Abstract
Plasmid R27 contains two independent partitioning modules, designated Par1 and Par2, within transfer region 2. Par1 is member of the type I partitioning family (Walker-type ATPase), and Par2 is a member of the type II partitioning family (actin-type ATPase). Stability tests of cloned Par1 and Par2 and insertional disruptions of Par1 and Par2 within R27 demonstrated that Par1 is the major stability determinant whereas Par2 is the minor stability determinant. Creation of double-partitioning mutants resulted in R27 integrating into the chromosome, suggesting that at least one partitioning module is required for R27 to exist in the extrachromosomal form. Using the lacO/LacI-green fluorescent protein (GFP) system, we labeled and visualized R27 and R27 partitioning mutants (Par1− and Par2−) under different growth conditions in live Escherichia coli cells. Plasmid R27 was visualized as the discrete GFP foci present at the mid- and quarter-cell regions in >99% of the cells. Time lapse experiments demonstrated that an increase in R27 plasmid foci resulted from focus duplication in either the mid- or quarter-cell regions of E. coli. Both R27 Par− variants gave a high percentage of plasmidless cells, as suggested by a uniform GFP signal, and cells with GFP patterns scattered throughout the entire cell, suggesting that plasmid molecules are randomly distributed throughout the cytoplasm. Those cells that did contain R27 Par− with one or two discrete foci had localization patterns that were statistically different from those formed with wild-type R27. Therefore, these results suggest that partitioning-impaired plasmids are characterized by individual and clustered plasmids that are randomly located within the host cytoplasm.
Partitioning modules are found on many low-copy-number plasmids and facilitate the faithful segregation of bacterial plasmids to daughter cells at cell division. Partitioning modules consist of a centromere region adjacent to two coregulated genes that code for an ATPase and a centromere-specific DNA-binding protein (11). Gerdes et al. (8) have proposed the existence of two partitioning family modules based on the type of ATPase encoded. Type I modules encode Walker-type ATPases and are exemplified by the ParA/SopA systems encoded by P1 and F, respectively. Type II modules encode actin-type ATPases and are exemplified by ParM of R1. Partitioning protein homologs are also involved in chromosome segregation in Bacillus subtilis (13), Caulobacter crescentus (27), and Pseudomonas putida (9, 25).
DNA-binding partitioning proteins are typified by ParB/SopB of P1/F and ParR of R1 and bind to repeated DNA sequences within the centromere region. The ParB- and ParR-centromere complexes are referred to as the partitioning complex and mediate the pairing of plasmid molecules (3, 18). Binding of ParR to the centromere region also serves to regulate the expression of parM-parR, as the promoter is within the centromere region (15). Binding of ParB to the centromere region of P1, found downstream of the parA-parB operon, results in ParB spreading into the surrounding sequences and promotes silencing of this DNA (33). The partitioning complex is localized to the mid- (and quarter-) cell positions of Escherichia coli, and after duplication, complexes move rapidly to the quarter-cell positions, the mid-cell of the next generation (10, 29). It is not known what maintains the partitioning complex at the mid- and quarter-cell positions; however, it has been proposed that a host-encoded factor may be involved (11, 12).
The partitioning ATPases are cytoplasmic proteins whose ATPase activity is essential for partitioning (1, 16, 40). The ParA/SopA proteins are also involved in the autoregulation of the partitioning operons (1). The ATPases interact with their cognate DNA-binding proteins (16, 32), suggesting that ATP hydrolysis may be linked to movement of the partitioning complex from the mid- to quarter-cell positions. Recently, it has been shown that ParM of R1 forms actin-like filaments with ATP-dependent polymerization properties. Polymerization of ParM into filaments appears to begin at the partitioning complex and probably provides the necessary motive force to actively push the partitioning complexes from the mid- to quarter-cell positions (28).
IncHI1 plasmids frequently encode multiple-antibiotic resistance in Salmonella enterica serovar Typhi (14, 26, 30). R27, the prototypical IncHI1 plasmid, is a large plasmid, temperature sensitive for conjugative transfer, which encodes resistance to tetracycline and possesses a mosaic genetic organization of backbone components (i.e., conjugation, replication, and partitioning), suggesting a complex evolutionary history (20, 35). For example, the conjugative transfer system of R27 is a chimera of IncF- and IncP-like systems (21, 22). Replication is determined by one of two replicons, RepHI1A or RepHI1B (5, 6), maintaining a copy number of one or two copies per chromosomal origin (36). A third nonfunctional IncFIA replicon is also present on R27 and is responsible for one-way incompatibility with the F factor (6). Sequence analysis has identified two partitioning modules on R27 (35). The goal of this study was to determine, using genetic analysis and a LacI-green fluorescent protein (GFP) probe to label R27::lacO, how each partitioning module contributes to the stability of R27 and to study unstable partitioning mutants that have cellular localizations that are distinct from those of wild-type R27.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The following E. coli strains were used: DY330Rts (W3110 ΔlacU169 gal490 λcI857 Δ cro-bioA) (43), DY330R and DY330N (rifampin- and nalidixic acid-resistant, temperature-sensitive revertants of DY330ts) (23), and DH5α [supE44 lacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (34). The following plasmids with relevant characteristics were used: drR27 (IncHI1 htdA::TnlacZ; Tcr and Kmr) (35), pSG25 (cassette delivery vector, Kmr Apr) (10), pSG20 (GFP-LacI expression vector, arabinose-inducible promoter, Apr) (10), pJP124 (Tn7 transposase vector, Cmr; gift from Nancy Craig), pOU82 (unstable test vector, Apr; gift from Michael Yarmolinsky) (7). The chloramphenicol acetyltransferase (Cmr) and trimethoprim resistance (Tpr) cassettes for insertional mutagenesis were amplified from pNK20 (19) and R751 (39), respectively.
Growth conditions.
