Abstract
We cloned, expressed, and purified the Escherichia coli YggH protein and show that it catalyzes the S-adenosyl-l-methionine-dependent formation of N7-methylguanosine at position 46 (m7G46) in tRNA. Additionally, we generated an E. coli strain with a disrupted yggH gene and show that the mutant strain lacks tRNA (m7G46) methyltransferase activity.
About 30 different modified nucleosides have been identified in Escherichia coli tRNA. Methylation is one of the most common modifications, and several mutants affected in tRNA methylation have been obtained (5). However, only a few E. coli tRNA methyltransferase (MTase) genes have been cloned and characterized: trmA, trmD, and trmH are involved in the formations of m5U54, m1G37, and Gm18, respectively (8, 15, 16). Several tRNA MTases have been purified, and the corresponding genes have been mapped on the E. coli chromosome (5, 13), but it has not been convincingly shown which open reading frame (ORF) encodes a given enzyme. On the other hand, evolutionary relationships among various RNA MTase families have been studied and predictions of novel specificities for uncharacterized ORFs have been made (3). Nevertheless, there are still missing links between many known enzymatic activities and predicted RNA MTase genes.
As part of a large-scale project aimed at the identification and classification of novel RNA MTases among the uncharacterized or putative proteins in sequence databases, we analyzed the product of the E. coli yggH ORF. This protein exhibits similarity to S-adenosyl-l-methionine (AdoMet)-dependent MTases in the predicted cofactor-binding region but shares no specific amino acid signatures with other families of RNA MTases in the predicted catalytic region, suggesting that it may encode an RNA MTase with a novel specificity. Thus, we selected it for experimental characterization.
Amplification and cloning of the yggH ORF.
The yggH ORF was PCR amplified from E. coli genomic DNA (strain XL1-Blue) by using Pfu DNA polymerase (Promega). The primers (Table 1) were designed to amplify the yggH ORF with its ribosome binding site. Primers LDB1 and LDB3 were used for the production of a recombinant YggH protein bearing a C-terminal His tag (YggHH6). Primers LDB1 and LDB2 were used for the production of the untagged YggH.
TABLE 1.
Oligonucleotides
| Oligonucleotide | Sequence | Target gene |
|---|---|---|
| LDB1 | CCTCTAGAAATTAAGAAGGAGATATACATATGAAAAACGACGTCATTTCACCGG | yggH |
| LDB2 | GGCCTCGAGTTATTATTTCACCCTCTCGAAC | yggH |
| LDB3 | GGCCTCGAGTTTCACCCTCTCGAACATTAAG | yggH |
| LDB4 | CGCCCCGCAACGCCGATAAGGTATC | metT |
| LDB5 | ATCTGGTGCGTCTACCAATTTCGCC | metT |
| LDB6 | CGCTAATACGACTCACTATAGGCTACGTAGCTCAGTTGGTTAGAG | metT |
| LDB7 | CCTGGTGGCTACGACGGGATTCGAACCTGTGAC | metT |
| LDB8 | CATAATGATGGGATCACAGGTTCGAATC | metT |
| LDB9 | GATTCGAACCTGTGATCCCATCATTATG | metT |
| LDB10 | ATGAAAAACGACGTCATTTCACCGGAATTTGATGAAAACGCATTCAAATATGTATCCGCTC | bla |
| LDB11 | TACTCCGTGACCAAGACGATGACCACGTTGTTCAAATTTCAGAGTTGGTAGCTCTTGATC | bla |
| LDB12 | GCGACGCTTGCATGGTC | yggH promoter |
| LDB13 | ACCTGAACGATACGGCG | yggH |
| LDB14 | TGTTGAGATCCAGTTCG | bla |
The PCR products were cloned into the pCR-BluntII-TOPO vector (Invitrogen) according to the manufacturer's instructions, generating the pCR-yggHH6 and pCR-yggH plasmids (the strains and plasmids used are shown in Table 2). The XbaI/XhoI insert of the pCR-yggHH6 plasmid was subcloned into the corresponding sites of the pET30b overexpression vector (Novagen), generating the pET-yggHH6 plasmid.
