Abstract
Microtubule-associated protein 2 (MAP2), a neuron-specific protein, stabilizes microtubules and is critical for neurite outgrowth and dendrite development. Although MAP2 is widely used as a marker of neuronal differentiation, regulation of its transcription has not been investigated. We showed that MAP2 is frequently activated in human cutaneous melanoma. Here, we identified a 2.2 kb region that is sufficient for neuronal-specific expression in vitro and in vivo. Comparative analysis of the mouse, rat and human MAP2 promoter sequences showed the presence of a conserved bHLH factor binding sites. Electrophoretic mobility shift analysis, promoter mutagenesis and co-transfection experiments showed that NeuroD, a pro-neuronal differentiation factor, and Hairy and Enhancer of Split (HES1), a transcription repressor, are involved in the regulation of MAP2 promoter activity. Melanoma cells express both NeuroD and HES1. Chromatin immunoprecipitation showed that in metastatic melanoma cells N-box region of the MAP2 promoter is occupied by endogenous HES1. We show that the inhibition of Notch signaling, a regulator of HES1 gene expression, and/or shRNA knockdown of HES1 results in the upregulation of MAP2 promoter activity. Thus, our data suggest that Notch signaling, which is implicated in melanoma progression, and HES1 play a role in MAP2 gene regulation during melanoma progression.
INTRODUCTION
Microtubule dynamics is regulated by a family of proteins known as microtubule-associated proteins (MAPs). MAP family includes several isoforms of MAP1, MAP2 and tau, which are expressed primarily in neurons. Among neuronal MAPs, MAP2 expression is considered a hallmark of neuronal differentiation (1,2). MAP2 is found primarily in the dendritic extensions of post-mitotic, terminally differentiated neurons. MAP2 plays a critical role in neurite outgrowth and dendrite development (3–5). Change in the expression of MAP2 is a helpful diagnostic and prognostic feature in various neurological disorders (6–8). Despite its extensive use as a marker of neuronal differentiation, and its role in morphological and functional differentiation of neurons, little is known about the regulation of MAP2 gene expression.
Interestingly, MAP2 gene is activated in certain human non-neuronal cancers that exhibit characteristics of neural differentiation (2,9,10). We showed that cutaneous primary melanoma, a cancer that arises from neural crest-derived melanocytes, often express abundant MAP2 protein (11). Since the only function of MAP2, known to date, is to stabilize microtubules, we investigated whether MAP2 expression in rapidly proliferating cancer cells could interfere with microtubule dynamics, inhibit mitosis and, therefore, delay or inhibit tumor progression. We showed, in a five-year clinical follow-up study, that patients whose primary tumors express abundant MAP2 had longer disease-free survival rate than those patients with weak or no MAP2 expression (12). Moreover, we showed that reactivation of endogenous MAP2 gene transcription by treatment with pharmacological compounds or expression of exogenous MAP2 by adenovirus in metastatic melanoma cells in vitro leads to cell cycle arrest, growth inhibition and apoptosis (11,12). These observations suggested that reactivation of MAP2 gene transcription is potentially a useful strategy for the treatment of metastatic melanoma.
In this study, we show that a 3.3 kb 5′ genomic region of human MAP2 gene that contains several E- and N-boxes, which are binding sites for basic helix–loop–helix (bHLH) transcription factors, is sufficient for neuronal-specific expression of a reporter gene both in vitro and in vivo. We also show that MAP2 promoter is activated by neurogenic bHLH factor NeuroD/BETA2, preferentially, in melanoma cells and repressed by the bHLH repressor Hairy and Enhancer of Split 1 (HES1), a regulator of neuronal differentiation. Although both NeuroD and HES1 are expressed in metastatic melanoma cells, results of co-transfection and chromatin immunoprecipitation experiments suggest that MAP2 gene is regulated by the relative levels of these factors, and predominantly by the level of HES1 protein. Using HES1shRNA knockdown and a pharmacological inhibitor of HES1 activator Notch1, we show that MAP2 promoter activity is regulated by Notch1. We also show that Notch1 signaling, which is implicated in melanoma progression (13–15), differentially regulates MAP2 promoter activity in primary and metastatic melanoma cells.
MATERIALS AND METHODS
Cell culture
Mouse neuroblastoma cell line, Neuro2a, and human embryonal carcinoma cell line NT2-D1 were purchased from American Type Culture Collection (Manassas, VA). Neuro2a cells were grown in minimal essential medium (MEM). The human glioblastoma cell lines U251, U138 and rat PC12 cells were cultured in Rosewell Park Memorial Institute (RPMI) medium and DMEM, respectively. The human breast cancer cell line MCF-7 was cultured in MEM supplemented with bovine insulin. Human fibroblast cell line NIH3T3 was cultured in DMEM with high glucose, and human embryonic teratocarcinoma cell line NT2-D1 was cultured in DMEM with high glucose and supplemented with NaHCO3 (1.5 g/l). Neonatal foreskin melanocytes, primary melanoma cell lines WM35 and WM75, and metastatic melanoma cell lines SK-MEL-19, 451Lu and SK-MEL-23 cl.22a were cultured as described previously (16). All culture media were supplemented with 10% fetal bovine serum (FBS). Medium for PC12 cells were supplemented with 5% FBS and 10% horse serum, 1% l-glutamine, and 1% penicillin and streptomycin. All cell culture media, l-glutamine, penicillin and streptomycin, non-essential amino acids were from GIBCO (Grand Island, NY), and FBS was from Sigma (St Louis, MO) or Atlanta Biologicals (Lawrenceville, GA).
Differentiation of PC12 cells
Before differentiation, cells were grown in 1% horse serum containing media on collagen-coated tissue culture dishes. After the cells got attached, they were treated with 100 ng/ml nerve growth factor (NGF 2.5S; Promega, Madison, WI). Twenty-four hours after NGF treatment, cells were transfected with promoter constructs and cultured for additional 48 h in medium containing NGF. For MAP2 immunostaining, PC12 cells were treated with 100 ng/ml NGF for 6 days (17,18).
Plasmids and antibodies
pCI-Hes1, a mouse Hes1 expression plasmid and Hes1 antibody (19) were gifts from Prof. R. Kageyama (Institute for Virus research, Kyoto University, Kyoto, Japan). Rabbit polyclonal antibody directed against the C-terminal region of rat Hes1 for supershift experiments was kindly provided by Dr Y. N. Jan (University of California, San Francisco, CA). NeuroD expression plasmid pCDNA3-NeuroD1-FLAG (pNeuroD) (20) was provided by Dr Haeyoung Suh-Kim (Ajou University School of Medicine, Suwon, Korea). Goat polyclonal anti-NeuroD antibody for western blot analysis (N-19) and for supershift experiments and chromatin immunoprecipitation assay (N-19X) and anti-HES1 antibody for ChIP (N-17X) were purchased from Santa Cruz Biotech (Santa Cruz, CA). Affinity purified anti-MAP2ab was obtained from Zymed Laboratories (San Francisco, CA), anti-α-tubulin antibody was obtained from Sigma (St Louis, MO). Horseradish peroxidase (HRP) conjugated goat anti-mouse IgG was from Jackson Laboratories (West Grove, PA). Anti-mouse IgG–FITC was purchased from Dako Corp. (Carpentaria, CA).