All E. coli strains were grown in Luria-Bertani (LB) medium (Difco Laboratories) or morpholinepropanesulfonic acid (MOPS) minimal medium (MOPS buffer with 20 amino acids (aa) and 0.4% glucose or 0.4% glycerol) (10). When DH5α was grown in MOPS minimal medium, biotin and thiamine were included. During the construction of R27 parR::Tp, when trimethoprim resistance was utilized, strains were grown with Mueller-Hinton medium. When necessary, the following antibiotics were added to the growth medium: ampicillin (100 μg/ml), kanamycin (50 μg/ml), tetracycline (10 μg/ml), chloramphenicol (16 μg/ml), and trimethoprim (50 μg/ml). E. coli strain DH5α was used for the stability assay, which was performed at 30°C and gave a 40-min generation time when cells were grown in LB medium and a 120-min generation time when cells were grown in MOPS medium. Strain DY330 was used for the fluorescence microscopy experiments, where cells from overnight cultures were diluted 1/1,000 and grown at room temperature (22°C) to mid-log phase prior to the induction of GFP-LacI. When strain DY330 was grown in LB medium, the generation time was 60 min, and when it was grown in MOPS medium, the generation time was 180 min. GFP-LacI was induced by adding 0.2% arabinose to the growth medium. Expression was then repressed after 30 to 40 min by adding 0.4% glucose to the medium and continuing growth for an additional 30 min.
Nucleotide sequence and statistical analysis.
Laser gene software (DNASTAR Inc., Madison, Wis.) was used for nucleotide sequence analysis. Repeated nucleotide sequences were identified with GeneQuest. The predicted protein sequence for each open reading frame was compared to those in the GenBank nonredundant database by using PSI-BLAST. Statistical analysis was performed with the SigmaStat software package (Jandel Scientific).
Cloning of Par1 and Par2.
The Par1 and Par2 regions were amplified by using Pfx high-fidelity DNA polymerase (Life Technologies, Inc., Rockville, Md.) with primers par1-f (5′ ATATGGATCCCGTTTTAAGTTACTGGTTACC), par1-r (5′ ATATGGATCCCACTTCTAGGCCCCAATC), par2-f (5′ ATATGGATCCTACTACCGGATGAAAGTCATC), and par2-r (5′ ATATGGATCCGTGATAACATTCAGTCAGCC). Primers incorporated BamHI sites into each end of the amplified DNA. Par1 and Par2 regions were digested with BamHI and cloned into the BamHI site of pOU82 (7) to create pPar1 and pPar2, respectively.
Construction of parB and parR mutants.
The parB and parR genes were mutated by inserting a Cmr or trimethoprim resistance cassette by using the E. coli (DY330) recombination system (43). DNA substrates were generated through PCR with primers (∼60 nucleotide) that produced a linear Cmr or trimethoprim resistance cassette with 40-bp terminal arms homologous to the desired target site. Primers used were as follows: to create DNA templates to disrupt parB with a Cmr cassette, trev204 (5′ ACTGAAGTAATCCATCGCTCCGGGCTTCAAGGCCTGAAAGCTGTGACGGAAGATCACTTC) and trev205 (5′ GCCTCAAGCTTCCGCCCATTGTGTAATGTAAAAAGTTTCTTTATTCAGGCGTAGCACCAG); to disrupt parR with a Cmr cassette, trev206 (5′ ACATCGTGGCGAGCAGATTTCTCTAATAAGATCTGCAATCCTGTGACGGAAGATCACTTC) and trev207 (5′ CATTAATAAAACTAGATAACTCGGGGAAGAGATTATTTAGTTATTCAGGCGTAGCACCAG); to disrupt parR with a Tpr cassette, trev535 (5′ ACATCGTGGCGAGCAGATTTCTCTAATAAGATCTGCAATCTTCGCTGCTGCCCAAGGTG) and trev536 (5′ CATTAATAAAACTAGATAACTCGGGGAAGAGATTATTTAGGTGCACTCAACCGTGAATTC). DNA substrates were introduced by electroporation into DY330Rts harboring R27::lacO (or R27::lacO parB::Cm) that was grown according to Yu et al. (43). Cells were plated on agar plates containing both tetracycline, to select for R27::lacO, and chloramphenicol or trimethoprim, to select for the desired insertions. To screen presumptive colonies, the target gene was PCR amplified and analyzed by 1% agarose gel electrophoresis. An increase in the size of the open reading frame by 700 bp (Tpr cassette) or 900 bp (Cmr cassette) demonstrated that the resistance cassette had been inserted into the target gene.
Pulsed-field gel electrophoresis.
Preparation of agarose-embedded E. coli strain DY330 and restriction endonuclease digestion, as well as electrophoresis and image processing, are described elsewhere (38), except that NotI was used to digest the DNA.
Stability assay.
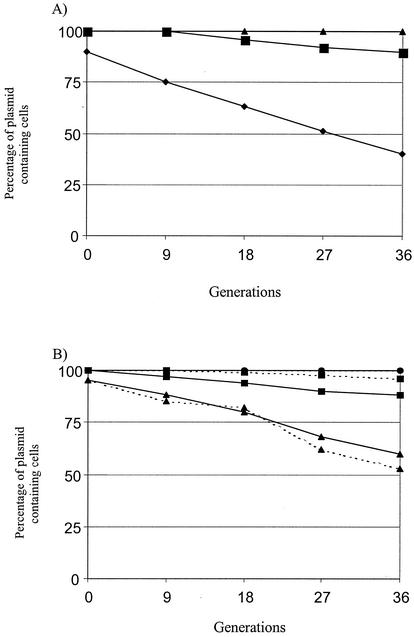
DH5α containing the test plasmids (each of which contained the entire β-galactosidase gene, pOU82, or coded for the α fragment of β-galactosidase, drR27) was grown overnight in LB broth with selection at 37°C and shaken at 200 rpm. Overnight cultures were diluted 1/100,000 into fresh LB broth without antibiotics and grown at 30°C with shaking (200 rpm). Every 12 h each culture was reinoculated at 1/100,000 into fresh LB broth to maintain exponential growth. Aliquots were removed at 6 or 12 h (times indicated on the graphs) (Fig. 2), serially diluted and plated on nutrient plates containing 50 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/liter. Blue plasmid-containing colonies and white plasmidless colonies were counted, and the percentage of plasmid-containing cells was plotted against time. Each stability test was performed twice, and the average values are presented.
