shown). Placement of the telomeres in the carboxy terminus of bbc03 (PF 49) restored the copy number, but the vector remained unstable. This suggests that the DNA sequence near bbc03 significantly impacts replication or partitioning functions of linear and circular plasmids.
Although the DNA sequence encompassing PF 49 members (such as bbc03 and bbe18) appears to be necessary for both circular and linear plasmid maintenance, apparently the PF 49-encoded proteins are not. While constructing derivatives of the cp9-based shuttle vector, pBSV2, we noted that an IR and upstream sequence could substitute for a member of PF 49 (BBC03) (Fig. 4A), suggesting that both the IR and the DNA sequence encompassing bbc03 contain the same required binding site for plasmid maintenance. The IR plus upstream sequence of cp9 is less than 300 bp long and does not contain any ORFs of significant size. Eggers et al. reported similar results with a cp32-based shuttle vector and proposed candidate binding sites for the DnaA protein, which is required for initiation of plasmid replication in some systems (8). The requirement for a PF 49 member in linear plasmid replication was determined by inactivating bbe18 (on pBSV25) with an antibiotic marker, creating the vector pE18::gnt (Fig. 4B). Our data support the view that a functional PF 49 protein is not required for circular or linear plasmid replication. However, the sequence, either within bbe18 or downstream of it, appears to be necessary. Most likely, this region of DNA provides a binding site for a plasmid maintenance protein. PF 49 members are widely distributed among B. burgdorferi plasmids, are highly conserved, and are usually located near the origin of replication, suggesting a role in plasmid maintenance. Our results indicate that the PF 49 protein is not required for plasmid replication, but it may contribute to plasmid stability, perhaps as a functional component of the partitioning system (8, 12). However, the data do not exclude the possibility that a family 49 protein is supplied in trans.
The data presented here suggest that B. burgdorferi linear and circular plasmid maintenance functions are conserved, differing mainly in the resolution of replicated linear plasmids. Apparently, linear and circular plasmids in B. burgdorferi do not require a PF 49 protein for plasmid replication, but they do require the DNA sequence, perhaps serving as a protein binding site. Finally, the shuttle vectors reported here provide the tools necessary to dissect the components and functions of linear plasmid maintenance in B. burgdorferi.
Acknowledgments
We thank Sandy Stewart and Gail Sylva for excellent technical assistance. We gratefully acknowledge Jay Carroll and Kit Tilly for thoughtful discussions and manuscript comments. We thank Frank Gherardini, Joe Hinnebusch, James Musser, and Tom Schwan for critical review of the manuscript.
REFERENCES
- 1.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 2.Casjens, S. 1999. Evolution of the linear DNA replicons of the Borrelia spirochetes. Curr. Opin. Microbiol. 2:529-534. [DOI] [PubMed] [Google Scholar]
- 3.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 4.Chaconas, G., P. E. Stewart, K. Tilly, J. L. Bono, and P. Rosa. 2001. Telomere resolution in the Lyme disease spirochete. EMBO J. 20:3229-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, P., E. Kim, and S. N. Cohen. 1996. Streptomyces linear plasmids that contain a phage-like, centrally located, replication origin. Mol. Microbiol. 22:789-800. [DOI] [PubMed] [Google Scholar]
- 6.Chang, P.-C., and S. N. Cohen. 1994. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science 265:952-954. [DOI] [PubMed] [Google Scholar]
- 7.Dunn, J. J., S. R. Buchstein, L.-L. Butler, S. Fisenne, D. S. Polin, B. N. Lade, and B. J. Luft. 1994. Complete nucleotide sequence of a circular plasmid from the Lyme disease spirochete, Borrelia burgdorferi. J. Bacteriol. 176:2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eggers, C. H., M. J. Caimano, M. L. Clawson, W. G. Miller, D. S. Samuels, and J. D. Radolf. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for expression of fluorescent reporters in the Lyme disease spirochaete. Mol. Microbiol. 43:281-295. [DOI] [PubMed] [Google Scholar]
- 9.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferdows, M. S., P. Serwer, G. A. Griess, S. J. Norris, and A. G. Barbour. 1996. Conversion of a linear to a circular plasmid in the relapsing fever agent Borrelia hermsii. J. Bacteriol. 178:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 12.Gerdes, K., J. Moller-Jensen, and R. B. Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 13.Grigoriev, P. S., and M. B. Lobocka. 2001. Determinants of segregational stability of the linear plasmid-prophage N15 of Escherichia coli. Mol. Microbiol. 42:355-368. [DOI] [PubMed] [Google Scholar]