Construction of human MAP2 promoter-luciferase plasmids
Human MAP2 genomic sequence (GenBank accession no. NT_005403) was identified using BLAST search with MAP2 cDNA sequence, and a 2.2 kb region corresponding −1854 to +368 was amplified by PCR using forward primer 5′-CTGGCCTTTTTGGTTCTCAT and reverse primer 5′-TAGTCTAAGCTTAGC TGAGAATCTACCGA containing HindIII sites and subcloned into the HindIII site of pGL3 basic vector. This plasmid is designated as phMAP2-2. Since attempts to amplify the longer genomic DNA fragment containing region upstream of −1854 were unsuccessful, we first PCR amplified a region corresponding to −2965 to −106 using a forward primer 5′-GACGACAAGCTTCAGAAGAAGGGTAAGGCAAGCATCA and a reverse primer 5′-GACGACAAGCTTCGCAGTGGCGAACAGGAAAA, and constructed the longer phMAP2-1 plasmid by joining the 5′ −2965/−263 fragment with −263/+369 fragment at the internal KpnI site. Deletion constructs phMAP2-3 (−263/+369) were made by splicing KpnI/KpnI (−1854/−268) fragment out from phMAP2-2 and re-ligating the plasmid. phMAP2-4 (−269/+30) was made by digestion of phMAP2-3 with PstI (+31) and HindIII (+368), followed by fill-in reaction with DNA polymerase (Klenow fragment) and blunt end ligation.
E- and N-box mutant MAP2 promoters
mutE1 (−2888/−2883), mutE2 (−2799/−2794), mutE3(−2304/−2299), mutE4 (−1593/−1588) individual mutations and various combination of E-box mutations (E1,2,3,4; E1,2,3; E2,3) were generated in phMAP2-1, mutN1 (−961/−956), mutN2 (−63/−58), mutE9 (−358/−353), mutE11 (−279/−274), mutE12 (+67/+72) and mutE13 (+267/+272) and combinations of these mutations (E12,13; E12,13,N1) were generated in phMAP2-2 using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. The following primers were used for mutagenesis (only sense sequence is given and mutated nucleotides within the consensus sequence are underlined):
mutE1: 5′-CACAGAAGTATCTACATGTTCGGTTGTCTTG; mutE2: 5′-CTCCATGTTGGAAAGCATGCGGAAAAAAAGAAAAG; mutE3: 5′-GATGATGGTGGCAGGTACACTCTTTTAGACATC; mutE4: 5′-GGGAAAGAAAGTGTCTACGTGAGAGAAGAAAGG; mutE9: 5′-CACAAAGGAGACGCGAGGATGACATTCATAG; mutE11: 5′-GCCTGTCAGGGCTTCCGTACCAGAAG; mutE12: 5′-GTGGTATTGCTGAATATTCGCTGGTAATGG;mutE13: 5′-GAGTATAAAGAATGGGCTTGCCTTACTGGTGAG; mutN1: 5′-CACTTAACCTCCCTAACCAGAGGCACTGTAAAATAG; mutN2: 5′-GGCACACACAGAGCCGCCCTGGTGGCTTG CAG.
Transient transfection and luciferase assays
Transient transfections with various promoter constructs were performed using Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Cells in 24- or 6-well tissue culture dishes, in triplicates, were transfected with 0.6 or 3 μg, respectively, of empty vector pGL3 or promoter-reporter constructs (phMAP2-1, phMAP2-2, phMAP2-3 and phMAP2-4) and 0.07 or 0.5 μg Renilla luciferase plasmid pRL. In co-transfection experiments, different amounts of NeuroD and Hes1 expression plasmids were added. For Notch1 inhibition and HES1 knockdown experiments, cells were either pre-treated with indicated amount of γ-secretase inhibitor-DAPT (Calbiochem, La Jolla, CA) and/or transfected with indicated amount of lentiviral HES1shRNA plasmid (The RNAi Consortium collection, a library of short hairpin DNAs cloned in pLKO.1 lentiviral vector; Open biosystem, Huntsville, AL) 1 day before transfection with MAP2 promoter constructs. After 48 h of transfection, cells were washed with pre-chilled PBS and lysed in passive lysis buffer (Dual Luciferase kit, Promega). Firefly luciferase and Renilla luciferase activities in the cell lysates were measured according to the manufacturer's instructions using TD 20/20-luminometer (Turner Biosystems, Sunnyvale, CA). Firefly luciferase activity was normalized to the Renilla luciferase activity and reported as relative luciferase activity (RLA).
Transient transgenic expression
The 3.3 kb MAP2 genomic fragment from phMAP2-1 and 2.2 kb fragment from phMAP2-2 was isolated by digestion with HindIII and subcloned into the HindIII site upstream of Hsp68-lacZ gene in pHSF5 vector (21). The orientation and sequences were confirmed by restriction digestion and sequencing. Transgenic embryos were generated by Biotechnology Center (University of Wisconsin, Madison, WI). Embryos harvested on day 11.5 were stained for β-galactosidase activity essentially as described by Shashikant et al. (21).
Immunostaining
Control embryos (11.5 days old) were fixed in 4% formaldehyde overnight at 4°C and dehydrated using a graded series of methanol, and embedded in paraffin. Sections (9 μm thick) were cut and stained for MAP2 with anti-MAP2ab (Sigma) using citrate buffer antigen retrieval method (22) followed by anti-mouse IgG–HRP and the antigen was detected with HRP substrate diaminobenzidene.
Electrophoresis mobility shift assay (EMSA)
Wild type E- and N-boxes containing oligonucleotides were labeled with [γ-32P]ATP (3000 Ci/mMol; ICN Biomedicals, Inc., Irvine, CA) with T4-polynucleotide kinase (Promega Corp., Madison, WI). Unincorporated label was removed using NAP5 columns (Amersham Pharmacia Biotech, Piscataway, NJ). Radioactivity in the eluted fractions was measured and ∼20 000 c.p.m were used for each binding reaction. The following reverse phase purified oligonucleotides were annealed and used as probes and competitors. Core sequences of E- and N-box are underlined: WtN1, 5′-CCTCCCTACACAAGGGCACTGT; mutN1, 5′-CCTCCCTAACCAGAGGCACTGT; WtN2, 5′-CACACACAGACACGAGCTGGTGGCTT; mutN2, 5′-CACACACAGAACCGGACTGGTGGCTT; mutN2-scrambled, 5′-ACCAACCAGAACCGGACTGGTGGCTT; WtE6E7, 5′-GGGATATTCAACTGCTAATTTCAGTTGCTAACATG; mutE6E7, 5′-GGGATATTACACGTCTAATTTACGTGTCTAACATG; WtE8, 5′-AACATGGCCAAGTGCATAATTA; mutE8, 5′-AACATGGCACAGGTCATAATTA; wtE9, 5′-AAGGAGACCAGATGATGACATT; mutE9, 5′-AAGGAGACACGAGTATGACATT; wtE10, 5′-TGGGGACACACTTGGATG GAGG; mutE10, 5′-TGGGGACAACCTGTGATGGAGG; wtE11, 5′-GCCTGTCAGGCATTTGGTACCAGAAG; and mutE11, 5′-GCCTGTCAGGGCTTCCGTACCAGAAG. Nuclear extracts of COS-7 or cl.22a melanoma cells transfected with Hes1 expression plasmid or with NeuroD expression plasmid were prepared essentially as described previously (16). Labeled probes were incubated with 5–10 μg of nuclear extract at room temperature for 30 min. For supershift experiments, anti-Hes1 or anti-NeuroD antibody or unrelated polyclonal antibody were added to the reaction mixture and incubated at room temperature for additional 30 min. DNA–protein complexes were analyzed by electrophoresis on non-denaturing 5% polyacrylamide gels run at constant 200 V for 2 h in 0.5× TBE. The gels were dried and exposed to X-ray film (Eastman Kodak Company, Rochester, NY).