FIG. 2.
(A) Partitioning stability assay for Par1 and Par2 cloned into the unstable test vector pOU82. ▴, pPar1; ▪, pPar2; •, pOU82. (B) Stability assay for wtR27 (•), R27 Par1− (parB:: Cm) (▴), and R27 Par2− (parR:: Cm) (▪). Solid lines represent strains grown in LB medium, and broken lines represent strains grown in MOPS minimal medium. The lines for strains containing wtR27 grown in both LB and MOPS media are overlapping at 100%. Strains were grown in E. coli at 30°C without selection. See text for details.
R27::lacO construction.
To produce a R27::lacO construct that is stable and mating proficient, pSG25 was first transformed into E. coli strain Stbl-2 and then R27 was mated into these cells by using the methods previously described (37). Once the presence of both plasmids was confirmed by using plasmid isolation procedures, electroporation was used to introduce pJP124 into the strain. Plasmid pJP124 contains the Tn7 transposase genes cloned into pACYC that promote random transposition of Tn7 into conjugating plasmids (42). Cells containing all three plasmids were collected from agar plates in 1 ml of phosphate-buffered saline, and 0.1 ml of this cell suspension was used to inoculate 10 ml of fresh LB medium plus ampicillin, trimethoprim, and chloramphenicol. Cells were grown to midexponential phase and were used as donors for mating. Transposition of the cassette into R27 was achieved by mating R27 to DY330R and selecting with tetracycline, for R27, and kanamycin, for the lacO cassette. To ensure that the cassette did not transpose into transfer genes, transconjugants were pooled and mated into DY330N (pSG20; cells containing pSG20 express GFP-LacI). Several colonies were then individually screened to ensure stability and mating efficiency, one of which was used for further analysis. Within this construct, the cassette was found to be inserted into an intergenic region between genes orf183 and orf184, as determined by sequencing the region flanking Tn7 with primer NLC95 (5′ ATAATCCTTAAAAACTCCATTTCCACCCCT) (42).
Microscopy and photography.
For microscopy experiments, E. coli strain DY330 was grown as indicated (Fig. 5) and for time lapse experiments cells were grown in LB broth (Fig. 6). One-milliliter aliquots of samples were collected, pelleted and resuspended in 20 to 50 μl of MOPS medium. A 1-μl sample was added to a MOPS medium-1.5% agarose slab on a microscope slide. A coverslip was used to cover the sample and the edges sealed with vacuum grease. Fluorescence microscopy was performed with a Leica UV microscope equipped with a charge-coupled device camera (Cooke SensiCam) and a standard fluorescein isothiocyanate filter set (Chroma). Samples were illuminated with a UV (Leica HB100) source, and images were collected and processed using SensiControl 4.0 and PhotoPaint (Corel). Quantitative measurements (i.e., number of foci, focus location, and cell size) were performed as described previously (10, 23).
FIG. 5.
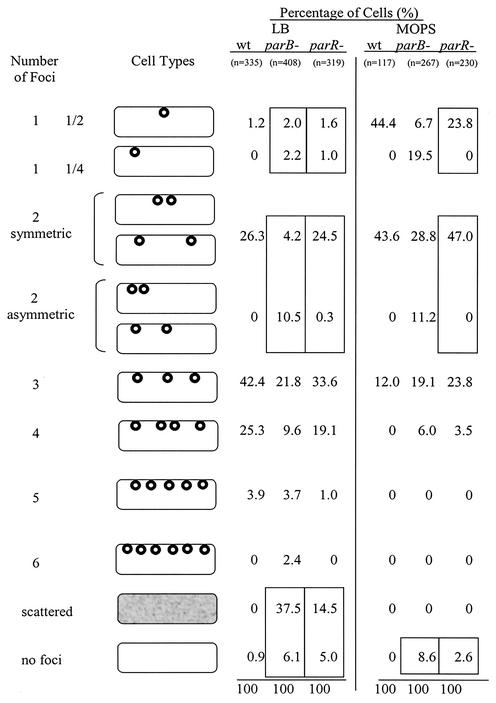
Summary of the localization of wild-type R27 and R27 partitioning mutants Par1− (parB-) and Par2− (parR-) in E. coli grown in MOPS (slow growth) and LB (fast growth) media. Cell types represent the number and location of GFP foci. The one-focus and two-focus patterns have been categorized according to their cellular location. Values are given as a percentage of the total number of cells analyzed for each plasmid type under each growth condition. Sample sizes are included. Boxed values highlight (i) the localization patterns for one-focus and two-focus mutants that are statistically different from R27, (ii) the scattered GFP patterns seen in cells containing partitioning mutants but not R27, and (iii) the high percentage of cells with a uniform GFP signal, indicating plasmidless cells.
FIG. 6.
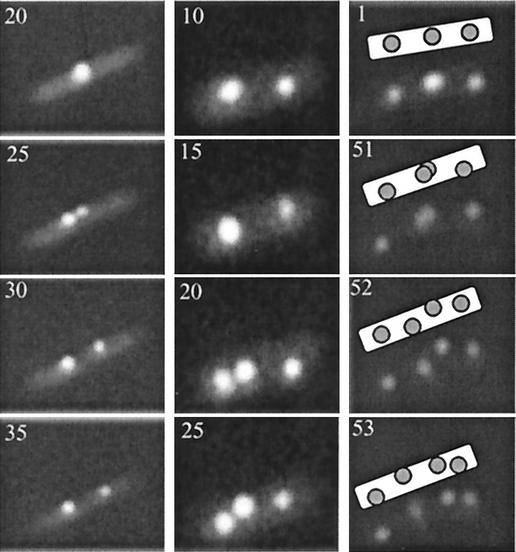
Time lapse fluorescence microscopy of R27 plasmid foci duplicating at either the mid- or quarter-cell positions of E. coli. The time in minutes at which image was collected is indicated in the top lefthand corner.
