Abstract
RNA polymerase III contains seventeen subunits in yeasts (Saccharomyces cerevisiae and Schizosaccharomyces pombe) and in human cells. Twelve of them are akin to the core RNA polymerase I or II. The five other are RNA polymerase III-specific and form the functionally distinct groups Rpc31-Rpc34-Rpc82 and Rpc37-Rpc53. Currently sequenced eukaryotic genomes revealed significant homology to these seventeen subunits in Fungi, Animals, Plants and Amoebozoans. Except for subunit Rpc31, this also extended to the much more distantly related genomes of Alveolates and Excavates, indicating that the complex subunit organization of RNA polymerase III emerged at a very early stage of eukaryotic evolution. The Sch.pombe subunits were expressed in S.cerevisiae null mutants and tested for growth. Ten core subunits showed heterospecific complementation, but the two largest catalytic subunits (Rpc1 and Rpc2) and all five RNA polymerase III-specific subunits (Rpc82, Rpc53, Rpc37, Rpc34 and Rpc31) were non-functional. Three highly conserved RNA polymerase III-specific domains were found in the twelve-subunit core structure. They correspond to the Rpc17-Rpc25 dimer, involved in transcription initiation, to an N-terminal domain of the largest subunit Rpc1 important to anchor Rpc31, Rpc34 and Rpc82, and to a C-terminal domain of Rpc1 that presumably holds Rpc37, Rpc53 and their Rpc11 partner.
INTRODUCTION
Three major DNA-dependent RNA polymerases I, II and III transcribe the nuclear genome of eukaryotes. They have been mostly investigated in the budding yeast Saccharomyces cerevisiae, where the twelve-subunit structure of RNA polymerase II is known at a resolution of 3.8 Å (1,2). Eleven of these subunits are akin to subunits of the archaeal enzyme (3,4) and five are also related to bacterial subunits (5). Rpb8, on the other hand, is only found in eukaryotes and is common to RNA polymerase I, II and III (6). RNA polymerase I and III have a closely related twelve-subunit structure, but the former is endowed with two additional enzyme-specific subunits (7) whilst RNA polymerase III has five additional specific subunits (8) forming two functionally distinct groups, Rpc31-Rpc34-Rpc82 and Rpc37-Rpc53 (9,10).
With its seventeen distinct subunits and a predicted MW of about 0.7 MDa, RNA polymerase III is undoubtedly one of the more elaborated proteins of the eukaryotic cell. This enzyme transcribes about 5% of the S.cerevisiae genome, producing all tRNAs, the 5S ribosomal RNA and a handful of small nuclear RNA (snRNA's) involved in various aspects of RNA and protein synthesis (11). The corresponding genes are bound by pre-initiation complexes formed by the TATA-box binding protein (TBP), the monomeric factors Brf1 and Bdp1 and the six-subunit protein TFIIIC, and also containing a specialized transcription factor (TFIIIA) in the case of the 5S rDNA genes. TBP is common to the RNA polymerase I and II pre-initiation complexes and Brf1 is paralogous to the TFIIB pre-initiation factor of RNA polymerase II, but Bdp1 and TFIIIC are entirely specific of RNA polymerase III (10,12,13).
The human and fission yeast (Schizosaccharomyces pombe) RNA polymerase III have the same subunit composition as in S.cerevisiae (14–17). We found here that the five enzyme-specific subunits of fission yeast cannot replace their S.cerevisiae counterparts in vivo, contrasting with the in vivo complementation seen for ten of twelve core enzyme subunits. Nevertheless, homology search in all other eukaryotic genomes revealed significant conservation to these enzyme-specific subunits, thus showing that the evolution of RNA polymerase III combines a conserved twelve-subunit core with an ancient but fast evolving set of five enzyme-specific subunits. These data shed some new light on the genetic mechanisms that generated and maintained the three distinct nuclear transcription systems of eukaryotes.
MATERIALS AND METHODS
cDNA library screening
Dedicated primers (Supplementary Data S1) were devised for the PCR amplification of cDNA libraries prepared from the Sch.pombe wild-type strain 972 h− (18). cDNAs were extracted for the rpc17, rpc25, rpc31, rpc34, rpc37, rpc53, rpc82, rpc2 and rpc1 genes and cloned by appropriate restriction endonucleases in the pGEN multi-copy vector (Table 1). The corresponding inserts were subjected to sequencing of alkaline-treated double-stranded DNA templates with Sequenase 2.0 (United States Biochemicals) or to automated sequence analysis on MegaBACE 500 Sequencing System (Amersham Biosciences). Both DNA strands were sequenced and checked for spurious mutations due to PCR amplification or plasmid construction. This identified two rpc25 mutants (rpc25-F72I and rpc25-W201R) creating single-site replacements at highly conserved positions, that were unable to complement the corresponding S.cerevisiae null allele.
Table 1.
pGEN-derived plasmids constructed in this study
| Name | Sch.pombe cDNA | Inserted fragment | Length (bp) |
|---|---|---|---|
| pGP-1 | rpc1+ | SmaI/NheI | 4269 |
| pESH-2 | rpc2+ | BglII/EcoRI | 3588 |
| pESH-82 | rpc82+ | BamHI/EcoRI | 1818 |
| pGP-53 | rpc53+ | EcoRI/XhoI | 1107 |
| pGP-34 | rpc34+ | BglII/KpnI | 926 |
| pGP-31 | rpc31+ | BglII/EcoRI | 724 |
| pGP-25 | rpc25+ | BamHI/EcoRI | 639 |
| pGP-17 | rpc17+ | BamHI/EcoRI | 492 |
Yeast strains and plasmids
S.cerevisiae strains and plasmids are listed in Table 2. The Yeast extract/peptone/dextrose (YPD) and CAUW (minimal SD medium supplemented with 0.1% casein hydrolysate and 0.002% adenine sulphate, uracil and tryptophan) were described previously (19). CAUW without tryptophan or uracil were used as omission media. 5-Fluoro-orotic acid (FOA) medium was used to counterselect URA3+ plasmids. It corresponds to the CAUW medium with 1 g/l of 5-FOA, solidified by 2.5% agar (19). To express the Sch.pombe cDNAs in S.cerevisiae, they were sub-cloned in the pGEN multi-copy expression vector of S.cerevisiae, where transcriptional expression depends on the strong PGK1 promoter and ADH1 terminator (19).