Chromatin immunoprecipitation assay (ChIP assay)
Assay was performed using ChIP-IT kit (Active Motif, Carlsbad, CA) following the manufacturer's protocol. Briefly, SK-MEL-23 cells were cross linked, chromatin was prepared and immunoprecipitated with anti-HES1 antibody (Santa Cruz) or with control IgG. Then, immunoprecipitated DNA was eluted and PCR amplified using appropriate primers. For PCR amplification of N1 (36 cycles) and N2 (29 cycles) box region in MAP2 promoter region, the following primers were used: N1-forward, 5′-CCCAGGAAATAAATGCAGGA; N1-reverse, 5′-GGGCCGATATGTGATTTCTG; N2-forward, 5′-CCACTCGCCTTATTTTCCTG; and N2-reverse, 5′-CGCATATGCAGCAAACAC. For PCR amplification of E9–E11 boxes (36 cycles) in promoter region the following primers were used: forward, 5′-TAAGCGGTGTGTGTGTGTGC; and reverse, 5′-ATGATGACAAGCCACTCAGC.
RT–PCR
Total RNA was isolated from different melanoma and non-melanoma cells using RNeasy Mini Kit (Qiagen, Valencia, CA) and amplified by using one-step RT–PCR Kit (Qiagen) as per manufacturer's instructions using 100 ng of total RNA and MAP2 forward primer 5′-GCAGTTCTCAAAGGCTAGAC (nt 31–50 in MAP2ab cDNA in exon 3) and reverse primer 5′-TTGATCGTGGAACTCCATCT (nt 721–740 in MAP2ab cDNA in exon 9).
SDS–PAGE and western blotting
Total cell lysates of normal melanocytes, different melanoma and non-melanoma cell lines were prepared using lysis buffer [1% Triton X-100 in PBS containing cocktail of protease inhibitors (Roche Diagnostics Corp., Indianapolis, IN)]. Protein was estimated using BCA protein assay kit (Pierce Biotechnology Inc., Rockford, IL) and 50–100 μg protein was separated on 9% SDS–PAGE, transferred on to Polyscreen membrane (NEN Life Sciences, Boston, MA) and the membranes were blocked and then incubated overnight at 4°C with the primary antibodies diluted (anti-Hes1 at 0.6 μg/ml; anti-NeuroD at 1:250; anti-α-tubulin at 1:2000 and anti-β-actin at 1:2000) in 5% non-fat dry milk in TBST, followed by incubation with the appropriate HRP-conjugated secondary antibodies (at 1:5000 to 1:10 000 dilution) for 1 h at room temperature. Protein bands were detected by chemiluminescence using ECL kit (Amersham Biosciences, Piscataway, NJ).
RESULTS
Nucleotide sequences upstream of MAP2 gene are conserved in mouse, rat and human
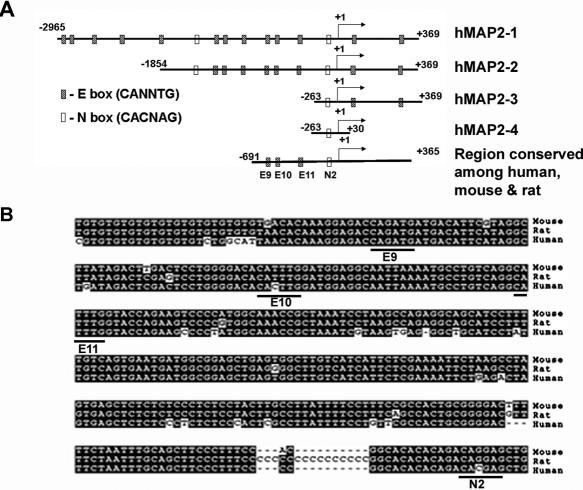
Using BLAST search, we identified the human genomic sequences upstream of MAP2 cDNA in a chromosome 2 contig (GenBank accession no. NT_005403) and amplified, by PCR, a 3334 bp genomic DNA fragment consisting 2965 nt upstream of MAP2 transcription start site (+1), exon 1 (78 bp) and a part of intron 1 (291 bp). We sequenced this fragment (GenBank accession no. DQ 386449), and cloned it into pGL3 luciferase reporter vector (phMAP2-1; −2965/+369). First, we analyzed this MAP2 genomic sequence for transcription factor binding sites and compared the human sequence with mouse and rat MAP2 genomic sequences using CLUSTALW program (Figure 1A and B). This analysis showed the following:
Mouse, rat (GenBank accession nos: mouse, NT_039170; and rat, NW_047816) and human MAP2 genomic sequences show 80% sequence identity in 1 kb region corresponding to −691/+365 of human MAP2 genomic sequence. In addition, an 87 bp rat genomic sequence showed 85% identity to human sequences between −1388/−1302. There were no homologous sequences to this 87 bp in the mouse.
Inspection of the 3.3 kb human MAP2 5′ genomic sequence showed the presence of 13 (E1–E13) putative E boxes (CANNTG) and two (N1 and N2) putative N-boxes (CACNAG), which are potential sites for binding bHLH proteins including neuronal transcriptional activators and repressors (23,24).
The proximal cluster of three E boxes (E9–E11) at −358/−353, −313/−308 and −279/−274, and a N2 box −63/−58 found in the human MAP2 sequence are conserved among mouse, rat and human sequences; additional upstream (E1–E8) and downstream (E12 and E13 within exon 1 and at +267/+272 within intron 1) E boxes and N1 box at −961/−956 within the human MAP2 promoter sequence are not present in the mouse and the rat MAP2 genomic sequences.
No canonical TATA box could be identified in the MAP2 proximal 5′ genomic sequences, suggesting that MAP2 promoter is a TATA-less promoter (20,25,26).
Figure 1.
(A) Schematic diagram of the 3.3 kb human MAP2 genomic DNA fragment consisting 5′ regulatory region, the first exon and partial sequence of the first intron, and different promoter constructs used in this study. The relative location (not drawn to scale) of putative E- and N-boxes, and the transcription start site (arrow at +1) are shown. (B) Mouse and rat genomic sequences homologous to the 3.3 kb human MAP2 promoter region were retrieved from GenBank database by BLAST search, conserved E- and N-boxes region was aligned using CLUSTALW sequence alignment program and shown by shading using Boxshade program. Dashes show gaps in the sequences. Conserved E boxes and N2 box sequences are highlighted.
MAP2 promoter activity
To investigate the promoter activity of MAP2 5′ genomic sequence, we generated, in addition to the 3.3 kb luciferase reporter plasmid (phMAP2-1), 5′ deletion constructs phMAP2-2 (−1864/+369), phMAP2-3 (−263/+369) and a 3′ deletion construct phMAP2-4 (−263/+30) that lacks intron 1 sequence and part of exon 1. We tested the activity of these MAP2 promoter plasmids in rat PC12 cells and human teratocarcinoma cell line NT2/D1 by transient transfection followed by luciferase assays. PC12 cells treated with NGF are known to differentiate into neuron-like cells with neurite outgrowths and express MAP2 (Figure 2A, inset) (17,18). NT2/D1 is an inducible model system for neuronal differentiation. Continuous culture of NT2/D1 cells in complete serum containing retinoids or the addition of retinoic acid to the steroid free culture medium is known to induce neuronal differentiation and MAP2 expression (as shown by RT–PCR, inset in Figure 2B) in these cells (27).
Figure 2.