RESULTS
Nucleotide sequence analysis of Par1 and Par2.
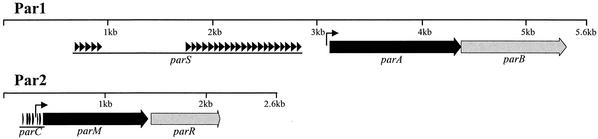
Two partitioning modules within the conjugative transfer region 2 (Tra2) of R27 which are separated by 2.7 kb and transcribed in the same direction were identified (Fig. 1) (22, 35). Partitioning module 1 (Par1) contains two genes, arranged in a putative operon, whose gene products are similar to the ParA/SopA and ParB/SopB protein families exemplified by P1 and the F factor (8). ParAR27 (417 aa) shares 20% identity with SopAF and contains a WalkerA ATPase motif (GTGGKS). ParBR27 (335 aa) shares 27% identity with ParBP1. Upstream of parA are the putative −10 (tagaat) and −35 (aataca) promoter sequences; 240 bp upstream of the parA start are 26 direct repeats of 34 bp (cccCctTAaTcGcCAg--ccATGG-gg-a--c-g [upper case, conserved; lower case, majority; dash, nonconserved]). These repeats likely serve as the centromere region and have been designated parS.
FIG. 1.
Genetic organization of Par1 and Par2 modules of R27. The parA and parM genes code for ATPases belonging to the Walker- and actin-type ATPases, respectively. The parB and parR genes code for centromere-binding proteins. The parS and parC regions are proposed centromere regions. The region shown indicates the region cloned for the stability assay. Black arrows indicate positions of direct repeats, and raised arrows indicate putative promoters. The GenBank accession number is AF250878.
The partitioning module 2 (Par2) contains two genes, arranged in a putative operon, whose gene products are homologous to ParM and ParR of R1 (8). ParMR27 (344 aa) is 33% identical to ParMR1 and contains the actin-like ATPase motifs characteristic of these proteins (8). ParRR27 contains no detectable similarity to ParRR1. ParR-like proteins share few conserved residues and vary in size and have been proposed to be functionally analogous and homologous based on their role in partitioning and genetic location (8). Upstream of parM are the putative −10 (tataaa) and −35 (ttgacc) promoter sequences. The promoter sequences are flanked by two sets of 6-bp direct repeats (dr1, GtTtaa, and dr2, AaAaCA) such that dr1 is present seven times upstream of the promoter and dr2 is present five times downstream. These repeats likely represent the centromere region and have been designated parC.
Stability of Par1 and Par2 clones and R27 parB and parR mutants.
To determine if both partitioning modules are functional, Par1 and Par2 were cloned into pOU82, creating pPar1 and pPar2. Plasmid pOU82 is an unstable R1 derivative used for stability assays and is lac+, so the frequency of plasmid loss can be monitored with X-Gal, where plasmid-containing colonies are blue and plasmidless colonies are white (7). Stabilization of pOU82 by either Par1 or Par2 would demonstrate that the partitioning module is functional. Clones pPar1 and pPar2 and pOU82 alone were grown overnight in DH5α with selection and then diluted 1/100,000 in fresh LB broth without selection and grown for 24 h. Samples were taken every 6 h, serially diluted, and plated on X-Gal-containing LB plates. The frequency of plasmid retention was plotted versus generation time (generation time is 40 min) (Fig. 2A). After 36 generations, pPar1 was present in 100% of the cells and pPar2 was present in 90% of the cells whereas pOU82 was present in 40% of the cells. These results suggest that both Par1 and Par2 are functional partitioning modules. In addition, Par1 stabilized pOU82 more efficiently than Par2.
To test the ability of Par1 and Par2 in stabilizing R27, insertional mutations of Par1− (parB mutant) and Par2− (parR mutant) were created in drR27, a derepressed derivative of R27 containing TnlacZ inserted into htdA (41). Stability assays were performed as described above except that stability was tested in both LB medium and MOPS minimal medium (Fig. 2B). After 36 generations without selection, wild-type R27 was present in 100% of the cells, regardless of the growth medium. When cells were grown in LB medium, the Par1 mutant (parB::Cm) was present in 60% of the cells and the Par2 mutant (parR::Cm) was present in 88% of the cells after 36 generations. When the cells were grown in MOPS minimal medium, the Par1 mutant (parB::Cm) was present in 53% of the cells and the Par2 mutant (parR::Cm) was present in 98% of the cells after 36 generations. These results suggest that both Par1 and Par2 are involved in stabilizing R27 at higher growth rates and that only Par1 is required to stabilize R27 at lower growth rates. Overall, the contribution of Par1 towards R27 stability is greater than that of Par2 under the conditions tested.
Double-partitioning mutant of R27.