Table 2.
Plasmids and yeast strains
| Name | Genotype | Origin |
|---|---|---|
| Yeast strains | ||
| MW409 | MATa ade2-1 lys2-801 ura3-52 trp1- Δ63 his3- Δ200 rpc160- Δ1::HIS3 / pC160-6 (URA3 CEN4 RPC160) | (46) |
| SRY31 | MATα ura3-52 his3- Δ200 trp1- Δ1 lys2-801 ade2-1 ret1 Δ::HIS3 / Yep24-RET1 (2μ URA3 RET1) | (44) |
| SC116 | MATα rpc82::HIS3-1 ura3-52 trp1- Δ1 his3-Δ200 ade2-1 lys2-801 leu2-Δ / pRS314 (CEN4 TRP1 rpc82-6) | (C. Mann) |
| SC116-WT | MATα rpc82::HIS3-1 ura3-52 trp1- Δ1 his3-Δ200 ade2-1 lys2-801 leu2-Δ / Scp80 (URA3 CEN4 ARS1 RPC82) | This work |
| CMY356 | MATa rpc53::HIS3-1 ura3-52 trp1- Δ1 his3- Δ200 ade2-1 lys2-801 / pEMBLYc32-RPC53 (URA3 CEN4 ARS1 RPC53) | (C. Mann) |
| YMR2 | MATa rpc37::KanMX4 ade2-1 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 lys2-801 / pRS316-RPC37 (CEN4 URA3 RPC37) | (47) |
| D57-9A | MATa ade2-1 lys2-801 ura3-52 his3- Δ200 trp1- Δ1 rpc34-Δ::HIS3 / pYS34 (URA3 CEN4 RPC34) | (42,53) |
| CD8326 | MATa ade2-1 ura3-53 lys2-801 his3 trp1- Δ800 leu2-3rpc31- Δ::LEU2 / pC326 (2 μ URA3 RPC31) | (43) |
| FYBL3-2B | MATα ura3- Δ851 his3- Δ200 leu2- Δ1 trp1- Δ63 rpc25- Δ::KANMX4 / pFL44-RPC25 (2 μ URA3 RPC25) | (48) |
| YGP-25 | MATα ura3- Δ851 his3- Δ200 leu2- Δ1 trp1- Δ63 rpc25- Δ::KANMX4 / pGP-25 (2μ TRP1 rpc25+ Sch. pombe) | This work |
| YOL14 | MATa ade2 ura3 trp1 leu2 lys2 his3 rpc17::HIS3 / pRS316-RPC17 (URA3 CEN4 RPC17) | (49) |
| YGP-17 | MATa ade2 ura3 trp1 leu2 lys2 his3 rpc17::HIS3 / pGP-17 (2μ TRP1 rpc17+ Sch. pombe) | This work |
Heterospecific complementation in S.cerevisiae
pGEN expression vectors bearing the Sch.pombe open reading frame corresponding to individual RNA polymerase III subunits were transformed into S.cerevisiae host strain deleted for the relevant subunit and bearing the wild-type gene on a replicative URA3+ plasmid. Heterospecific complementation was monitored by the counter-selection of URA3+ plasmids on 5-FOA, thereby testing the ability of the Sch.pombe subunit to rescue the S.cerevisiae null-mutant. Constructions that failed to complement were carefully re-sequenced to check for spurious mutations, and the transcriptional expression of the corresponding cDNAs was assayed by RT–PCR (Supplementary Data S2). Inspection of the coding sequence showed that rarely used codons were not over-represented in the Sch.pombe cDNAs, compared to the corresponding S.cerevisiae genes.
Sequence accession and alignments
New Sch.pombe cDNA sequences were deposited in the GenBank/EMBL databases with the following accession nos: DQ156227 (rpc1+), DQ156226 (rpc2+), DQ156220 (rpc17+), DQ156221 (rpc25+), DQ156222 (rpc31+), DQ156223 (rpc34+), NM001022708 (rpc37+), DQ156224 (rpc53+) and DQ156225 (rpc82+). A comparison to the Sch.pombe genome showed that the rpc25 gene contained two introns, that rpc34, rpc82 and rpc1 had one intron and that rpc17, rpc31, rpc37, rpc53 and rpc2 were intron-less. These intron positions exactly matched those predicted in the Sch.pombe database maintained by the Sanger Institute. The predicted protein sequences were compared to the eukaryotic protein database kept at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), using the iterative homology search software PSI-Blast (20). Sequence alignments were then optimized by hand, using the following groups of amino acid conservation: DE, NQ, DN, EQ, KR, FWY, AGST, ILVM.
Genetic nomenclature
Roman letters were used to designate protein subunits (e.g. Rpc53) and italics for gene names, with lower-case (rpc53) and upper case (RPC53) symbols for Sch.pombe or S.cerevisiae, respectively. RNA polymerase II subunits are called Rpb1 to Rpb12, where Rpb5, Rpb6, Rpb8, Rpb10 and Rpb12 designate five subunits that are common to all three nuclear polymerases (http://www.yeastgenome.org/). RNA polymerase III subunits are designated by the Rpc acronym followed by a number referring to the apparent MW of the corresponding S.cerevisiae subunit, except for the two largest subunits, called here Rpc1 (alias Rpc160) and Rpc2 (alias Rpc128) as in Sch.pombe (http://www.genedb.org/genedb/pombe/index.jsp).