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Relevant property | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]c | Stratagene |
| P4X-SB25 | met mutant thr mutant relA Hfr | R. Lavallée |
| DY330 F′ | W3110 ΔlacU169gal-490 λcI857 Δ(cro-bioA) F′ (pro-lac) | D. Bregeon |
| RDB1 | DY330 F′ ΔyggH | This study |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pCR-Blunt II-TOPO | Vector for cloning of PCR fragments | Invitrogen |
| pCR-yggH | yggH ORF cloned into pCR-Blunt II-TOPO | This study |
| pCR-yggHH6 | yggHH6 ORF cloned into pCR-Blunt II-TOPO | This study |
| pET30b | Vector for recombinant protein overexpression | Novagen |
| pET-yggHH6 | yggHH6 ORF cloned into pET30b | This study |
| pYL6 |
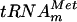 in vitro transcription in vitro transcription |
This study |
| pMet(G46A) |
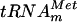 (G46A) in vitro transcription (G46A) in vitro transcription |
This study |
Expression and purification of the YggHH6 recombinant protein.
The YggHH6 protein was expressed in E. coli strain BL21(DE3). Transformed cells were grown at 37°C in Luria broth (supplemented with kanamycin at 30 μg/ml) to an optical density at 660 nm of 0.7. At this stage, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.5 mM to induce recombinant protein expression. Cells were harvested after 3 h of incubation at 37°C, resuspended in buffer A (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 10% glycerol), and lysed by sonication. The lysate was cleared by centrifugation (20,000 × g for 10 min) and was applied to a column of Chelating Sepharose Fast Flow (Pharmacia Biotech) charged with Ni2+. The column was washed with buffer A supplemented with 5 mM imidazole, and the adsorbed material was eluted with a linear gradient (0.05 M up to 0.4 M) of imidazole. Eluted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). At this stage, the YggHH6 preparation contained several minor contaminants (data not shown).
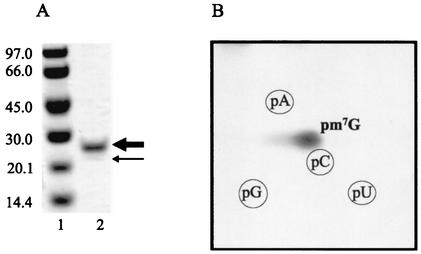
YggHH6 was further purified by gel filtration chromatography. The partially purified enzyme was dialyzed against buffer A supplemented with 200 mM imidazole to keep the protein soluble and was applied on a Superdex 200 column (Pharmacia Biotech) equilibrated with the same buffer. SDS-PAGE analysis of the fractions containing YggHH6 showed two discrete bands (Fig. 1A), both of which corresponded to the YggH protein as demonstrated by mass spectrometry fingerprint analysis. A similar mass fingerprint was obtained for both bands, except for the C-terminal tag tryptic peptide, which was absent for the lower band (result not shown). Thus, the lower band most probably corresponds to a degradation product of YggHH6, lacking the C-terminal His tag. Gel filtration chromatography revealed that the apparent molecular mass of the YggHH6 protein is about 27 kDa. This shows that the protein exists as a monomer.
FIG. 1.
The product of the E. coli yggH ORF catalyzes the formation of m7G in tRNA. (A) SDS-PAGE of the purified YggHH6 protein. Lane 1, molecular mass markers in kilodaltons (Pharmacia Biotech); lane 2, purified protein. The thick and thin arrows indicate the recombinant YggHH6 protein and its minor contaminant, respectively (see the text for details). (B) Autoradiography of a two-dimensional chromatogram of 5′ phosphate nucleotides on a thin-layer cellulose plate. Total tRNA (100 μg) from the methionine-starved P4X-SB25 strain was incubated in a 200-μl reaction mixture containing 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]-Na (pH 7.0), 4 mM MgCl2, 10 μM [methyl-14C]AdoMet (53 mCi/mmol), and 0.4 μg of the purified YggHH6 protein. After a 30-min incubation at 37°C, the tRNA was recovered and digested by nuclease P1, and the resulting nucleotides were analyzed as described previously (12).
yggH encodes an MTase responsible for the formation of m7G46 in the variable loop of tRNA.