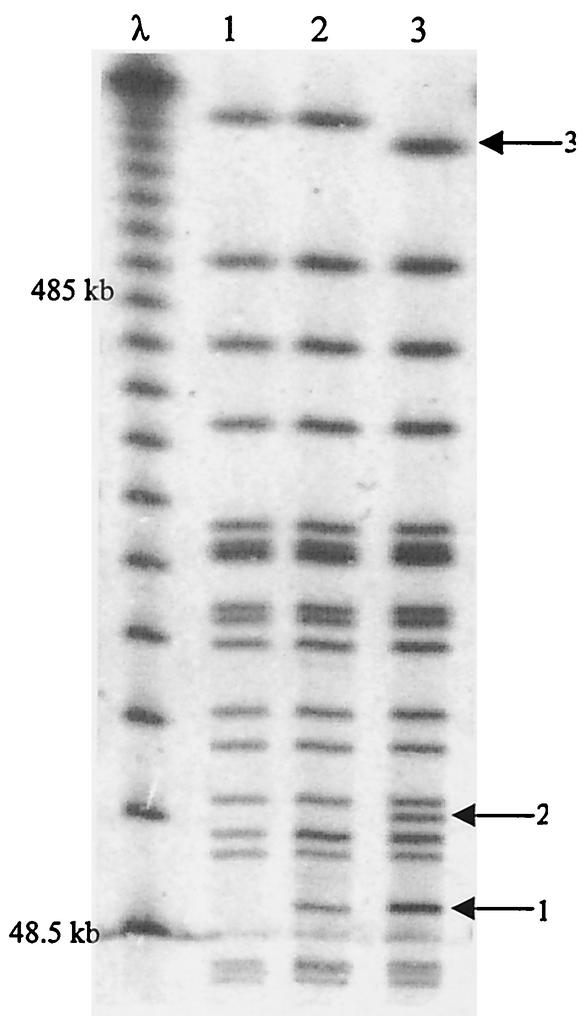
A double-partitioning mutant of R27 was created by inserting a trimethoprim resistance cassette into parR of the R27 parB::Cm mutant. Several mutants were isolated but were unable to be transferred by conjugation from DY330Rts into DH5α in order to perform the stability assay. By using resistance markers to monitor the presence of R27 in cells, it was found that the doubly par-defective mutant was 100% stable after 24 h (data not shown). E. coli cells containing the doubly Par-defective mutant allowed the formation of plaques when infected with Hgal, an H-pilus-specific phage (21), and also mobilized a cloned origin of transfer (21), suggesting that the conjugation apparatus of R27 parB::Cm parR::Tp was functional. Since mutations in neither parB nor parR affected plasmid transfer ability, neither of these gene products is essential for conjugation (data not shown). Given the above observations, we hypothesized that the double-partitioning mutant of R27 may have integrated into the chromosome. To determine if R27 parB::Cm parR::Tp was present in the extrachromosomal form or was inserted into the chromosome, we performed pulsed-field gel electrophoresis on the DNA of the E. coli strains in which the doubly Par-defective plasmid had been created. Figure 3 shows a pulsed-field gel banding pattern of DNA from DY330R (lane 1), the host strain, DY330R(R27 parB::Cm) (lane 2) and DY330R(R27 parB::Cm parR::Tp) (lane 3) digested with the restriction endonuclease NotI. Both lanes 2 and 3, representing R27-containing strains, contained bands that were not present in lane 1, the host strain, and were therefore specific to R27 (arrow 1). These bands (∼60 kb) are likely NotI fragments from R27. Lane 3, representing the double mutant, contained a band not present in either of the other lanes (arrow 2) and a decrease in a band size (arrow 3). These two differences in the banding patterns would be expected if R27 parB::Cm parR::Tp did insert into the chromosome and therefore created a banding pattern different from that for DY330R (lane 1) and DY330R(R27 parB::Cm) (lane 2). These results suggest that creating a double-partitioning mutant of R27 results in R27 inserting into the chromosome, preventing the conjugation of R27 DNA out of the donor. Since DY330R is recA+ and both E. coli and R27 are known to contain several insertion sequences (i.e., IS1, IS2, IS30) (35), it is possible that R27 parB::Cm parR::Tp integrated into the chromosome via homologous recombination, where it would be stably maintained.
FIG. 3.
Pulsed-field gel electrophoresis to illustrate the chromosomal integration of the R27 double-partitioning mutant. The banding patterns are of NotI-digested DNA from DY330R (lane 1), DY330R(R27 parB::Cm) (lane 2), and DY330R(R27 parB::Cm parR::Tp). The left lane is a phage ladder with 48.5-kb increments. Arrows to the right of the gel indicate the R27-specific band (1), the R27-chromosome-specific band (2), and decrease in chromosomal band size due to R27 insertion into the chromosome (3).
Localization of R27 in E. coli cells.
The lacO/LacI-GFP system was used to visualize the cellular location of R27::lacO in live E. coli-expressing LacI-GFP. To compare plasmid localization under different growth rates, R27::lacO was visualized when cells were grown in LB medium (fast growth; 60-min generation time) and MOPS minimal medium (slow growth; 180-min generation time). Representative fluorescence images are shown in Fig. 4, and these results are summarized in Fig. 5. When E. coli-expressing LacI-GFP and -containing R27::lacO were grown in LB and MOPS media, >99% of the cells contained GFP foci. Cells grown in LB medium containing R27::lacO had one focus (1.2%), two foci (26.3%), three foci (42.4%), four foci (25.3%), or five foci (3.9%) (Fig. 5). When grown in MOPS medium, cells containing R27::lacO had one focus (44.4%), two foci (43.6%), or three foci (12%). These results demonstrate that R27 localizes at the mid- and quarter-cell regions of live E. coli.
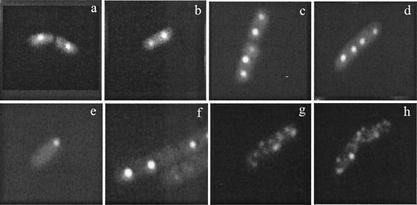
FIG. 4.
Representative micrographs of R27 (a through d) and R27 partitioning mutants (e through h) localized in live E. coli cells. (a) one focus at mid-cell; (b) two foci at quarter-cells; (c) three foci at mid- and quarter-cells; (d) four foci at mid- and quarter-cells; (e) one focus at pole; (f) two asymmetric foci at mid- and quarter-cell; (g and h) scattered GFP signal. The GFP patterns of the R27 partitioning mutants (e through h) are representative of either the parB mutant (Par1−) or the parR mutant (Par2−) as summarized in the legend to Fig. 5.
Time lapse observations of R27::lacO focus duplication.
Figure 6 shows representative experiments that demonstrate the kinetics of R27 focus duplication in E. coli growing on a nutrient agarose surface. Each type of duplication event, that is, one focus to two foci, two foci to three foci, and three foci to four foci, was documented twice. In the cells with one focus, the focus duplicated at the mid-cell region and the two foci moved apart rapidly, within 5 min, to the quarter-cell regions. In cells containing two foci, one focus duplicated at the quarter-cell region and subsequently one focus remained at the quarter-cell region while the second focus moved to the mid-cell region. In three-foci cells, the mid-cell focus duplicated and the foci moved to either side of the division plane. Since the focus patterns before and after focus duplication and movement resembled the localization patterns of wild-type R27 in E. coli containing two, three, or four foci, these observations likely represent the progression of a plasmid-partitioning cycle, where foci can duplicate at either the mid- or quarter-cell regions.