RESULTS
Complementation between Sch.pombe and S.cerevisiae is limited to the core RNA polymerase III
RNA polymerase III has seventeen subunits in S.cerevisiae, all essential for growth (9,10,21), and there is substantial evidence for a similar subunit composition of RNA polymerase III in Sch.pombe (16,17). Twelve of these subunits are identical (Rpb5, Rpb6, Rpb8, Rpb10 and Rpb12) or paralogous (Rpc11, Rpc17, Rpc25, Rpc19, Rpc40, Rpc1 and Rpc2) to the entire RNA polymerase II (22), and are also identical or paralogous to twelve of the fourteen RNA polymerase I subunits (21,23–26). Rpc31, Rpc34, Rpc37, Rpc53 and Rpc82, on the other hand, are strictly specific of RNA polymerase III (10).
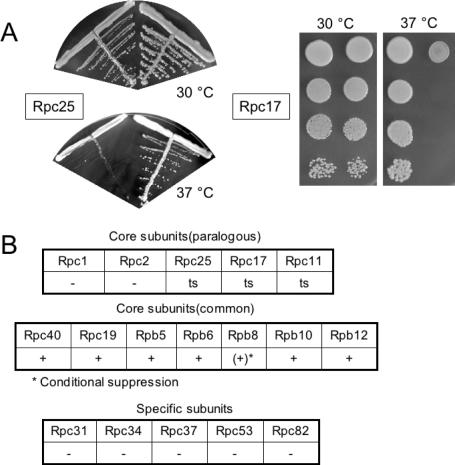
Complementation was reported for eight of the fission yeast subunits (Rpb5, Rpb6, Rpb8, Rpb10, Rpb12, Rpc11, Rpc19 and Rpc40) when expressed in the corresponding S.cerevisiae null-mutant (27–34). We extended this analysis by cloning the remaining nine Sch.pombe cDNAs (encoding Rpc1, Rpc2, Rpc17, Rpc25, Rpc31, Rpc34, Rpc37, Rpc53 and Rpc82) from a previously described cDNA library (18) and by sub-cloning them in the high copy S.cerevisiae vector pGEN, between the PGK1 promoter and ADH1 terminator. These subunits were examined for there ability to replace their S.cerevisiae counterparts in strains deleted for the corresponding host gene, using a previously described plasmid-shuffle assay (19). Two new complementing subunits were identified (Rpc17 and Rpc25, see Figure 1A), but the largest subunits (Rpc1 and Rpc2) and the five enzyme-specific components of RNA polymerase III (Rpc31, Rpc34, Rpc37, Rpc53 and Rpc82) failed to complement the corresponding null allele in S.cerevisiae. Control experiments (Supplementary Data S2) showed that their corresponding cDNAs were effectively transcribed in S.cerevisiae. Full-length transcripts were produced, except for the Rpc1 subunit where our data do not rule out the production of incomplete mRNAs.
Figure 1.
In vivo complementation of Sch.pombe cDNAs expressed in S.cerevisiae. (A) Rpc17. Strain YGP-17, containing the rpc17-Δ::HIS3 null allele and the multi-copy plasmid pGP-17 expressing the Sch.pombe rpc17 cDNA (Table 1) was streaked on YPD and compared to the isogenic wild-type strain YOL14 after three days at the temperatures indicated. (B) Rpc25. Strain YGP-25, containing the S.cerevisiae rpc25-Δ::kanMX4 null allele and the multi-copy plasmid pGP-25 expressing the Sch.pombe rpc25 cDNA was serially diluted on YPD and compared to the isogenic wild-type strain FYBL3-2B after three days at the temperatures indicated. (C) Recapitulation of the heterospecific complementation pattern of the Sch.pombe core subunits (two upper panels) and enzyme-specific subunits (lower panel), based on previous reports (29,30,34) and on this study (subunits Rpc1, Rpc2, Rpc17, Rpc25, Rpc31, Rpc34, Rpc37, Rpc53 and Rpc82). Note that complementation by Rpb8 is only seen when over-expressing the largest subunit of RNA polymerase III (31).
These in vivo complementation data are summarized in Figure 1. Heterospecific complementation roughly correlated with sequence conservation. This, however, was not an absolute rule, since complementation did not occur for the highly conserved Rpc1 and Rpc2 subunits (60 and 70% of identity, respectively), but was observed with the much less conserved Rpc17 of Sch.pombe (18% of identity) as well as its human counterpart (21). Taken together, our data show that ten of the twelve Sch.pombe core subunits were functional in S.cerevisiae, but not the two largest ones (Rpc1 and Rpc2), a complementation pattern that closely paralleled the one observed for the twelve core subunits of RNA polymerase II (30). However, no complementation at all was observed for the five enzyme-specific subunits Rpc31, Rpc34, Rpc37, Rpc53 and Rpc82.
The subunit composition of RNA polymerase III is widely conserved across eukaryotes
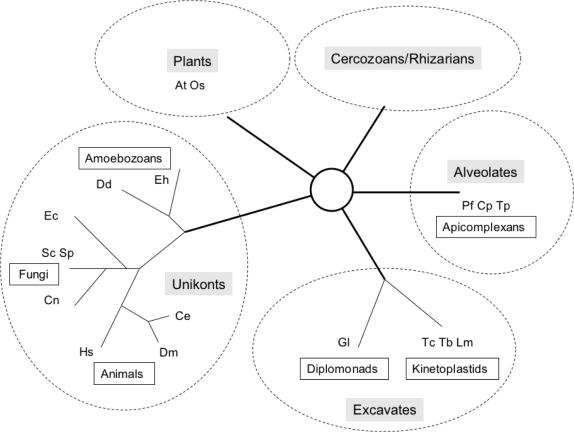
Our complementation data between the RNA polymerase III of S.cerevisiae and Sch.pombe point to a rapid divergence of their enzyme-specific subunits, as opposed to the much slower evolution of their core subunits. These two yeast species, though quite distant in terms of sequence homology, are ascomycetous fungi and therefore belong to the monophyletic ensemble of Unikonts, that combines all Fungi, Animals and Amoebozoans. Plants, Alveolates and Excavates, on the other hand, are excluded from Unikonts and define the three other large monophyletic groups of eukaryotes that are now recognized by most current phylogenetic trees. A fifth major group corresponds to the poorly characterized but probably monophyletic taxon formed by Cercozoans and Rhizarians (35,36). All these groups are very distant from each other in terms of amino acid sequence conservation, with no convincing root taxon identified so far. Their phyletic relations are summarized in Figure 2.