To determine whether the product of the yggH ORF was an MTase acting on tRNA, the purified YggHH6 protein was incubated with 14C-radiolabeled AdoMet (S-adenosyl-l-[methyl-14C]methionine) and total tRNA was extracted from a methionine-starved P4X-SB25 strain (an E. coli met mutant relA strain). After incubation, the tRNA was hydrolyzed by nuclease P1 and the resulting nucleotides were analyzed by bidimensional cellulose thin-layer chromatography (2D-TLC) followed by autoradiography. The result shown in Fig. 1B revealed the formation of a single radioactive compound with migration characteristics similar to those of 7-methylguanosine 5′-phosphate (pm7G). Examination of the tRNA sequence database (http://www.uni-bayreuth.de/departments/biochemie/trna/) revealed that m7G is found only at position 46 (in the variable loop) in 23 E. coli tRNA species, including 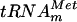 (Fig. 2A). Therefore, an in vitro transcribed
(Fig. 2A). Therefore, an in vitro transcribed 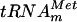 was tested as a substrate for the purified YggHH6 protein. The metT gene, encoding
was tested as a substrate for the purified YggHH6 protein. The metT gene, encoding 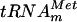 , was PCR amplified from E. coli genomic DNA by using primers LDB4 and LDB5. These primers were designed to amplify a 150-bp fragment containing the metT gene. A second PCR was performed on this 150-bp fragment by using primers LDB6 and LDB7. The second couple of primers was designed to introduce a T7 RNA polymerase-dependent promoter at the 5′ end of the metT gene and an MvaI restriction site at the 3′ end. The use of two consecutive PCRs was necessary, since on the E. coli chromosome, the metT gene is in tandem with the metU gene that also encodes
, was PCR amplified from E. coli genomic DNA by using primers LDB4 and LDB5. These primers were designed to amplify a 150-bp fragment containing the metT gene. A second PCR was performed on this 150-bp fragment by using primers LDB6 and LDB7. The second couple of primers was designed to introduce a T7 RNA polymerase-dependent promoter at the 5′ end of the metT gene and an MvaI restriction site at the 3′ end. The use of two consecutive PCRs was necessary, since on the E. coli chromosome, the metT gene is in tandem with the metU gene that also encodes 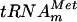 . The final PCR product was cloned into the SmaI site of the pUC18 vector, generating the pYL6 plasmid. Transcripts of
. The final PCR product was cloned into the SmaI site of the pUC18 vector, generating the pYL6 plasmid. Transcripts of 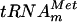 were generated by using T7 RNA polymerase and MvaI-digested pYL6 as the template as described previously (17). Full-length transcripts were purified by 10% PAGE.
were generated by using T7 RNA polymerase and MvaI-digested pYL6 as the template as described previously (17). Full-length transcripts were purified by 10% PAGE.
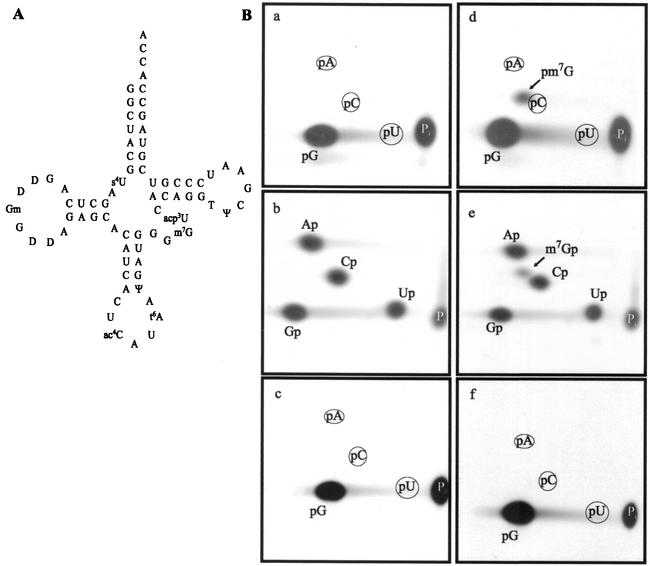
FIG. 2.