MAP2 promoter activity: activity of MAP2 promoter in PC12 (A), NT2/D1 (B) and melanoma cells (C). (A) Control and NGF-treated rat PC12 cells and HeLa cells were transfected in triplicates in 24-well plates with the indicated MAP2 promoter-luciferase plasmids and the Renilla luciferase plasmid. Firefly and Renilla luciferase activities were measured using Dual Luciferase Reporter Assay System (Promega). Data from a representative of at least three experiments are shown as fold induction compared to the activity of promoter without NGF. Panels on right in (A) show immunostaining of control and NGF-treated PC12 cells with anti-MAP2 mAb. (B) Human teratocarcinoma cell line NT2/D1 (grown long term in medium containing serum) and HeLa cells were transfected with the indicated MAP2 promoter constructs and luciferase activity was measured as described above. Data are shown as RLA (RLA = 1). Error bars: means ± SEM. Inset in (B) shows RT–PCR analysis of MAP2 mRNA expression in HeLa and NT2/D1 cells. Total RNA (100 ng) was used to perform RT–PCR (OneStep RT–PCR kit; Qiagen) using primers that amplify a 709 bp fragment that spans human MAP2 exons 3–9. (C) Activity of phMAP2-2 in human primary and metastatic melanoma cell lines. Luciferase activity was measured and data (means ± SEM) are shown as RLA.
In NGF-treated PC12 cells, phMAP2-1 and phMAP2-2 promoter constructs showed 6- to 7-fold higher luciferase activity compared with untreated cells. Although promoter construct lacking sequences between −1854/−263 (phMAP2-3) showed ∼2-fold higher basal activity compared with phMAP2-1 and phMAP2-2 in untreated PC12 cells, NGF treatment did not produce a significant increase in the luciferase activity. In non-neuronal HeLa cells, all three promoter constructs produced only weak luciferase activity and treatment with NGF produced a 2-fold increase only in cells transfected with the 3.3 kb phMAP2-1 promoter construct (Figure 2A). We found that luciferase activity driven by the 3.3 kb MAP2-1 promoter was always lower than that of the shorter 2.2 kb MAP2-2 promoter fragment in every cell line tested, indicating the presence of negative regulatory sequences between nt −2965 and −1854. There are four putative E boxes (E1–E4) within this region of MAP2 DNA. To test whether these E boxes play a role in attenuating MAP2 promoter activity, we mutated E1, E2, E3 and E4 boxes individually or in various combinations including mutation of all four E boxes and tested the activity of the mutant promoters. Mutation of neither individual E boxes nor any combination of E boxes (including all four E boxes) altered the basal activity of the 3.3 kb phMAP2-1 promoter (data not shown). These data suggested that the distal E boxes do not contribute significantly to the basal activity of human MAP2 promoter and showed that the 2.2 kb genomic fragment −1854/+369 (phMAP2-2) was sufficient for maximal NGF-inducible, neuronal-selective MAP2 promoter activity.
In NT2 cells cultured in complete medium, phMAP2-2 produced 150- to 200-fold higher luciferase activity compared with the promoter-less pGL3-luc plasmid. Luciferase activity in transfected NT2 cells was 15- to 20-fold higher than that found in HeLa cells transfected with the same MAP2 promoter construct (Figure 2B) confirming the neuronal-specificity of the 2.2 kb MAP2 promoter. We then tested the activity of this MAP2 promoter in a representative panel of primary and metastatic melanoma cells lines, and human glioblastoma cell lines U138, U251 and a breast cancer cell line, MCF-7. In phMAP2-2 transfected melanoma cells, there was 20- to 30-fold higher luciferase activity compared to cells transfected with promoter-less pGL3-Luc (Figure 2C). Surprisingly, both primary and metastatic melanoma cells showed significant MAP2 promoter activity. Although MAP2 promoter activity in melanoma cells was lower than that found in NT2 cells, it was 4- to 6-fold higher than that observed in HeLa cells or MCF-7 cells (data not shown). MAP2 promoter activity was comparable to the activity in glioblastoma cell lines (data not shown). These data show that the neuronal-specific MAP2 gene promoter is active in melanoma cells and are consistent with the well-documented molecular plasticity and neural differentiation of cutaneous melanomas (28,29).
MAP2 promoter activity in vivo
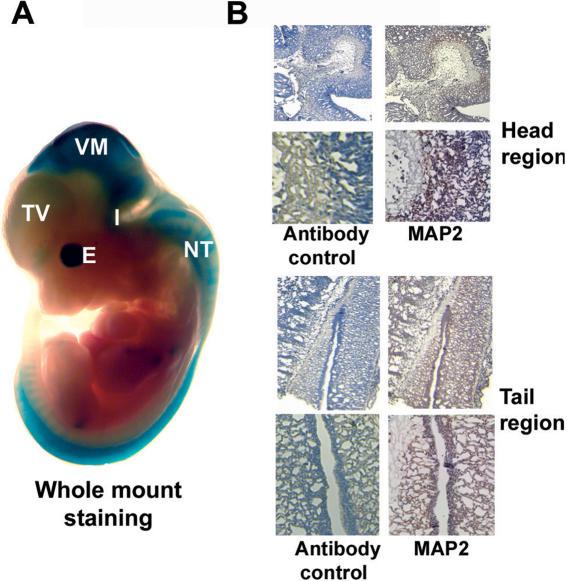
To verify that the cloned MAP2 genomic DNA fragments show neuronal-specific expression in vivo, we tested their activity by transient expression in transgenic mouse embryos. Promoter fragments hMAP2-1 and hMAP2-2 were cloned into β-galactosidase reporter construct pHHSF (21) and transgenic embryos were generated. Two independent sets of transgenic embryos were created for each construct. Staining of 11.5-day-old embryos with X-gal showed β-galactosidase activity in 6 of 11 and 8 of 25 embryos produced by microinjection of hMAP2-1 and hMAP2-2, respectively. The X-gal staining was restricted to developing central nervous system including a weak staining of telencephalic vesicle and strong staining of ventral mesencephalon, midbrain and hindbrain junction (isthmus), rhombic lip and along the length of the neural tube, developing eyes (Figure 3A). Both hMAP2-1 and hMAP2-2 transgenic embryos showed similar pattern of staining. Staining of sections of paraffin embedded, 11.5-day-old littermate control embryos with anti-MAP2ab-specific mAb M13 showed expression of endogenous MAP2 protein in the same regions corresponding to those where human MAP2 promoter driven β-galactosidase activity was expressed (Figure 3A and B). These data show that the 2.2 kb human MAP2 promoter also contains sequences necessary for neuronal-selective expression in vivo.
Figure 3.
Human MAP2 promoter activity in vivo. (A) Whole-mount in situ staining of 11.5-day-old pHSF-6/MAP2-1/lacZ promoter transgenic embryo with X-gal for β-galactosidase gene expression. The β-galactosidase expression is seen mostly in the developing central nervous system. TV, telencephalic vesicle; VM, ventral mesencephalon; I, isthmus (midbrain–hind brain junction) and rhombic lip; and NT, neural tube. Areas of the head and the tail shown in (B) are indicated by red colored squares. (B) Paraffin sections of head region and tail regions of 11.5-day-old littermate control mouse embryos were stained with anti-MAP2ab mAb M13 (Zymed) (right panels) or control IgG (left panels) using citrate buffer antigen retrieval method followed by anti-mouse IgG–HRP and AEC detection method. Endogenous MAP2ab protein expression is seen in regions of the embryo that correspond to the region of X-gal staining. Upper panels: low magnification; lower panels: high magnification of the areas indicated by red squares in the upper panels.
Regulation of MAP2 promoter
Since MAP2 is a marker of neuronal differentiation, we investigated the possibility that MAP2 promoter is regulated by the key neuronal transcription factors. NeuroD/BETA2 is a critical bHLH neurogenic factor that binds to a core 6 bp E box element (CANNTG) as a heterodimer with a ubiquitous partner E47 (30). HES1 is an important negative regulator of neurogenesis (24). HES1 represses neuronal genes actively by binding to N-box (CACNTG) as a homodimer or in a dominant-negative fashion by forming heterodimers with other bHLH factors that bind to E boxes (24,31).