Figure 2.
Phylogenic positions of eukaryotes with fully sequenced genomes. The allocation of eukaryotes to five major phyla (surrounded by ovals) is based on two recent syntheses (35,36). No assumption is made as to the root of these eukaryotic phyla. Seventeen species with fully sequenced genomes were taken into account, and are designated by the following symbols. Fungi: Sc (S.cerevisiae), Sp (Sch.pombe); Cn (Cryptococcus neoformans), Ec (Encephalitozoon cuniculi). Animals: Hs (H.sapiens), Dm (Drosophila melanogaster), Ce (Caenorhabditis elegans). Amoebozoans: Dd (D.discoideum), Eh (E.histolytica). Plants: At (A.thaliana), Os (O.sativa). Alveolates: Pf (P.falciparum), Cp (Cryptosporidium parvum), Tp (Theileria parva). Excavates: Tc (T.cruzi), Tb (T.brucei), Lm (L.major), Gl (G.lamblia).
More than thirty eukaryotic genomes have been sequenced in the past few years. Most of them correspond to unikonts (twelve fungi, six animals and two amoebozoans, Dictyostelium discoideum and Entamoeba histolytica). However, fully sequences genomes also include two angiosperm plants (Arabidopsis thaliana and Oryza sativa) and three apicomplexans (Plasmodium falciparum, Theileria sp. and Cryptosporidium sp.) that belong to the large phylum of Alveolates. Excavates are represented by the genomes of several kinetoplastids (Leishmania major and Trypanosoma sp.) and of one diplomonad (Giardia lamblia). Except for Cercozoans and Rhizarians, this list corresponds to all of the main eukaryotic phyla currently identified (35,36).
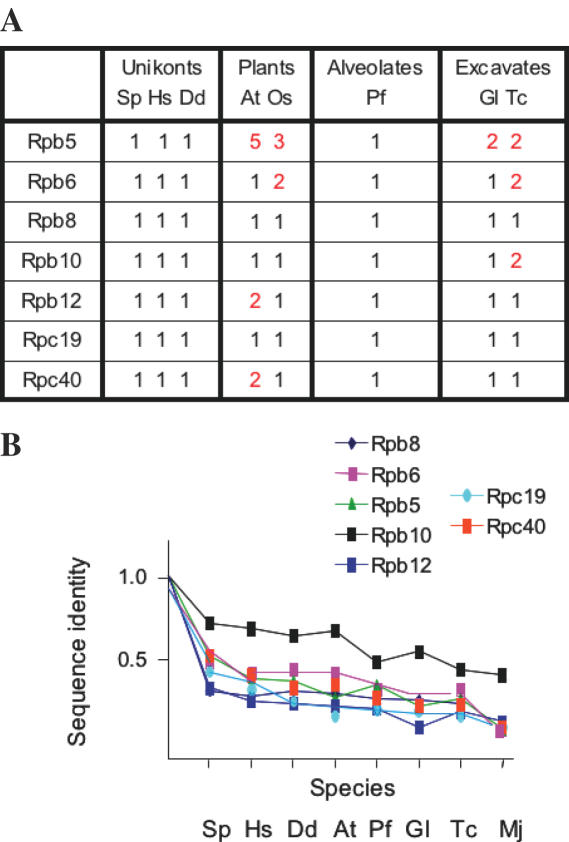
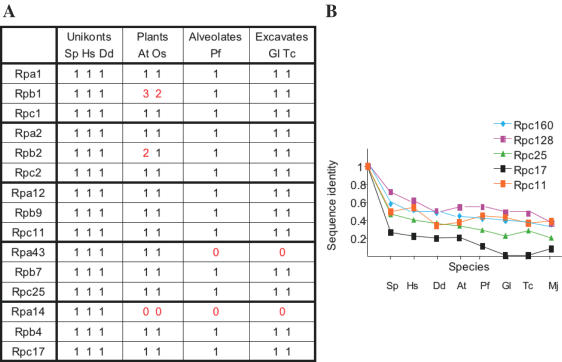
Homology to the RNA polymerase III subunits of S.cerevisiae was investigated in these eukaryotic genomes by iterative techniques (20) completed when necessary by manual comparison. Significant homology to the seventeen yeast subunits was detected in all eukaryotic phyla, except Rpc31, which is only found in Plants and Unikonts, thus demonstrating the very ancient origin of RNA polymerase III, including its enzyme-specific subunits (Figures 3–5). Moreover, subunits Rpa1, Rpa2 and Rpa12 (RNA polymerase I), Rpb1, Rpb2 and Rpb9 (RNA polymerase II) and Rpc1, Rpc2 and Rpc11 (RNA polymerase III) existed in all eukaryotic genomes sequenced so far, thus implying a very ancient separation of the three nuclear transcription systems (Figure 4). In contrast, the RNA polymerase I counterpart of Rpb7 and Rpc25, Rpa43, was only detected in Plants and Unikonts (24,26), and its Rpa14 partner (akin to Rpb4 and Rpc17) was not found outside Ascomycetes (37).
Figure 3.
Conservation of the seven RNA polymerase III subunits shared with RNA polymerase II and/or I. (A) Number of homologous coding sequences identified by iterative homology search. Closely related forms generated by alternative splicing or differing by 1% of their amino acid sequences were not listed. (B) The ratios of identical amino acids were derived from sequence alignments with the appropriate full-length S.cerevisiae subunit, using Psi-Blast iterative homology search (20). Species symbols are as in Figure 2, with one archaeal (Mj = Methanocaldococcus jannaschii) sequence added as external reference.
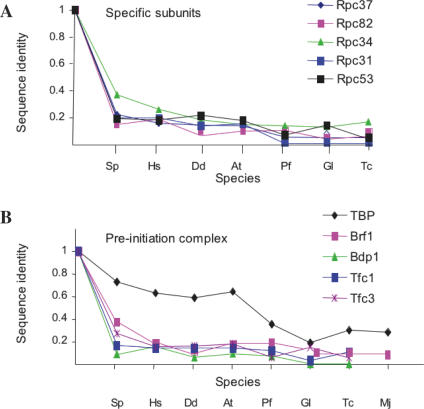
Figure 5.