In vitro-transcribed E. coli 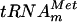 is a substrate of the YggH MTase. (A) Cloverleaf representation of the nucleotide sequence of E. coli
is a substrate of the YggH MTase. (A) Cloverleaf representation of the nucleotide sequence of E. coli 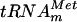 (9). (B) Autoradiography of two-dimensional chromatograms of 5′ and 3′ phosphate nucleotides on thin-layer cellulose plates. [α-32P]GTP-labeled (a and d) or [α-32P]UTP-labeled (b and e) in vitro-transcribed
(9). (B) Autoradiography of two-dimensional chromatograms of 5′ and 3′ phosphate nucleotides on thin-layer cellulose plates. [α-32P]GTP-labeled (a and d) or [α-32P]UTP-labeled (b and e) in vitro-transcribed 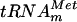 and [α-32P]GTP-labeled in vitro-transcribed
and [α-32P]GTP-labeled in vitro-transcribed 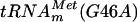 (c and f) (106 cpm) were incubated in the presence (d, e, and f) or absence (a, b, and c) of the YggHH6 protein. The reaction mixture contained 50 mM PIPES-Na (pH 7.0), 4 mM MgCl2, 50 μM AdoMet, and 0.4 μg of the purified YggHH6 protein. After 30 min of incubation at 37°C, the tRNA was recovered and digested by nuclease P1 (a, c, d, and f) or RNase T2 (b and e), and the resulting nucleotides were analyzed as described in the legend to Fig. 1.
(c and f) (106 cpm) were incubated in the presence (d, e, and f) or absence (a, b, and c) of the YggHH6 protein. The reaction mixture contained 50 mM PIPES-Na (pH 7.0), 4 mM MgCl2, 50 μM AdoMet, and 0.4 μg of the purified YggHH6 protein. After 30 min of incubation at 37°C, the tRNA was recovered and digested by nuclease P1 (a, c, d, and f) or RNase T2 (b and e), and the resulting nucleotides were analyzed as described in the legend to Fig. 1.
The purified YggHH6 protein was incubated with AdoMet and [α-32P]GTP-labeled in vitro-transcribed 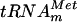 . After incubation, the tRNA was hydrolyzed using nuclease P1, and the resulting 5′ phosphate nucleotides were analyzed by 2D-TLC and autoradiography. The results showed the formation of m7G in the incubated tRNA (Fig. 2B). To further confirm that m7G formation occurs at position 46, a similar experiment was performed using [α-32P]UTP-labeled
. After incubation, the tRNA was hydrolyzed using nuclease P1, and the resulting 5′ phosphate nucleotides were analyzed by 2D-TLC and autoradiography. The results showed the formation of m7G in the incubated tRNA (Fig. 2B). To further confirm that m7G formation occurs at position 46, a similar experiment was performed using [α-32P]UTP-labeled 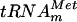 . After incubation in the presence of AdoMet and purified YggHH6, the tRNA was hydrolyzed by RNase T2. The analysis of the resulting 3′ phosphate nucleotides revealed the formation of m7G in the tRNA (Fig. 2B), demonstrating that the m7G produced by YggHH6 is 5′ adjacent to a uridine. In the
. After incubation in the presence of AdoMet and purified YggHH6, the tRNA was hydrolyzed by RNase T2. The analysis of the resulting 3′ phosphate nucleotides revealed the formation of m7G in the tRNA (Fig. 2B), demonstrating that the m7G produced by YggHH6 is 5′ adjacent to a uridine. In the 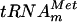 , several guanosines are 5′ adjacent to a uridine. To further confirm that m7G formation occurs at position 46, a mutant
, several guanosines are 5′ adjacent to a uridine. To further confirm that m7G formation occurs at position 46, a mutant 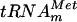 in which G46 was mutated into A
in which G46 was mutated into A 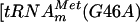 ] was generated by site-directed mutagenesis on the pYL6 plasmid by using primers LDB8 and LDB9. Transcripts of tRNAmMet(G46A) were obtained as described above. The purified YggHH6 protein was incubated with AdoMet and [α-32P]GTP-labeled in vitro-transcribed
] was generated by site-directed mutagenesis on the pYL6 plasmid by using primers LDB8 and LDB9. Transcripts of tRNAmMet(G46A) were obtained as described above. The purified YggHH6 protein was incubated with AdoMet and [α-32P]GTP-labeled in vitro-transcribed 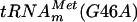 . After incubation, the mutant tRNA was hydrolyzed using nuclease P1, and the resulting 5′ phosphate nucleotides were analyzed by 2D-TLC and autoradiography. The results showed that m7G was not found in the incubated mutant tRNA (Fig. 2B). Altogether, these results strongly suggest that the purified YggHH6 recombinant protein catalyzes the formation of m7G at position 46 in E. coli tRNA.