Localization of R27 partitioning mutants in E. coli.
When E. coli expressing LacI-GFP containing the R27::lacO parB::Cm mutant (Par1−) was grown in LB and MOPS media, 93.9 and 91.4% of the cells contained GFP signals (discrete foci or scattered foci), respectively. The remainder of the cells gave a uniform fluorescence signal, indicating the absence of R27::lacO. Of the LB medium-grown cells, 37.5% contained a scattered GFP pattern that was clearly distinguishable from discrete foci and not seen in cells containing wild-type R27 (Fig. 4 and 5). Scattered foci patterns occupied the entire cell and were dynamic when viewed in real time, resulting in images that gave a smeared appearance. None of the MOPS medium-grown cells contained scattered GFP patterns. Cells grown in LB medium that displayed discrete foci contained one focus (4.2%), two foci (14.7%), three foci (21.8%), four foci (9.6%), five foci (3.7%), or six foci (2.4%). Cells grown in MOPS medium that displayed discrete foci contained one focus (26.2%), two foci (40%), three foci (19.1%), or four foci (6%).
When E. coli expressing LacI-GFP containing the R27::lacO parR::Cm mutant (Par2−) was grown in LB and MOPS media, 95 and 97.4% of the cells contained GFP signals (discrete foci or scattered pattern), respectively. The remainder of the cells gave a uniform fluorescence signal, indicating the absence of a lacO cassette. Of the LB medium-grown cells, 14.5% contained a scattered GFP pattern that was clearly distinguishable from discrete foci (Fig. 5). None of the MOPS medium-grown cells contained scattered patterns. Cells grown in LB which displayed discrete foci contained one focus (2.6%), two foci (24.8%), three foci (33.6%), four foci (19.1%), or five foci (1.0%). Cells grown in MOPS medium which displayed discrete foci contained one focus (23.8%), two foci (47.0%), three foci (23.8%), or four foci (3.5%).
Statistical analysis of R27 localization.
The localization patterns of the R27 partitioning mutants contained both scattered GFP patterns, which were not seen with R27, and discrete foci. To determine if there is a significant difference in discrete focus localization patterns between R27 and either of the partitioning mutants, we statistically analyzed the localization data of the one-focus and two-focus cells grown in MOPS and LB media. For the two-focus data, we averaged the focus positions to create a data set that represents the midpoint between the foci. The Mann-Whitney test (critical value set at α = 0.05) indicated that there is a statistically significant difference between the localization patterns of cells containing R27 parB::Cm with one focus grown in MOPS medium (P < 0.001), the averaged two-focus cells grown in MOPS medium (P = 0.015), the one-focus cell grown in LB medium (P = 0.006), and the average of two-focus cells grown in LB medium (P < 0.001) and the R27 grown under the corresponding conditions. Significant differences between the localization pattern of the R27 parR::CAT mutant one-focus cells grown in LB medium (P = 0.004) and the average of two-focus cells grown in LB medium (P = 0.006) and the R27 grown under the corresponding conditions were found. There was no significant difference between the localization patterns of the R27 parR::Cm mutant one focus cells grown in MOPS medium (P = 0.817) and two focus cells grown in LB medium (P = 0.417) and R27 grown under the corresponding conditions. The boxed values of the Par mutants in Fig. 5 indicate the localization data, which were significantly different from those of wild-type R27.
DISCUSSION
In this work, we demonstrate that R27 contains two independent, functional partitioning modules, Par1 and Par2, which belong to the type I and type II partitioning families, respectively (8). In both the cloning stability test and the mutational stability tests, Par1 was shown to be the major stability determinant whereas Par2 contributes to stability in a minor yet significant manner. Each of the R27 Par mutants is therefore referred to as being partitioning impaired, as they each maintain some stabilizing abilities. The inability of the doubly Par− R27 to exist in the extrachromosomal form illustrates the extreme instability of such a mutant. As partitioning modules are responsible for positioning plasmids at the mid- and quarter-cell regions of E. coli (see below) (10, 17, 23, 29, 31), the same regions where the host replisome is located (24), it is likely that the doubly Par− R27 is unable to replicate efficiently due to a lack of positioning near the replisome. The recovery of doubly Par− R27 integrated into the chromosome is likely due to the antibiotic selection of R27 double-partitioning mutants which have undergone homologous recombination with the chromosome, as this is the only efficient means of R27 replication.
The redundant partitioning modules provide extra stability to R27, as both partitioning modules contribute to the stability of R27 and likely play a key role in the persistence of IncHI1 plasmids within S. enterica serovar Typhi (14, 26, 30). Under both growth conditions tested during the stability assay, that is, fast growth and slow growth, the R27 Par1 mutant was very unstable, as it was lost from nearly half of the cells within 36 generations. The R27 Par2 mutant was moderately unstable under both growth conditions and appeared to contribute slightly more to R27 stability at a faster growth rate. One reason for the differential contribution of Par1 and Par2 to R27 stability could be the differential expression of the Par operons. However, GFP fusions to both ParB and ParR within R27, and therefore under the control of the native promoter, resulted in bright GFP signals under a variety of growth conditions, including those discussed here, suggesting that both of the Par operons are expressed (T. D. Lawley and D. E. Taylor, unpublished data). As some partitioning modules have been found to be important under only certain growth conditions, such as during the transition from the exponential to stationary growth phase (9, 25), it is possible that the conditions for the optimal function of Par2 have not been identified.
In addition to R27, a 90-kb virulence plasmid from enteropathogenic E. coli, pB171, also contains two Par modules (2), as does pHCM1, a 218-kb IncHI1 plasmid from S. enterica serovar Typhi (30) and R478, a 286-kb IncHI2 plasmid from Serratia marcescens (M. W. Gilmour and D. E. Taylor, unpublished data). As the majority of well-known partitioning-proficient plasmids contain only one Par module, the possession of two Par modules by these plasmids is of interest. The possession of two Par modules by these plasmids would provide a means of avoiding competition with plasmids containing only one homologous Par module through plasmid-mediated incompatibility and therefore enhance the survivability of plasmids with two Par modules within bacterial communities. If initially both Par modules functioned optimally under the same physiological conditions, perhaps one Par module might have diverged in specificity in a manner analogous to gene duplication and divergence of function (4), resulting in plasmid stability under a wider range of conditions.