A role of NeuroD in the regulation of MAP2 gene promoter is indicated by the following observations: (i) in neuroepithelial cells, induction of cell cycle arrest and neuronal differentiation is associated with the activation of NeuroD and MAP2 expression (32). (ii) MAP2 promoter contains a consensus NeuroD-binding E-box (CAGATG) found in other NeuroD target gene (33).
To test whether NeuroD activates MAP2 promoter, we transfected HeLa cells and melanoma cells with MAP2 promoter plasmids phMAP2-1 or phMAP2-2 and increasing amounts of NeuroD expression plasmid pNeuroD. As shown in Figure 4A, co-transfection with pNeuroD produced a dose-dependent increase in luciferase activity in both phMAP2-1 and phMAP2-2 promoter transfected melanoma cells. In HeLa cells, co-transfection of pNeuroD appears to suppress the MAP2 promoter activity at lower dose but stimulates it slightly at a higher dose. However, NeuroD-stimulated MAP2 promoter activity in HeLa cells was significantly lower than its basal activity seen in melanoma cells (Figure 4A).
Figure 4.
Role of NeuroD/BETA2 in regulation of MAP2 promoter. (A) HeLa cells and metastatic melanoma cells c22a were transfected with indicated MAP2 promoter–reporter plasmids and either 25 or 100 ng of NeuroD expression plasmid pcDNA3-NeuroD1-FLAG or 100 ng of pcDNA3. Equal amount of total DNA was used in all transfection. Luciferase activities were measured and RLA was calculated as described in the legend to Figure 1. Data shown are means ± SEM. (B) Electrophoretic mobility shift analysis of E-box sequence. 32P-labeled double-stranded 22mer oligonucleotides with core sequence of E9 box CAGATG (see Materials and Methods) were incubated with nuclear extract (5–10 μg protein) of c22a metastatic melanoma cells transfected with pNeuroD either alone or in the presence of 5–50 M excess cold E9, E10 or mutant E9 oligonucleotides. Lanes 8 and 9 show mobility shift pattern after the addition of control rabbit IgG (lane 8) or rabbit anti-NeuroD antibody (lane 9) to the E8 oligonucleotide–protein complexes.
Since the consensus NeuroD-binding E-box, E9 is conserved between human and rodent MAP2, we first tested binding of NeuroD to E9 box sequences by electrophoretic mobility gel shift assay. Incubation of nuclear extracts of pNeuroD transfected (to enhance the sensitivity of the assay) melanoma cells with 32P-labeled oligonucleotides (22mers with a core E-box sequence CANNTG) showed that the oligomer containing the E9 box (Figure 4B, lane 1), but not E10 or E11 box (data not shown), produce two DNA–protein complexes. Appearance of these complexes, especially the slower migrating complex, was inhibited in the presence of molar excess of wild-type cold E9 oligonucleotide (Figure 4B, lanes 2 and 3). Addition of molar excess of mutant E9 oligonucleotide with a core sequence ACGAGT (lanes 4 and 5) or wild-type E10 oligonucleotide (lanes 6 and 7) produced only a slight inhibition in the formation of slow migrating E9–protein complex. Addition of anti-NeuroD antibody (lane 9) to the reaction mixture completely prevented the formation of these complexes, similar to the reported effect of this antibody on binding of NeuroD to E boxes of other target genes (34,35). These data show that NeuroD binds to E9 box (−366/−345) of MAP2 promoter.
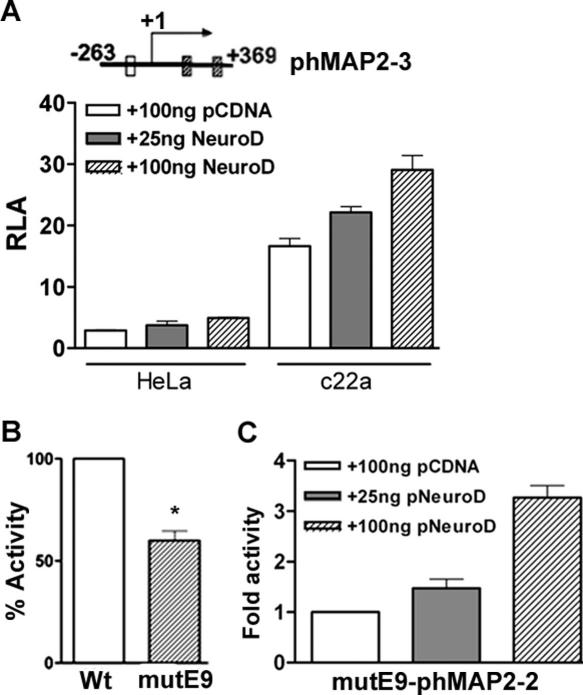
We observed that a MAP2 promoter deletion construct of pMAP2-3 (−263/+369) that lacks all proximal E boxes, including E9, still produced significant luciferase activity that was less than the maximal activity seen in cells transfected with the 2.2 kb phMAP2-2 promoter plasmid. To test whether this promoter constructs lacking the proximal E boxes could still be activated by NeuroD, we transfected HeLa cells and melanoma cells with phMAP2-3 plasmid and with increasing amounts of pNeuroD. Surprisingly, co-transfection with pNeuroD resulted in a dose-dependent increase in luciferase activity of this MAP2 promoter in melanoma cells but not in HeLa cells (Figure 5A). These data suggest that although NeuroD can activate MAP2 gene transcription by binding to E9 box, other factors that are constitutively expressed in melanoma cells and those that may be activated by NeuroD contribute to the transactivation of MAP2 promoter. To test this, we generated 2.2 kb phMAP2-2 promoter with mutations in the core sequence of E9 box (CAGATG→GCGAGG), and compared its basal activity with that of the wild-type promoter. As shown in Figure 5B, mutation of E9 box resulted in nearly 50% decrease in the basal luciferase activity. Additionally, this E9 mutant phMAP2-2 promoter continued to exhibit a dose-dependent activation by NeuroD (Figure 5C). Thus, NeuroD appears to upregulate MAP2 promoter activity by both direct (by binding to E9 box) and indirect mechanisms (presumably by activation or induction of other trans-activators).
Figure 5.
Indirect activation MAP2 promoter by NeuroD. (A) HeLa cells and metastatic melanoma cells c22a were co-transfected with phMAP2-3 promoter-reporter plasmid lacking the E9 box and NeuroD expression plasmid pcDNA3-NeuroD-FLAG (pNeuroD) (25 and 100 ng) or 100 ng of control pcDNA3 vector. (B) Effect of mutation of the core E9 box sequence (CAGATG→ACGAGT) on MAP2 promoter activity. Asterisk indicates P-value (Student's t-test, P < 0.001). (C) Dose-dependent activation of mutE9- phMAP2-2 promoter by NeuroD. c22a melanoma cells were co-transfected with mutE9 phMAP2-3 and increasing amount of pNeuroD plasmid. Equal amounts of total DNA were used in all transfections. Luciferase activities were measured and RLA (means ± SEM) was calculated as described in the legend to Figure 1.