Conservation of the Rpc31, Rpc34, Rpc37, Rpc53 and Rpc82 RNA polymerase III-specific subunits and of components of the RNA polymerase III initiation complex. (A) Enzyme-specific subunits: data and species symbols are as in Figure 2. (B) Components of the pre-initiation complex. Several eukaryotic species have multiple forms of TBP and Brf1, but we only considered the most strongly conserved one in each case. Data and species symbols are as in Figure 2.
Figure 4.
Conservation of the five RNA polymerase III core subunits (Rpc1, Rpc2, Rpc11, Rpc17 and Rpc25) (A) Number of homologous coding sequences identified by iterative homology search, using Rpc1, Rpc2, Rpc11, Rpc17 and Rpc25 and their paralogues in RNA polymerase I and II. The additional Rpb1-like and Rpb2-like subunits of plants correspond to the recently discovered RNA polymerase IV enzyme (54). (B) Data and species symbols are as in Figure 2.
Rpb5, Rpb6, Rpb8, Rpb10 and Rpb12 are identical in all three RNA polymerases of S.cerevisiae (5), whilst Rpc19 and Rpc40 are common to RNA polymerase I and III but paralogous to Rpb3 and Rpb11 in RNA polymerase II (38). The strong conservation of these common subunits was expected, since all but one (Rpb8) have a clear pre-eukaryotic ancestry (3). These subunits were generally encoded by single genes and are thus presumably shared by all three RNA polymerases in most of the genomes analysed. There were some exceptions to this rule, especially in the case of Rpb5, with two distinct forms clearly identified in Trypanomosoma sp. (39) and with possibly up to five Rpb5-like gene products in A.thaliana (40).
A recent survey of the G.lamblia genome failed to detect gene products related to the five enzyme-specific subunits Rpc31, Rpc34, Rpc37, Rpc53 and Rpc82 (41). Yet, our own data (Figure 5) leave no doubt as to the very significant homology relating Rpc34, Rpc37, Rpc53 and Rpc82 to gene products encoded in all eukaryotic phyla (the relevant alignments, including those of G.lamblia, are reproduced in Supplementary Data S3). Rpc31-like sequences, on the other hand, were only detected in Plants and Unikonts. Enzyme-specific subunits are, on the whole, less stringently conserved than the core subunits. In each case, however, large domains of fifty to hundred amino acids, with invariant positions forming unmistakable signatures, were conserved in all eukaryotes. In Rpc34, the importance of some of these invariant amino acids was underscored by previously described mutants impairing the two-hybrid interaction of that subunit with Brf1 (42). In Rpc31, homology included the acidic C-end of the subunit, important for transcription initiation (43). No mutant is currently available for the conserved Rpc37, Rpc53 and Rpc82 domains.
In S.cerevisiae, RNA polymerase III is directed to its cognate genes by a multi-protein complex in which the highly conserved TBP protein is shared with the two other RNA polymerases, whilst Brf1 is akin to TFIIB in RNA polymerase II. Bdp1 and the six-subunit TFIIIC factor, on the other hand, are entirely specific of the RNA polymerase III system (10,12,13). Brf1, Bdp1 and TFIIIC-like factors are well documented in Sch.pombe and Homo sapiens (12,13,17,44). As summarized Figure 5, a limited but significant homology to Brf1 and the Tfc1 and Tfc4 subunits of TFIIIC was detected in all eukaryotes. This was also true for the Bdp1 factors (except in the case of Excavates), where homology essentially corresponded to a previously recognized SANT domain (see also Supplementary Data S4). As for the enzyme-specific RNA polymerase III subunits, the overall homology of the pre-initiation complex was rather low (except for the highly conserved TBP protein), but invariant signature motifs were clearly identifiable for Brf1, Bdp1, Tfc1 and Tfc4 in all eukaryotes investigated.
Two domains of the largest subunit may be anchoring sites for the enzyme-specific subunits
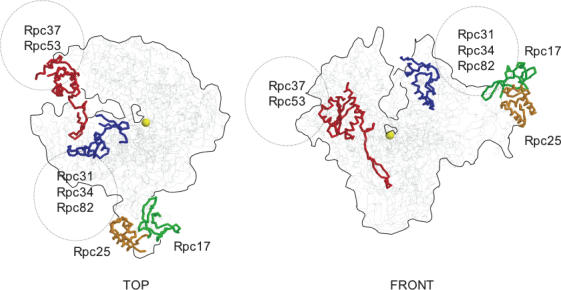
The Rpc31, Rpc34 and Rpc82 subunits of S.cerevisiae bind each other and are important for transcription initiation (42,43,45,46). Rpc37 and Rpc53, on the other hand, form a dimer (9) that co-operates with Rpc11 during elongation and termination (47). We hypothesized that these two groups of subunits may bind the twelve-subunit core enzyme at conserved but RNA polymerase III-specific docking sites. Three such domains, all longer than fifty amino acids, were identified by iterative homology searches (20) to the core RNA polymerase III of S.cerevisiae. One of these domains corresponds to the C-halves of Rpc25 (positions 103–212) and Rpc17 (positions 71–161), and is consistent with the probable role of the Rpc25-Rpc17 dimer in transcription initiation (48,49). The two others were detected near the N- and C-end of Rpc1 (positions 117–227 and 1170–1292 in S.cerevisiae), respectively. They are flanked by conserved amino acid motifs shared with Rpb1 (see Supplementary Data S5) and their spatial position can thus be confidently extrapolated from the RNA polymerase II crystal structure (22), as illustrated Figure 6.
Figure 6.