. After incubation, the mutant tRNA was hydrolyzed using nuclease P1, and the resulting 5′ phosphate nucleotides were analyzed by 2D-TLC and autoradiography. The results showed that m7G was not found in the incubated mutant tRNA (Fig. 2B). Altogether, these results strongly suggest that the purified YggHH6 recombinant protein catalyzes the formation of m7G at position 46 in E. coli tRNA.
The E. coli yggH gene is not essential for growth.
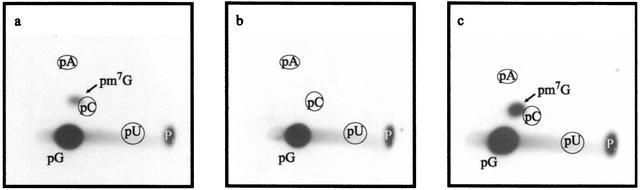
The E. coli yggH gene was inactivated by the insertion of an ampicillin resistance (Apr) cassette. This was achieved by homologous recombination, depending on bacteriophage λ recombination functions present in the host strain (18). A linear DNA fragment in which the β-lactamase gene is flanked by 40 bp corresponding to the 5′ and 3′ ends of the yggH gene was obtained by PCR using the oligonucleotides LDB10 and LDB11 as primers and plasmid pUC18 as the template. The PCR product was used to transform the DY330 F′(pro-lac) strain, and transformants were selected for ampicillin resistance. The presence of the Apr cassette in the yggH gene in the resulting RDB1 strain was checked by PCR using oligonucleotides LDB12, LDB13, and LDB14 as primers (result not shown). To determine whether m7G46 formation was affected in the RDB1 strain, crude extracts of the DY330 F′(pro-lac) and RDB1 strains were incubated with AdoMet and [α-32P]GTP-labeled in vitro-transcribed 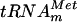 . After incubation, tRNA was hydrolyzed by nuclease P1 and the nucleotides were analyzed by 2D-TLC and autoradiography. The results shown in Fig. 3 revealed the absence of m7G formation in RDB1 extract. Moreover, when the RDB1 strain was transformed with plasmid pCR-yggH, an extract of the resulting strain allowed m7G formation (Fig. 3). Also, total (crude) tRNA extracted from the wild-type strain DY330 F′(pro-lac) was not a substrate for the purified YggH enzyme, while tRNA from the RDB1 strain was an excellent substrate for this enzyme (data not shown). All these data further confirm the role of the YggH protein in the formation of m7G in tRNA and show that the yggH gene is not essential for growth.
. After incubation, tRNA was hydrolyzed by nuclease P1 and the nucleotides were analyzed by 2D-TLC and autoradiography. The results shown in Fig. 3 revealed the absence of m7G formation in RDB1 extract. Moreover, when the RDB1 strain was transformed with plasmid pCR-yggH, an extract of the resulting strain allowed m7G formation (Fig. 3). Also, total (crude) tRNA extracted from the wild-type strain DY330 F′(pro-lac) was not a substrate for the purified YggH enzyme, while tRNA from the RDB1 strain was an excellent substrate for this enzyme (data not shown). All these data further confirm the role of the YggH protein in the formation of m7G in tRNA and show that the yggH gene is not essential for growth.
FIG. 3.
The E. coli RDB1 strain with an inactivated yggH gene lacks tRNA (m7G46) MTase activity. The panels show autoradiography of two-dimensional chromatograms of 5′ phosphate nucleotides on thin-layer cellulose plates. [α-32P]GTP-labeled in vitro-transcribed 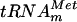 (106 cpm) was incubated with a crude extract of the DY330 F′ strain (wild type) (a), of the RDB1 strain (b), or of the RDB1/pCR-yggH strain (c). The reaction mixture contained 50 mM PIPES-Na (pH 7.0), 4 mM MgCl2, 50 μM AdoMet, and 100 μg of total protein. After 30 min of incubation at 37°C, the tRNA was recovered and digested by nuclease P1, and the resulting nucleotides were analyzed as described in the legend to Fig. 1.