MAP2 promoter is inhibited by HES1
As mentioned earlier, HES1 is an important regulator of neurogenesis and neuronal differentiation, Hes1 null mice express both early and late neuronal markers including MAP2 prematurely (36). To test whether MAP2 promoter is regulated by HES1, we co-transfected NT2 cells and melanoma cells (SK-MEL-19) with the 2.2 kb phMAP2-2 promoter and increasing amounts of mouse Hes1 expression plasmid pCI-Hes1. In both NT2 cells (data not shown) and melanoma cells, co-transfection with Hes1 resulted in a dose-dependent inhibition of luciferase activity with a maximal (84%) inhibition at the highest dose of pCI-Hes1 plasmid (Figure 6A). These data suggest a role for HES1 in the regulation of MAP2 promoter activity.
Figure 6.
HES1 (hairy and enhancer of split) represses MAP2 promoter activity. (A) Human metastatic melanoma cells were transfected with 2.2 kb phMAP2-2 promoter and increasing amounts (0.1, 0.3 and 0.6 μg) of mouse Hes1 expression plasmid pCI-Hes1 or 0.6 μg the empty vector pcDNA3. Forty-eight hours after transfection luciferase activities were measured. RLA were calculated and MAP2 promoter activity in Hes1 expressing cells is shown as percent (means ± SEM) of pcDNA3 co-transfected cells. (B) Mutational analysis of the role of N-boxes in repression of MAP2 promoter by Hes1. Metastatic melanoma cells were transfected with wild-type (wt) and N-box mutated (mutated nucleotides shown underlined) phMAP2-2 promoter plasmids mutN1 (ACCAGA), mutN2 (GCCGCC) and mutN1N2 and luciferase activity in cells transfected with mutant plasmids is shown as percent of wild-type transfected cells (*P < 0.05 compared with wt; one-way ANOVA). (C) Metastatic melanoma cells were transfected with wt and N-box mutant plasmids together with 1.5 μg empty vector pcDNA or pCI-Hes1 plasmid and luciferase activities in cell lysates was measured 48 h after the transfection. Repression of the wild-type and N-box mutant promoter activity by Hes1 is shown as percent activity of wild-type and the mutant plasmids in cells transfected with the pcDNA empty vector (*P < 0.05; **P < 0.01 compared with wt; one-way ANOVA). (D) Repression of the phMAP2-4 (−263/+30) promoter by Hes1. Melanoma cells in 24-well plate were transfected, in triplicates, with 0.6 μg phMAP2-4 plasmid and 0.6 μg of empty vector pcDNA or indicated amounts of pCI-Hes1 plasmid. After 48 h transfection, cells were lysed and RLA was determined. MAP2 promoter activity in the presence of Hes1 is shown as percent of empty vector transfected cells. Equal amount of total DNA was used in all transfection. Data (mean ± SEM) shown are representative of least three experiments. (E) Electrophoretic mobility shift analysis of N-box. 32P-labeled N2 box double-stranded oligonucleotides were incubated with nuclear extract (5–10 μg protein) of pCI-Hes1 transfected c22a metastatic melanoma cell either alone or in the presence of 5–50 M excess cold N1, N2 or mutant N2 box oligonucleotides or a polyclonal Hes1 antibody or a control rabbit IgG. Left arrow indicates the specific N2 box DNA–protein complex and the right arrow indicates the Hes1 supershifted complexes. Asterisks indicate non-specific complexes.
The proximal N-box is necessary for HES1-mediated repression
In phMAP2-2 promoter sequence, there are two putative HES-binding motifs: N1 at −961/−956 (CACAAG) and N2 at −63/−58 (CACGAG). To investigate the role of these N-boxes in repression of MAP2 promoter activity by HES1, we tested whether mutation of either one or both N-boxes increases basal activity of the promoter and/or makes it less sensitive or refractory to repression by HES1. In melanoma cells transfected with the mutant phMAP2-2 promoter plasmids, there was a modest but significant increase (26% over wild type) in the luciferase activity driven by the mutN2 (CACGAG→GCCGCC) and mutN1N2 promoters (P < 0.05), and a smaller increase (10% over wild type) in the activity driven by the mutN1 (CACAAG→ACCAGA) MAP2 promoter (Figure 6B). In addition, co-transfection with pCI-Hes1, which resulted in 60% inhibition of luciferase activity driven by the wild-type and mutN1 promoters, produced significantly less inhibition of mutN2 and mutN1N2 promoter activities (Figure 6C, P < 0.05 and P < 0.01, respectively). These data suggest that N2 box is involved in HES1-mediated repression of MAP2 promoter. In addition to its action by direct binding to N2 box, HES1 is also known to act indirectly by making non-functional heterodimers with transcription activators that bind to E boxes (37). Whereas the 2.2 kb MAP2 minimal promoter contains two potential HES-binding N-boxes, the 293 bp phMAP2-4 promoter deletion construct that we generated contains only the proximal N2 box. Transfection of melanoma cells with phMAP2-4 promoter-luciferase plasmid and Hes1 expression plasmid showed that Hes1 was able to repress the activity of phMAP2-4 promoter containing only the proximal N-box (Figure 6D), confirming that the regulation of MAP2 promoter by HES1 is mediated primarily by this proximal N2 box at −63/−58.
To test whether HES1 binds to N2 sequence, we performed electrophoretic mobility shift assay. As shown in Figure 6E, Hes1 transfected melanoma nuclear extracts retarded the mobility of N2 box oligonucleotides. Addition of a molar excess of cold wild-type N2 box DNA resulted in a complete inhibition of the mobility shift of the labeled N2 box oligonucleotide. In some experiments, we also observed a second, faster migrating and non-specific complex (Figure 6E, asterisk on the left), which could not be competed out by molar excess of cold N2 box DNA. The presence of molar excess wild-type N1 box or mutant N2 box oligonucleotides in the reaction resulted in only a slight decrease in the formation of this N2 DNA–protein complex (Figure 6E). The addition of anti-Hes1 antibody, but not control rabbit IgG, to the reaction caused further retardation of the N2 box–protein complex. However, addition of these antibodies to N2 DNA–protein complex showed two additional non-specific complexes (Figure 6E, asterisks on the right). These data showed that HES1 represses MAP2 promoter activity by binding preferentially to the proximal N2 box sequence.
HES1 is a dominant regulator of MAP2 promoter activity
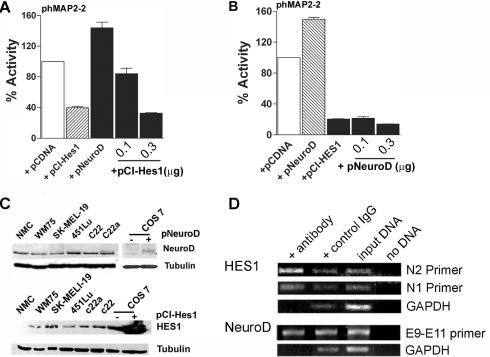
Neuronal differentiation is known to be regulated by the relative levels of pro-neuronal transcription factors and repressors neuronal-specific genes (38). To investigate the relative contribution of NeuroD and HES1 in the regulation of MAP2 promoter activity, we co-transfected melanoma cells with phMAP2-2 and (i) constant amount of NeuroD expression plasmid and increasing amounts of pCI-Hes1 plasmid or (ii) a constant amount of pCI-Hes1 plasmid and increasing amounts of pNeuroD plasmid. In melanoma cells, co-transfection with pNeuroD alone resulted in ∼50% increase in luciferase activity driven by phMAP2-2 promoter, and transfection of the promoter with pCI-Hes1 resulted in 60% inhibition in promoter activity. Transfection with increasing amounts of pCI-Hes1 along with constant amount of pNeuroD that produces 50% increase in promoter activity, not only prevented the pNeuroD-dependent increase but also produced a dose-dependent decrease in the promoter activity (Figure 7A). In contrast, co-transfection of increasing amounts of pNeuroD, which upregulated MAP2 promoter activity when present alone, was unable to relieve the inhibition of the promoter activity caused by HES1 (Figure 7B). These data suggest that HES1 is a dominant regulator of MAP2 promoter activity in human melanoma cells.