Conserved RNA polymerase III-specific domains. Top and front backbone views of RNA polymerase II in S.cerevisiae, prepared with the RASMOL software (http://www.umass.edu/microbio/rasmol/), using atomic coordinates 1WCM (22) at the PDB Protein Data Bank. The position of the three main RNA polymerase III-specific domains are shown as coloured thick lines on the RNA polymerase II structure. They correspond to the C-half of Rpc17, shown in green (positions 103–212, corresponding to 105–171 in Rpb7, see Supplementary Data S5), the C-half of Rpc25, shown in orange (positions 71–161, corresponding to 150–221 in Rpb4), and to N-terminal (red) and C-terminal (blue) motifs of Rpc1, delimitated by positions 117–227 and 1170–1292 (corresponding to 117–207 and 1148–1304 in Rpb1). The equivalence between Rpc1 and Rpb1 is based on the sequence alignments shown as Supplementary Data S5. The hypothetical localization of the Rpc37, Rpc53 and Rpc31, Rpc34 and Rpc82 groups of subunits is suggested by dashed ovals (see text for explanations).
Both domains are predicted to be on the external surface of the core enzyme, and are thus available for interactions with the enzyme-specific subunits of RNA polymerase III. Positions 117–227 of Rpc1 (corresponding to 117–207 on Rpb1) immediately follow a CX2C…CX2C Zn binding motif shared with all heteromultimeric RNA polymerases. A mutant of this motif was previously shown to dissociate Rpc31, Rpc34 and Rpc82 from the core RNA polymerase (46), thus strongly suggesting a direct role in the anchoring of these subunits. Positions 1170–1292 of Rpc1 correspond to 1148–1304 in Rpb1. In RNA polymerase II, this domain starts with a strong β-addition motif (corresponding to the Rpb1-β28 of positions 1145–1159) binding the Rpb9 paralogue of Rpc11 in RNA polymerase III. Rpc11 mediates transcript cleavage and termination (34) and recent evidence indicates that it closely co-operates with Rpc37 and Rpc53 in the termination/re-initiation process (47). We therefore predict that this domain determines the anchoring of a sub-complex formed by the Rpc37, Rpc53 and Rpc11 subunits.
DISCUSSION
The early biochemical characterization of yeast RNA polymerase III by Huet and her colleagues (8) revealed its surprisingly complex polypeptide composition, where seventeen distinct subunits contribute to a total MW of 0.7 MDa. Later studies established that all these polypeptides are bona fide and essential components of the RNA polymerase III system. Twelve of them are akin to the entire RNA polymerase II and to the RNA polymerase I core structure, with 42.5 and 46.3% identities, respectively but the existence of five additional polypeptides (Rpc31, Rpc34, Rpc37, Rpc53 and Rpc82) is a typical feature of RNA polymerase III (9,10). It recently emerged that the seventeen subunit composition of RNA polymerase III is entirely conserved not only between ascomycetous yeasts, such as S.cerevisiae and Sch.pombe (29,30,32,33,50), but also from yeasts to man (15,51).
Our heterospecific complementation data demonstrate that, except for Rpc1 and Rpc2, the core subunits of RNA polymerase III were readily exchangeable in vivo, thus showing a strong degree of functional conservation that is also consistent with a previous study of the RNA polymerase II enzyme (30). However, the enzyme-specific subunits diverged to a much stronger extent, as shown by their complete lack of heterospecific complementation. Despite this poor functional conservation, homology to all of the seventeen RNA polymerase III subunits including the enzyme-specific ones existed in currently sequenced eukaryotic genomes. The only notable exceptions concerned Rpc31, where homology was only detected in Unikonts and Plants, and Rpc17, the most variable of the core subunits, that may not exist in Excavates (G.lamblia, L.major, Trypanosoma cruzi and Trypanosoma brucei). Given that genome sequencing now extends to four of the five main eukaryotic phyla (35,36), the complex organization of RNA polymerase III can therefore be traced back in all present-day eukaryotes, although this could not be directly tested in Cercozoans and Rhizarians, where no fully sequenced genome is currently available. A more limited but still very significant conservation also applies to the RNA polymerase III-associated factors Brf1, Bdp1 and to at least two TFIIIC subunits, Tfc1 and Tfc4.
The early divergence of RNA polymerase III from RNA polymerase I and II is further supported by the fact that Rpc1, Rpc2 and Rpc11 co-existed with their paralogues in RNA polymerase I (Rpa1, Rpa2 and Rpa12) or II (Rpb1, Rpb2 and Rpb9) in all eukaryotic genomes investigated. The situation was somewhat different for Rpc25 and Rpc17. Orthologues to their RNA polymerase II counterparts (Rpb7 and Rpb4) were detected in all eukaryotic phyla but this was not so for RNA polymerase I, as Rpa43-like gene products were limited to Unikonts and Plants whilst Rpa14-like polypeptides were so far only found in Ascomycetes (37). The seven other core subunits are common to all three RNA polymerases (Rpb5, Rpb6, Rpb8, Rpb10 and Rpb12) or to RNA polymerase I and III (Rpc19 and Rpc40) in S.cerevisiae. They were encoded by single genes in most but not all of the eukaryotic genomes investigated so far, with some intriguing exceptions, such as the five Rpb5-like gene products predicted in A.thaliana (40). We also noticed that the human genome contains multiple retrocopies of the genes encoding some RNA polymerase subunits (e.g. Rpb4, Rpb5, Rpb10 and Rpb12), but about all of them accumulated stop codons in their coding sequences and probably are non-functional. Taken together, these data imply that the existence of three distinct RNA polymerases, with a common sub-set of core subunits shared by all three enzymes (or by RNA polymerase I and III in the case of Rpc19 and Rpc40) was established at a very early stage of eukaryotic evolution.
Rpc31, Rpc34 and Rpc82 form a cluster of enzyme-specific subunits that contribute to transcription initiation in S.cerevisiae (42,43,45,46) and H.sapiens (15,52). There is evidence that these subunits are anchored at or near the N-terminal Zn-fold of Rpc1 (46), itself prolonged by a highly conserved but RNA polymerase III-specific domain (this study). Rpc53 and Rpc37 interact in a two-hybrid assay (9) and, together with Rpc11, are involved in the termination and re-initiation of transcription in S.cerevisiae (47). We surmise that these subunits are anchored to the core polymerase by the conserved RNA polymerase III-specific domain found here near the C-end of Rpc1.