(106 cpm) was incubated with a crude extract of the DY330 F′ strain (wild type) (a), of the RDB1 strain (b), or of the RDB1/pCR-yggH strain (c). The reaction mixture contained 50 mM PIPES-Na (pH 7.0), 4 mM MgCl2, 50 μM AdoMet, and 100 μg of total protein. After 30 min of incubation at 37°C, the tRNA was recovered and digested by nuclease P1, and the resulting nucleotides were analyzed as described in the legend to Fig. 1.
Interestingly, a trmB mutant (strain GM18) affected in the formation of m7G in tRNAs was obtained in the 1970s (14). Surprisingly, the trmB mutation has been mapped at 6 min and the yggH ORF maps at 66 min on the E. coli chromosome. Because of this discrepancy, yggH cannot yet be renamed trmB. A possible explanation for the absence of tRNA (m7G46) MTase activity in the GM18 strain would be that trmB encodes a factor influencing yggH expression. Alternatively, two tRNA (m7G46) MTases might exist in E. coli, as has been suggested previously (4). However, the fact that the inactivation of the yggH gene leads to a complete absence of tRNA (m7G46) MTase activity does not support this hypothesis. Further work is required to better characterize the trmB mutation.
Sequence analysis of the YggH MTase reveals a distinct family of m7G MTases.
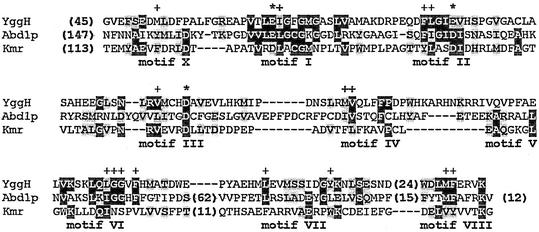
Searches of the sequence database by using PSI-BLAST (2) revealed that orthologs of the yggH gene are present in all completely sequenced bacterial genomes and in crown eukaryotes (animals, plants, and fungi), while they are absent from all archaea (data not shown; see also the National Center for Biotechnology Information's COG database at http://www.ncbi.nlm.nih.gov/cgi-bin/COG/palox?COG0220). This pattern of phylogenetic distribution is perfectly consistent with the observed presence or absence of m7G in tRNAs from these organisms (11). Analysis of the multiple sequence alignment (http://www.ncbi.nlm.nih.gov/COG/aln/COG0220.aln) revealed typical MTase motifs in the YggH family and allowed superimposition with the sequences of other m7G MTases acting on different RNAs: the Agr family specific for G1405 within bacterial 16S rRNA (7) and the Abd1 family specific for the cap structure in mRNA (6). The alignment of representative members of the three m7G MTase families (Fig. 4) revealed no striking similarities apart from the residues important for the stability of the common fold or forming the common cofactor-binding pocket. In particular, a tetrapeptide in motif IV, which typically harbors catalytic residues and is very similar in related MTases (10), exhibits completely different patterns of conservation in YggH, Abd1, and Agr, namely, PDPW, CLHY, and PCLE, respectively. It has been argued that the Agr and Abd1 families may use different mechanisms of guanine-N7 methylation, because the predicted substrate-binding regions and catalytic sites of these enzymes are dissimilar, even though they share a common structural core (7). Identification of the tRNA (m7G46) MTase activity of the yggH ORF suggests a third, considerably diverged class of enzymes that generate a similar product (m7G) within a distinct macromolecular context. It remains to be determined whether these three classes of enzymes exhibit any similarities in the m7G methylation mechanism other than the use of a common cofactor and whether they evolved from a common ancestor or independently from various lineages of the MTase superfamily.
FIG. 4.
Sequence alignment of the representative members of three m7G MTase families specific for tRNA, mRNA, and 16S rRNA: E. coli YggH, S. cerevisiae Abd1p (cap 0 MTase family), and Streptomyces kanamyceticus Kmr (Agr family). Conserved motifs are labeled according to the nomenclature used by Fauman et al. (10). The number of residues omitted for clarity is indicated in parentheses. Conserved AdoMet-binding carboxylate residues are indicated by asterisks, and conserved residues important for the stability of the MTase fold are indicated with pluses.
ADDENDUM IN PROOF
During the time in which this work was under review, the tRNA (m7G46) MTase from the yeast Saccharomyces cerevisiae was identified (1). Two proteins (Trm8p and Trm82p) forming a complex are required for m7G46 formation in yeast tRNA. Trm8p appears to be the yeast ortholog of the E. coli YggH protein.