Figure 7.
HES1 is a dominant repressor of MAP2 promoter activity. (A) Melanoma cells were transfected with phMAP2-2 promoter-luciferase plasmid alone, or with Hes1 expression plasmid pCI-Hes1 or NeuroD expression plasmid pcDNA3-NeuroD-FLAG. In addition, 0.1 and 0.3 μg of Hes1 expression plasmid pCI-Hes1 was included in cells co-transfected with NeuroD. (B) c22a melanoma cells were transfected with phMAP2-2 promoter-luciferase plasmid alone, or with Hes1 expression plasmid pCI-Hes1 or NeuroD expression plasmid pcDNA3-NeuroD-FLAG. Cells in additional wells transfected with pCI-Hes1 also received 0.1 or 0.3 μg of pNeuroD plasmid. Equal amount of total DNA was used in all transfection. Luciferase activities were measured and the promoter activity in cells co-transfected with either pcDNA3-NeuroD or pCI-Hes1 alone or with increasing amounts of pCI-Hes1 or pcDNA3-NeuroD1, respectively, is shown as percentage (means ± SEM) of activity in cells transfected with phMAP2-2 and pcDNA. (C) Expression of NeuroD and HES1 in melanocytes and melanoma cells. Western blot analysis of NeuroD expression in control and pcDNA3-Neuro-FLAG plasmid transfected COS cells, neonatal foreskin melanocytes (NMC), primary melanoma cell line WM75 and various metastatic melanoma cell lines. Seventy-five micrograms of detergent soluble total cellular proteins were separated by SDS–PAGE and immunoblotted with polyclonal goat anti-mouse NeuroD (upper panel) and anti-mouse Hes1 antibody (lower panel), followed by HRP-conjugated secondary antibody. Protein bands were detected by chemiluminescence. Western blotting for α-tubulin is shown as a control for protein loading. (D) Chromatin immunoprecipitation assay. Sheared chromatin from melanoma cells was immunoprecipitated (IP) with appropriate antibody (anti-HES1, anti-NeuroD or control IgG) and antibody bound DNA was isolated according to the manufacturer's protocol (Active Motif, Carlsbad, CA). Immunoprecipitated DNA was used as template in PCR using N1 and N2 primer (for HES1), E9–E11 primer (for NeuroD). GAPDH primers were used as control.
To test whether melanoma cells express these neuronal transcription factors and whether variable MAP2 promoter activity in different melanoma cell lines might be related to relative levels of these factors, we performed western blot analysis with anti-NeuroD and anti-Hes1 antibodies. As shown in Figure 7C, whereas variable amounts of NeuroD expression could be detected in both melanocytes and melanoma cells, HES1 expression is detectable only in melanoma cells. These observations support the RT–PCR data reported by Balint et al. (39), who showed that HES family mRNAs (specifically HES1) are expressed in melanoma cell lines but not in melanocytes. There was no clear relationship between the levels of NeuroD and HES1 among different melanoma cell lines.
To test whether endogenous NeuroD and HES1 proteins in melanoma cells bind to MAP2 promoter sequence, we performed chromatin immunoprecipitation assays (Figure 7D). Sheared chromatin was immunoprecipitated with anti-NeuroD and anti-Hes1 antibodies or with control IgG, followed by PCR amplification of the corresponding regions of DNA using specific primers. Analysis of amplified DNA (Figure 7D) showed that immunoprecipitation with anti-Hes1 antibody enriched (compared to control IgG precipitation) sequences that are amplified preferentially by primers flanking N2 box compared to primers flanking N1 box. Amplification of anti-NeuroD immunoprecipitated chromatin did not show any marked enrichment of sequences amplified by primers flanking E9–E11 region. These data show that endogenous HES1 protein in melanoma cells binds to MAP2 promoter region, and suggest that HES1 acts as a dominant factor in regulating MAP2 promoter activity.
Role of Notch in the regulation of MAP2 promoter activity in melanoma cells
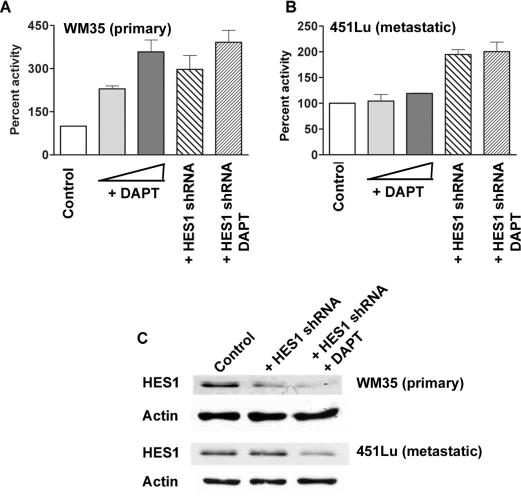
To test the effect of Notch1 signaling, a regulator of HES1 expression, on MAP2 promoter activity in melanoma cells, we treated primary and metastatic melanoma cells with γ-secretase inhibitor, DAPT (which inhibits the cleavage of intracellular domain of Notch1 and hence Notch1 signaling) and/or transfected with HES1shRNA. In primary melanoma cell line WM35, a dose-dependent increase in MAP2 promoter activity was observed with increasing dose of γ-secretase inhibitor (Figure 8A). Transfection with HES1shRNA or transfection followed by treatment with γ-secretase inhibitor also caused similar activation of MAP2 promoter. In contrast, treatment with γ-secretase inhibitor alone was not sufficient to activate MAP2 promoter in metastatic melanoma cell line 451Lu. Transfection with HES1shRNA alone or transfection with HES1 shRNA followed by treatment with γ-secretase inhibitor was able to upregulate MAP2 promoter activity in these cells (Figure 8B). Treatment with DMSO or transfection with control shRNA did not significantly affect MAP2 promoter activity and were used as controls. Western blot analysis showed that transfection with HES1shRNA alone or treatment of shRNA transfected cells with DAPT decreased the level of HES1 more effectively in primary melanoma cells than in metastatic cells (Figure 8C). Although we did not observe a marked reduction in HES1 levels in shRNA transfected 451Lu cells, there was ∼2-fold increase in MAP2 promoter activity indicating that MAP2 promoter may be sensitive to small changes in HES1 levels. These data suggest that primary melanoma cells are more sensitive to Notch1 inhibition and MAP2 promoter upregulation than metastatic melanoma cells and are consistent with earlier published observations (14).
Figure 8.
Notch1 signaling is involved in MAP2 promoter regulation in melanoma cells. Primary melanoma cells, WM35 (A) and metastatic melanoma cells, 451Lu (B) were either pre-treated with 1 or 2 μM γ-secretase inhibitor (DAPT) or transfected with 0.6 μg of HES1shRNA and then treated with 2 μM DAPT. After 24 h, the cells were transfected with phMAP2-2 luciferase-promoter construct. Cells were lysed and luciferase activity was measured 48 h after promoter transfection. RLA was calculated as described in the legend to Figure 2. Data are shown as percent activity of control DMSO-treated cells or control shRNA transfected cells, and are from a representative of two individual experiments with triplicates. (C) Western blotting analysis of primary (WM35) and metastatic (451Lu) melanoma cells transfected with HES1shRNA alone or in combination with the treatment of DAPT, show decreased level of endogenous HES1 protein. Western blotting of β-actin is shown as loading control.