The cellular location of R27::lacO was visualized with a GFP-LacI probe within live E. coli. R27 foci were located at the mid- and quarter-cell locations. Cells grown in MOPS medium (slow growth) predominantly contained one, two, or three foci, whereas cells grown in LB medium (fast growth) predominantly contained two, three, or four foci, suggesting that the number of foci per cell is related to the growth rate. Several larger plasmids, including F, P1, R1, RP4, and R751, are also known to be localized to the mid- and quarter-cell positions (10, 17, 23, 29, 31), and each focus consists of multiple plasmid molecules (11, 31). Time lapse experiments demonstrated that plasmid foci duplicate at either the mid- or quarter-cell regions of E. coli. These observations imply that the progression of the R27 plasmid cycle starts with one focus duplicating at the mid-cell position and the resulting two foci moving to the quarter-cell positions. Subsequently, a focus at the quarter-cell position duplicates and one focus moves to the mid-cell position while the another remains at the quarter-cell position, creating a cell with a mid-cell focus and two quarter-cell foci. In three-focus cells, the mid-cell focus duplicates and both foci are positioned on either side of the division plane. These movements and localizations result in the complete stability of R27, as R27 was not lost from any cells in a long-term stability assay.
Based on the work on other model-partitioning systems (F, P1, and R1), it is presumed that the positioning of R27 foci at the mid- and quarter-cell regions is determined by both ParB and ParR, bound to their respective centromeres, interacting with an unknown host factor. Likewise, the motive force that moves R27 foci between the mid- and quarter-cell positions is determined by the polymerization of ParM (28) and by ParA, by an unknown mechanism, with the partitioning complex serving as the nucleation point. This model would imply that the actions of both partitioning modules are coordinated such that the timing of partitioning would be coupled. It is unknown whether or not each of the partitioning complexes would be tethered to the same or different host factors, although our results do suggest that each Par module would interact with the host factor at the mid- and quarter-cell region.
The presence of two partitioning modules with different stability contributions provided a unique opportunity to relate plasmid localization to plasmid stability, where R27 is completely stable, R27 Par2− is moderately stable, and R27 Par1− is unstable. From our analysis, there are two types of partitioning-impaired localization patterns that are distinct from wild-type R27 localization and are therefore indicative of plasmid instability: (i) mislocalized discrete GFP foci (plasmid clusters) and (ii) randomly scattered GFP patterns (individual plasmids).
When partitioning mutants formed discrete foci, the localization patterns of both Par− plasmids in cells grown in LB medium (one and two foci) differed significantly from those of wild-type R27. In contrast, only the localization patterns of the Par1− plasmid in cells grown in MOPS medium (one and two foci) differed significantly with those of R27. It is interesting that the localization patterns of the Par2 mutant grown in MOPS medium did not differ significantly from that of R27. This suggests that the Par1 module alone is sufficient to stabilize R27 at slower growth rates, but not at faster growth rates, which is consistent with the stability assay results (Fig. 2B). Due to the nature of the partitioning mutants, it is expected that one cause of plasmid mislocalization resulted from the reduced ability of the R27 partitioning complex to become tethered to the mid-and quarter-cell positions of the host. The tethering deficiency of Par1 and Par2 mutants could explain both the instability and the mislocalized discrete focus patterns observed for Par1 and Par2 mutants. It is easy to explain the relationship between these mislocalized plasmid clusters and plasmid instability, since if the mislocalized plasmid clusters are contained within one half of the cell, then at cell division, one of the daughter cells will not inherit plasmids.
A large percentage of cells containing either Par1− (37.5%) or Par2− (14.5%) grown in LB medium contained scattered GFP signals, which were quite dynamic when viewed in real time, suggesting that the plasmids were randomly dispersed throughout the cytoplasm. The partitioning mutants were expected to be impaired in plasmid pairing, which could reduce the formation of plasmid clusters (discrete foci). Therefore, the scattered GFP patterns may represent individual plasmid molecules randomly dispersed in the cytoplasm. The fact that the scattered pattern was absent from cells containing partitioning mutants and grown in MOPS suggests that the pairing impairment would be suppressed at a slower growth rate. The presence of scattered GFP patterns in cells containing partitioning-impaired plasmids at faster growth rates, but not at slower growth rates, is likely a direct result of the increased replication. As a result, the number of plasmids per cell would increase and the carrying capacity of only one of the R27 partitioning modules would have been surpassed, resulting in the excessive individual plasmids becoming randomly dispersed within the cytoplasm.
Since the stability of the Par1 mutant was similar when cells were grown in LB or MOPS medium (Fig. 2B), but only cells containing the Par1 mutant gave scattered patterns when grown at a fast growth rate (Fig. 5), it appears that the scattered patterns are not a major source of plasmid instability. One explanation is that at cell division, the majority of daughter cells inherit the randomly distributed plasmids, as they would be present in all regions of the dividing cell. Alternatively, the scattered GFP signals may be obscuring the discrete GFP foci at the mid- and quarter-cell positions, which are properly localized and stable. Nevertheless, the mislocalized plasmid clusters are the main source of plasmid instability, as mislocalized plasmid clusters were correlated with plasmid instability in all cases (Fig. 2B and 5).
Acknowledgments
We are grateful to Qin Jiang for performing pulsed-field gel electrophoresis. We thank Jalene LaMontagne and George Mulvey for assistance in statistical analysis and Andrew Wright, Matt Gilmour, James Gunton, and Dobryan Tracz for critical reading of the manuscript.
This study was supported by grant MOP6200 to D.E.T. from the Canadian Institutes for Health Research (CIHR). T.D.L. is supported by a Studentship from the Alberta Heritage Foundation for Health Research (AHFMR) and a Doctoral Scholarship from the CIHR. D.E.T. is an AHFMR Scientist.