- 14.Guo, B. P., S. J. Norris, L. C. Rosenberg, and M. Hook. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 63:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellwage, J., T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, J. T. Seppälä, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch, J., and A. G. Barbour. 1992. Linear- and circular-plasmid copy numbers in Borrelia burgdorferi. J. Bacteriol. 174:5251-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howe, T. R., F. W. LaQuier, and A. G. Barbour. 1986. Organization of genes encoding two outer membrane proteins of the Lyme disease agent Borrelia burgdorferi within a single transcriptional unit. Infect. Immun. 54:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyde, F. W., and R. C. Johnson. 1984. Genetic relationship of Lyme disease spirochetes to Borrelia, Treponema, and Leptospira spp. J. Clin. Microbiol. 20:151-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobryn, K., and G. Chaconas. 2001. The circle is broken: telomere resolution in linear replicons. Curr. Opin. Microbiol. 4:558-564. [DOI] [PubMed] [Google Scholar]
- 20.Kobryn, K., and G. Chaconas. 2002. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol. Cell 9:195-201. [DOI] [PubMed] [Google Scholar]
- 21.Kraiczy, P., C. Skerka, M. Kirschfink, V. Brade, and P. F. Zipfel. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and Factor H. Eur. J. Immunol. 31:1674-1684. [DOI] [PubMed] [Google Scholar]
- 22.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrenz, M. B., H. Kawabata, J. E. Purser, and S. J. Norris. 2002. Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious Borrelia. Infect. Immun. 70:4851-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marconi, R. T., D. S. Samuels, and C. F. Garon. 1993. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol. 175:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margolis, N., D. Hogan, K. Tilly, and P. A. Rosa. 1994. Plasmid location of Borrelia purine biosynthesis gene homologs. J. Bacteriol. 176:6427-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picardeau, M., C. le Dantec, and V. Vincent. 2000. Analysis of the internal replication region of a mycobacterial linear plasmid. Microbiology 146:305-313. [DOI] [PubMed] [Google Scholar]
- 27.Picardeau, M., J. R. Lobry, and B. J. Hinnebusch. 2000. Analyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res. 10:1594-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picardeau, M., J. R. Lobry, and B. J. Hinnebusch. 1999. Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol. Microbiol. 32:437-445. [DOI] [PubMed] [Google Scholar]
- 29.Plasterk, R. H. A., M. I. Simon, and A. G. Barbour. 1985. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature 318:257-263. [DOI] [PubMed] [Google Scholar]
- 30.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redenbach, M., M. Bibb, B. Gust, B. Seitz, and A. Spychaj. 1999. The linear plasmid SCP1 of Streptomyces coelicolor A3(2) possesses a centrally located replication origin and shows significant homology to the transposon Tn4811. Plasmid 42:174-185. [DOI] [PubMed] [Google Scholar]
- 32.Rich, S. M., S. A. Sawyer, and A. G. Barbour. 2001. Antigen polymorphism in Borrelia hermsii, a clonal pathogenic bacterium. Proc. Natl. Acad. Sci. USA 98:15038-15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosa, P., D. S. Samuels, D. Hogan, B. Stevenson, S. Casjens, and K. Tilly. 1996. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J. Bacteriol. 178:5946-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rybchin, V. N., and A. N. Svarchevsky. 1999. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol. 33:895-903. [DOI] [PubMed] [Google Scholar]
- 35.Sadziene, A., B. Wilske, M. S. Ferdows, and A. G. Barbour. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwan, T. G., W. Burgdorfer, and C. F. Garon. 1988. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun. 56:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiffman, D., and S. N. Cohen. 1992. Reconstruction of a Streptomyces linear replicon from separately cloned DNA fragments: existence of a cryptic origin of circular replication within the linear plasmid. Proc. Natl. Acad. Sci. USA 89:6129-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869-1879. [DOI] [PubMed] [Google Scholar]
- 40.Stevenson, B., S. Casjens, R. van Vugt, S. F. Porcella, K. Tilly, J. L. Bono, and P. Rosa. 1997. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J. Bacteriol. 179:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson, B., N. El-hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdoferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson, B., S. F. Porcella, K. L. Oie, C. A. Fitzpatrick, S. J. Raffel, L. Lubke, M. E. Schrumpf, and T. G. Schwan. 2000. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect. Immun. 68:3900-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 44.Tilly, K., S. Casjens, B. Stevenson, J. L. Bono, D. S. Samuels, D. Hogan, and P. Rosa. 1997. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol. Microbiol. 25:361-373. [DOI] [PubMed] [Google Scholar]
- 45.Tilly, K., A. F. Elias, J. Erret, E. Fischer, R. Iyer, I. Schwartz, J. L. Bono, and P. Rosa. 2001. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183:5544-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, Y., C. Kodner, L. Coleman, and R. C. Johnson. 1996. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect. Immun. 64:3870-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, J.-R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:1-20. [DOI] [PubMed] [Google Scholar]
- 48.Zückert, W. R., and J. Meyer. 1996. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 178:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]