Acknowledgments
We thank D. Bregeon (Faculté de Médecine Necker Enfants Malades, Paris, France) for the gift of the DY330 F′(pro-lac) strain, R. Lavallée (Institut de Recherches Microbiologiques J.-M. Wiame) for the SB25 strain, and G. Doumont (Université Libre de Bruxelles) for constructing the pYL6 plasmid.
L.G.S.D.B. is a fellow of the F.R.I.A. (Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture), and L.D. and R.W. are research associates of the F.N.R.S. (Fonds National de la Recherche Scientifique). J.M.B. is an EMBO/HHMI Young Investigator. This work was supported by grants from the F.R.F.C. (Fonds pour la Recherche Fondamentale Collective), the French Community of Belgium (Actions de Recherches Concertées), and the Université Libre de Bruxelles (Fonds E. Defay).
REFERENCES
- 1.Alexandrov, A., M. R. Martzen, and E. M. Phizicky. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8:1253-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anantharaman, V., E. V. Koonin, and L. Aravind. 2002. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30:1427-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aschhoff, H. J., H. Elten, H. H. Arnold, G. Mahal, W. Kersten, and H. Kersten. 1976. 7-Methylguanine specific tRNA-methyltransferase from Escherichia coli. Nucleic Acids Res. 3:3109-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björk, G. R. 1995. Biosynthesis and function of modified nucleosides in tRNA, p. 165-205. In D. Söll and U. L. Rajbhandary (ed.), tRNA: structure, biosynthesis, and function. ASM Press, Washington, D.C.
- 6.Bujnicki, J. M., M. Feder, M. Radlinska, and L. Rychlewski. 2001. mRNA:guanine-N7 cap methyltransferases: identification of novel members of the family, evolutionary analysis, homology modeling, and analysis of sequence-structure-function relationships. BMC Bioinformatics 2:2. [DOI] [PMC free article] [PubMed]
- 7.Bujnicki, J. M., and L. Rychlewski. 2001. Sequence analysis and structure prediction of aminoglycoside-resistance 16S rRNA:m7G methyltransferases. Acta Microbiol. Pol. 50:7-17. [PubMed] [Google Scholar]
- 8.Byström, A. S., and G. R. Björk. 1982. Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA(m1G) methyltransferase in Escherichia coli K12. Mol. Gen. Genet. 188:440-446. [DOI] [PubMed] [Google Scholar]
- 9.Cory, S., and K. A. Marcker. 1970. The nucleotide sequence of methionine transfer RNA-M. Eur. J. Biochem. 12:177-194. [DOI] [PubMed] [Google Scholar]
- 10.Fauman, E. B., R. M. Blumenthal, and X. Cheng. 1999. Structure and evolution of AdoMet-dependent MTases, p. 1-38. In X. Cheng and R. M. Blumenthal (ed.), S-Adenosylmethionine-dependent methyltransferases: structure and functions. World Scientific Inc., Singapore.
- 11.Grosjean, H., M. Sprinzl, and S. Steinberg. 1995. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie 77:139-141. [DOI] [PubMed] [Google Scholar]
- 12.Keith, G. 1995. Mobilities of modified ribonucleotides on two-dimensional cellulose thin layer chromatography. Biochimie 77:142-144. [DOI] [PubMed] [Google Scholar]
- 13.Leung, H.-C. E., T. G. Hagervall, G. R. Björk, and M. E. Winkler. 1998. Genetic locations and database accession numbers of RNA-modifying and -editing enzymes, p. 561-567. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 14.Marinus, M. G., N. R. Morris, D. Söll, and T. C. Kwong. 1975. Isolation and partial characterization of three Escherichia coli mutants with altered transfer ribonucleic acid methylases. J. Bacteriol. 122:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ny, T., P. H. R. Lindström, T. G. Hagervall, and G. R. Björk. 1988. Purification of transfer RNA (m5U54)-methyltransferase from Escherichia coli. Association with RNA. Eur. J. Biochem. 177:467-475. [DOI] [PubMed] [Google Scholar]
- 16.Persson, B. C., G. Jager, and C. Gustafsson. 1997. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res. 25:4093-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampson, J. R., and O. C. Uhlenbeck. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA 85:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu, D., H. M. Ellis, E.-C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]