The present study identified widely conserved domains in each of the five enzyme-specific subunits of RNA polymerase III. Moreover, the existence of conserved domains in the TFIIIC, Brf1 and Bdp1 partners of RNA polymerase III and the probable functional connection relating Rpc25 and Rpc17 to Brf1 (48,49) suggest that a network of protein–protein contacts was evolutionarily preserved in all eukaryotes. Indeed, studies on Rpc34 have shown that mutants of its conserved domain affect an Rpc34-Brf1 interaction detected by a two-hybrid assay and leading to transcription initiation and promoter opening defects (42). There is some evidence that this interaction concerns the C-end of Brf1 (53), itself highly conserved in most eukaryotes. In Rpc31, mutants of the conserved acidic C-end also interfere with transcription initiation (43). Extending these studies to the conserved domains identified here on Rpc82, Rpc37 and Rpc53 should help to understand how the five RNA polymerase III-specific subunits collectively ensure the transcriptional specificity of this enzyme.
Acknowledgments
The authors thank Alexei Kayushin, Nikolai Bystrov and Dmitry Staroverov for oligonucleotide synthesis and automated DNA sequencing, and Christophe Carles, Olivier Lefebvre, Michel Riva and Michel Werner for useful discussions. This work was partly supported by the Molecular and Cellular Biology programme of the Russian Academy of Sciences (direction ‘Functional Genomics’), by the Russian Foundation for Basic Research (project No. 04-04-48987), by an ARC grant to C.Z. and by an INTAS Young Investigator grant to E.K.S. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bushnell D.A., Kornberg R.D. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc. Natl Acad. Sci. USA. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armache K.J., Kettenberger H., Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl Acad. Sci. USA. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langer D., Hain J., Thuriaux P., Zillig W. Transcription in Archaea: similarity to that in Eucarya. Proc. Natl Acad. Sci. USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werner F., Weinzierl R.O. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 5.Woychik N.A., Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 6.Briand J.F., Navarro F., Rematier P., Boschiero C., Labarre S., Werner M., Shpakovski G.V., Thuriaux P. Partners of Rpb8p, a small subunit shared by yeast RNA polymerases I, II and III. Mol. Cell. Biol. 2001;21:6056–6065. doi: 10.1128/MCB.21.17.6056-6065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadal O., Chédin S., Quémeneur E., Carles C., Sentenac A., Thuriaux P. A34.5, a non-essential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machinery. Mol. Cell. Biol. 1997;17:1787–1795. doi: 10.1128/mcb.17.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huet J., Riva M., Sentenac A., Fromageot P. Yeast RNA polymerase C and its subunits: specific antibodies as structural and functional probes. J. Biol. Chem. 1985;260:15304–15310. [PubMed] [Google Scholar]
- 9.Flores A., Briand J.F., Boschiero C., Gadal O., Andrau J.C., Rubbi L., Van Mullem V., Goussot M., Marck C., Carles C., et al. A protein–protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chédin S., Ferri M.L., Peyroche G., Andrau J.C., Jourdain S., Lefebvre O., Werner M., Carles C., Sentenac A. The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Proceedings of the Cold Spring Harbor Symposium on Quantitative Biology; Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. pp. 381–389. [DOI] [PubMed] [Google Scholar]
- 11.Harismendy O., Gendrel C.G., Soularue P., Gidrol X., Sentenac A., Werner M., Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geiduschek E.P., Kassavetis G.A. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 13.Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 14.Hu P., Wu S., Sun Y., Yuan C.C., Kobayashi R., Myers M.P., Hernandez N. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol. Cell. Biol. 2002;22:8044–8055. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Roeder R.G. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 16.Wood V., Gwilliam R., Rajandream M.A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y., Maraia R.J. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 2001;29:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fikes J.D., Becker D.M., Winston F., Guarente L. Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature. 1990;346:291–294. doi: 10.1038/346291a0. [DOI] [PubMed] [Google Scholar]
- 19.Shpakovski G.V., Acker J., Wintzerith M., Lacroix J.F., Thuriaux P., Vigneron M. Four subunits shared by the three classes of RNA polymerases are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaffer A.A., Aravind L., Madden T.L., Shavirin S., Spouge J.L., Wolf Y.I., Koonin E.V., Altschul S.F. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siaut M., Zaros C., Levivier E., Ferri M.L., Werner M., Callebaut I., Thuriaux P., Sentenac A., Conesa C. A Rpb4/Rpb7 like complex in yeast RNA Polymerase III contains the orthologue of mammalian CGRP-RCP. Mol. Cell. Biol. 2003;23:195–205. doi: 10.1128/MCB.23.1.195-205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armache K.J., Mitterweger S., Meinhart A., Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005;280:7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 23.Shematorova E.K., Shpakovski G.V. Structure and functions of eukaryotic nuclear DNA-dependent RNA polymerase I. Mol. Biol. (Moscow) 2002;36:1–17. [PubMed] [Google Scholar]
- 24.Shpakovskii G.V., Shematorova E.K. [Characteristics of the cDNA of the Schizosaccharomyces pombe rpa43+ gene: structural similarity of the Rpa43 subunit of RNA-polymerase I with the Rpc25 subunit of RNA-polymerase III] Bioorg. Khim. 1999;25:791–796. [PubMed] [Google Scholar]