REFERENCES
- 1.Davis, M. A., K. A. Martin, and S. J. Austin. 1992. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol. Microbiol. 6:1141-1147. [DOI] [PubMed]
- 2.Ebersbach, G., and K. Gerdes. 2001. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc. Natl. Acad. Sci. USA 98:15078-15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edgar, R., D. K. Chattoraj, and M. Yarmolinsky. 2001. Pairing of P1 plasmid partition sites by ParB. Mol. Microbiol. 42:1360-1370. [DOI] [PubMed] [Google Scholar]
- 4.Fryxell, K. J. 1996. The coevolution of gene family trees. Trends Genet. 12:364-369. [DOI] [PubMed] [Google Scholar]
- 5.Gabant, P., A. O. Chahdi, and M. Couturier. 1994. Nucleotide sequence and replication characteristics of RepHI1B: a replicon specific to the IncHI1 plasmids. Plasmid 31:1111-1120. [DOI] [PubMed] [Google Scholar]
- 6.Gabant, P., P. Newnham, D. Taylor, and M. Couturier. 1993. Isolation and location on the R27 map of two replicons and an incompatibility determinant specific for IncHI1 plasmids. J. Bacteriol. 175:7697-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes, K., and S. Molin. 1986. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J. Mol. Biol. 190:269-279. [DOI] [PubMed] [Google Scholar]
- 8.Gerdes, K., J. Moller-Jensen, and R. Bugge-Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 9.Godfrin-Estevenon, A. M., F. Pasta, and D. Lane. 2002. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol. Microbiol. 43:39-49. [DOI] [PubMed] [Google Scholar]
- 10.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, A. Straight, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. [DOI] [PubMed] [Google Scholar]
- 12.Hiraga, S. 2000. Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Microbiol. 34:21-59. [DOI] [PubMed] [Google Scholar]
- 13.Ireton, K., N. W. Gunther, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanoff, B., and M. M. Levine. 1997. Typhoid fever: continuing challenges from a resilient bacterial foe. Bull. Inst. Pasteur 95:129-142. [Google Scholar]
- 15.Jensen, R. B., M. Dam, and K. Gerdes. 1994. Partitioning of plasmid R1. The parA operon is autoregulated by ParR and its transcription is highly stimulated by a downstream activating element. J. Mol. Biol. 236:1299-1309. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, R. B., and K. Gerdes. 1997. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J. Mol. Biol. 269:505-513. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, R. B., and K. Gerdes. 1999. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 18:4076-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, R. B., R. Lurz, and K. Gerdes. 1998. Mechanism of DNA segregation in prokaryotes: replicon pairing by parC of plasmid R1. Proc. Natl. Acad. Sci. USA 95:8550-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 20.Lawley, T. D., V. Burland, and D. E. Taylor. 2000. Analysis of the complete nucleotide sequence of the tetracycline-resistance transposon Tn10. Plasmid 43:235-239. [DOI] [PubMed] [Google Scholar]
- 21.Lawley, T. D., M. W. Gilmour, J. E. Gunton, L. J. Standeven, and D. E. Taylor. 2002. Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 184:2173-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawley, T. D., M. W. Gilmour, J. E. Gunton, D. M. Tracz, and D. E. Taylor. 2003. Functional and mutational analysis of conjugative transfer region 2 (Tra2) from the IncHI1 plasmid R27. J. Bacteriol. 185:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawley, T. D., G. S. Gordon, A. Wright, and D. E. Taylor. 2002. Bacterial conjugative transfer: visualization of successful mating pairs and plasmid establishment in live Escherichia coli. Mol. Microbiol. 44:947-956. [DOI] [PubMed] [Google Scholar]
- 24.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, R. A., C. R. Bignell, W. Zeng, A. C. Jones, and C. M. Thomas. 2002. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology 148:537-548. [DOI] [PubMed] [Google Scholar]
- 26.Mirza, S., S. Kariuki, K. Z. Mamum, N. J. Beeching, and C. A. Hart. 2000. Analysis of plasmid and chromosomal DNA of multidrug-resistant Salmonella enterica serovar Typhi from Asia. J. Clin. Microbiol. 38:1449-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohl, D. A., and J. W. Grober. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675-684. [DOI] [PubMed] [Google Scholar]
- 28.Moller-Jensen, J., R. B. Jensen, J. Lowe, and K. Gerdes. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90:951-957. [DOI] [PubMed] [Google Scholar]
- 30.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 31.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radnedge, L., B. Youngren, M. Davis, and S. Austin. 1998. Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J. 17:6076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodionov, O., M. Lobocka, and M. Yarmolinsky. 1999. Silencing of genes flanking the P1 plasmid centromere. Science 283:546-549. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor, D. E., and E. C. Brose. 1988. Modified Birnboim-Doly method for rapid detection of plasmid copy number. Nucleic Acids Res. 16:9056.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor, D. E., and J. G. Levine. 1980. Studies of temperature-sensitive transfer and maintenance of H incompatibility group plasmids. J. Gen. Microbiol. 116:475-484. [DOI] [PubMed] [Google Scholar]
- 38.Taylor, D. E., M. Rooker, M. Keelan, L. Ng, I. Martin, N. T. Perna, V. Burland, and F. R. Blattner. 2002. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184:4690-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPbeta plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe, E., M. Wachi, M. Yamasaki, and K. Nagai. 1992. ATPase activity of SopA, a protein essential for active partitioning of F plasmid. Mol. Gen. Genet. 234:346-352. [DOI] [PubMed] [Google Scholar]
- 41.Whelan, K. F., D. Maher, E. Colleran, and D. E. Taylor. 1994. Genetic and nucleotide sequence analysis of the gene htdA, which regulates conjugal transfer of IncHI plasmids. J. Bacteriol. 176:2242-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolkow, C. A., R. T. DeBoy, and N. L. Craig. 1996. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 10:2145-2157. [DOI] [PubMed] [Google Scholar]
- 43.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]