- 25.Nogi Y., Yano R., Dodd J., Carles C., Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the 12.2 subunit of RNA polymerase I and is essential only at high temperature. Mol. Cell. Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peyroche G., Levillain E., Siaut M., Callebaut I., Schultz P., Sentenac A., Riva M., Carles C. The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits. Proc. Natl Acad. Sci. USA. 2002;99:14670–14675. doi: 10.1073/pnas.232580799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shpakovski G.V. The fission yeast Schizosacharomyces pombe rpb6 gene encodes the common phosphorylated subunit of RNA polymerase and complements a mutation in the corresponding gene of Saccharomyces cerevisiae. Gene. 1994;147:63–69. doi: 10.1016/0378-1119(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 28.Shpakovskii G.V., Lebedenko E.N., Thuriaux P. [Cloning of cDNA for RNA polymerase subunit from the fission yeast Schizosaccharomyces pombe by heterospecific complementation in Saccharomyces cerevisiae] Bioorg Khim. 1997;23:110–117. [PubMed] [Google Scholar]
- 29.Shpakovski G.V., Shematorova E.K. Rpc19 and Rpc40, two alpha like subunits shared by nuclear RNA polymerase I and III, are interchangeable between the fission and budding yeasts. Curr. Genet. 1999;36:208–214. doi: 10.1007/s002940050492. [DOI] [PubMed] [Google Scholar]
- 30.Shpakovski G.V., Gadal O., Labarre-Mariotte S., Lebedenko E.N., Miklos I., Sakurai H., Proshkin S.A., Van Mullem V., Ishihama A., Thuriaux P. Functional conservation of RNA polymerase II in fission and budding yeasts. J. Mol. Biol. 2000;295:1119–1127. doi: 10.1006/jmbi.1999.3399. [DOI] [PubMed] [Google Scholar]
- 31.Voutsina A., Riva M., Carles C., Alexandraki D. Sequence divergence of the RNA polymerase shared subunit ABC14.5 (Rpb8) selectively affects RNA polymerase III assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:1047–1055. doi: 10.1093/nar/27.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai K., Imazawa Y., Yao Y., Yamamoto K., Hisatake K., Muramatsu M., Nogi Y. The fission yeast rpa17+ gene encodes a functional homolog of AC19, a subunit of RNA polymerases I and III of Saccharomyces cerevisiae. Mol. Gen. Genet. 1999;261:364–373. doi: 10.1007/s004380050977. [DOI] [PubMed] [Google Scholar]
- 33.Imazawa Y., Imai K., Fukushima A., Hisatake K., Muramatsu M., Nogi Y. Isolation and characterization of the fission yeast gene rpa42+, which encodes a subunit shared by RNA polymerases I and III. Mol. Gen. Genet. 1999;262:749–757. doi: 10.1007/pl00013818. [DOI] [PubMed] [Google Scholar]
- 34.Chédin S., Riva M., Schultz P., Sentenac A., Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adl S.M., Simpson A.G., Farmer M.A., Andersen R.A., Anderson O.R., Barta J.R., Bowser S.S., Brugerolle G., Fensome R.A., Fredericq S., et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 36.Baldauf S.L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 37.Imazawa Y., Hisatake K., Mitsuzawa H., Matsumoto M., Tsukui T., Nakagawa K., Nakadai T., Shimada M., Ishihama A., Nogi Y. The fission yeast protein Ker1p is an ortholog of RNA polymerase I subunit A14 in Saccharomyces cerevisiae and is required for stable association of Rrn3p and RPA21 in RNA polymerase I. J. Biol. Chem. 2005;280:11467–11474. doi: 10.1074/jbc.M411150200. [DOI] [PubMed] [Google Scholar]
- 38.Lalo D., Carles C., Sentenac A., Thuriaux P. Interactions between three common subunits of yeast RNA polymerases I and III. Proc. Natl Acad. Sci. USA. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walgraffe D., Devaux S., Lecordier L., Dierick J.F., Dieu M., Van den Abbeele J., Pays E., Vanhamme L. Characterization of subunits of the RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 2005;139:249–260. doi: 10.1016/j.molbiopara.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Larkin R.M., Hagen G., Guilfoyle T.J. Arabidopsis thaliana RNA polymerase II subunits related to yeast and human RPB5. Gene. 1999;231:41–47. doi: 10.1016/s0378-1119(99)00090-6. [DOI] [PubMed] [Google Scholar]
- 41.Best A.A., Morrison H.G., McArthur A.G., Sogin M.L., Olsen G.J. Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia. Genome Res. 2004;14:1537–1547. doi: 10.1101/gr.2256604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brun I., Sentenac A., Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thuillier V., Stettler S., Sentenac A., Thuriaux P., Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995;14:351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozenfeld S., Thuriaux P. Genetic interactions within TFIIIC, the promoter-binding factor of yeast RNA polymerase III. Mol. Gen. Genomics. 2001;265:705–710. doi: 10.1007/s004380100467. [DOI] [PubMed] [Google Scholar]
- 45.Bartholomew B., Durkovich D., Kassavetis G.A., Geiduschek E.P. Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werner M., Hermann-Le Denmat S., Treich I., Sentenac A., Thuriaux P. Effect of mutations in a zinc binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol. Cell. Biol. 1992;12:1087–1095. doi: 10.1128/mcb.12.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landrieux E., Nazif A., Ducrot C., Acker J., Riva M., Carles C. A subcomplex of RNA polymerase III subunits is required for transcription termination and reinitiation. EMBO J. 2005;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaros C., Thuriaux P. Rpc25, a conserved RNA polymerase III subunit, is critical for transcription initiation. Mol. Microbiol. 2005;55:104–114. doi: 10.1111/j.1365-2958.2004.04375.x. [DOI] [PubMed] [Google Scholar]
- 49.Ferri M.L., Peyroche G., Siaut M., Lefebvre O., Carles C., Conesa C., Sentenac A. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol. 2000;20:488–495. doi: 10.1128/mcb.20.2.488-495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y., Hamada M., Maraia R.J. RNA polymerase III from the fission yeast, Schizosaccharomyces pombe. Meth. Enzymol. 2003;370:165–173. doi: 10.1016/S0076-6879(03)70014-7. [DOI] [PubMed] [Google Scholar]
- 51.Hu P., Wu S., Hernandez N. A minimal RNA polymerase III transcription system from human cells reveals positive and negative regulatory roles for CK2. Mol. Cell. 2003;12:699–709. doi: 10.1016/j.molcel.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Khoo B., Brophy B., Jackson S.P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;9:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 53.Andrau J.C., Sentenac A., Werner M. Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 1999;288:511–520. doi: 10.1006/jmbi.1999.2724. [DOI] [PubMed] [Google Scholar]
- 54.Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]