DISCUSSION
Human cutaneous melanoma is known to exhibit molecular plasticity and often express genes characteristic of neural cell lineages (11,28). Our observation that MAP2 is activated in vivo during early stages of melanoma tumor progression, and that NeuroD can upregulate MAP2 promoter activity preferentially in melanoma cells in vitro are consistent with a well-documented tendency of melanoma, a tumor that originates from the neural crest-derived melanocytes, to exhibit neural differentiation. It is now becoming clear that transdifferentiation of melanoma can alter the biology of the tumor and, hence, the clinical outcome in patients. We showed that aggressiveness of primary melanoma in vivo is inversely correlated with MAP2 expression and that overexpression of MAP2 in metastatic melanoma cells in vitro results in cell cycle arrest and growth inhibition (12). In these studies aimed at understanding the mechanisms involved in the activation of MAP2 gene in melanoma cells, we identified the 5′ regulatory region of human MAP2 gene and show that MAP2 promoter is regulated by neuronal bHLH factors NeuroD and HES1, and HES1 acts as a dominant regulator of MAP2.
Human MAP2 promoter
Transcriptional regulation of genes that encode members of the microtubule-associated proteins, MAP1A, MAP1B and tau, has been investigated (40–44). Interestingly, promoter sequence of MAP2 gene does not share any common features with other MAP genes. Our data show that MAP2 promoter that supports tissue-specific activity both in vitro and in vivo contains multiple bHLH binding sites. We also show that MAP2 promoter is regulated by the neuronal bHLH transcription factors consistent with the wide use of MAP2 expression as marker for neuronal differentiation. Neuron-specific activity of promoters of other MAP genes is less well defined. For example, a 2.4 kb 5′ region of mouse MAP1A, which contains two TATA-less promoters and one Sp1 site that regulate the expression of two alternative transcripts, was shown to be active in both neuronal and non-neuronal cells. Rat and human MAP1B promoter sequences, however, contain a consensus element known as ‘neuronal motif’ (common to several neuronal genes such as GAP43, perinephrin and neurofilament), a Sp1 site, a TCC repeat and a cAMP response element. However, none of these elements appears to be necessary for neuron-specific MAP1B promoter activity (42). Similarly, although promoters for mouse, rat and human tau genes have been isolated, no consensus elements for neuron-specific expression were defined.
A distinguishing feature of MAP2 promoter is the presence of multiple E boxes, which are known to be recognized by bHLH factors including those that participate in the regulation of cell and tissue-specific gene expression. Presence of E boxes is consistent with neuronal differentiation-specific expression of MAP2. Among the E boxes in the MAP2 promoter, a cluster of three proximal E boxes (E9–E11) are of particular interest because (i) they are strictly conserved among mouse, rat and human, including their relative distance from the transcription start site and spacing between the boxes and (ii) the distal E-box (E9) within this composite element contains a NeuroD-binding sequence CAGATG, identical to that found in well-characterized NeuroD target gene rat glucagon (33). Other known NeuroD target genes that also contain a cluster of three or four E boxes in their proximal promoter regions are insulin and insulinoma-associated antigen 1, Pax6 and sulfonylurea receptor I (20,30,35,45). A comparison of NeuroD-binding E boxes of these genes (including MAP2) showed that NeuroD preferentially binds to the sequence CAN(A/G/T)NTG. Other E-box-like elements (CACNTG) found within this cluster do not appear to be functional with respect to NeuroD (33,45). Mutational analysis of E9, E10 and E11 boxes of MAP2 promoter and gel-shift data presented in this study also support these earlier published observations.
Neuronal differentiation is known to be regulated not only by transcriptional activators but also by HES and Id family of repressors (46–48). Among these repressors, the role of HES1 has been extensively investigated. For example, neuronal precursor cells infected with HES1-transducing retrovirus do not differentiate into neurons and fail to activate expression of neuronal markers (49), and Hes1 null mice express both early and late neuronal markers, including MAP2, prematurely (36). HES family repressors can inhibit gene expression either directly by binding (as homodimers) to their cognate-binding elements known as N-boxes (CACNAG) or indirectly by dominant-negative regulation (by forming non-functional heterodimers with transcriptional activators). To our knowledge, rat and human achaete-scute homolog-1 (MASH1), HES1 and E2F1 are the only genes shown to be directly repressed by binding of HES1 to the respective promoters (50–53). In HES1 proximal promoter there are three N-boxes and all three have been shown to bind HES1 and contribute in the autoregulation of HES1 expression (51). E2F1 promoter has a single N-box and binding of HES1 to this element appears to inhibit estrogen and heregulin-mediated upregulation of E2F1 in breast cancer cells (52). On the other hand, hASH1 promoter contains a single HES1-binding class C site with a core sequence CACGCA (50). In human MAP2 promoter, we identified two putative N-boxes, a distal CACAAG (N1) and a proximal CACGAG (N2) box. We showed, by mutational and gel-shift analysis, that the proximal N2 box is involved in HES1-mediated regulation of the promoter activity. Interestingly, these N-boxes are not present in the mouse and rat MAP2 promoters, although a N-box-like CAGGAG element, conserved between the mouse and the rat MAP2, is found at a position similar to that of human MAP2 promoter. However, the role of this N-box-like sequence in the regulation of MAP2 expression in these species remains to be investigated.
Our data show that MAP2 gene contains both positive (E boxes) and negative (N-boxes) regulatory elements and suggest that MAP2 expression might be regulated by a balance between positive and negative bHLH factors NeuroD and HES1. This is consistent with the proposal that transition of neuronal precursor cells from proliferation to neurogenesis involves coordinate increase in the pro-neural bHLH factors and a decrease in the activity of HES and Id factors (38). It is tempting to speculate that in vivo MAP2 gene might be regulated by a balance between NeuroD and HES1. For example, in rat insulinoma InR1G9 cell line, insulin gene (a NeuroD target) has been shown to be regulated by the ratio of NeuroD and the transcription repressor Id1 (33). It is therefore possible that variable levels of MAP2 promoter activity in melanoma cell lines may be due to variable ratio of NeuroD and HES1, as both factors are expressed in all melanoma cell lines tested. In this context, it is of interest to note that whereas transfection with increasing amounts of Hes1 plasmid produced a dose-dependent decrease of NeuroD activation of MAP2 promoter, transfection with increasing amount of NeuroD was unable to relieve the repression of the promoter by Hes1. Thus, MAP2 promoter activity seems to be regulated predominantly by HES1 levels. This possibility is also supported by the results of the ChIP assay, which showed that in melanoma cells N-box region of MAP2 gene promoter is occupied by endogenous HES1 protein. Further, reducing endogenous HES1 protein level in melanoma cells using HES1shRNA was also able to increase the basal activity of MAP2 promoter.
It is also relevant to note that Notch1 signaling, which activates HES1 gene expression, has been shown to be elevated in melanoma cells (54,55). It has also been shown that the activation of Notch1 signaling is required for β-catenin-mediated human primary melanoma progression and that primary melanoma cells are more sensitive to Notch1 inhibition than metastatic cells (39). Our data, using Notch1 inhibitor, indicate an involvement of Notch1 signaling via HES1 in the regulation of MAP2 gene expression. In agreement with published data (15), we found that primary melanoma cells are more responsive than metastatic cells to inhibition of Notch1 signaling and MAP2 promoter upregulation. These data are also consistent with our earlier findings that MAP2 is expressed more frequently in primary lesions (11,12). Thus, blocking of Notch1 signaling and/or HES1 may be considered as useful therapeutic strategy for melanoma treatment. However, our preliminary findings on induction of endogenous MAP2 gene expression suggest that in addition to regulation by Notch signaling, MAP2 gene is also regulated by epigenetic mechanisms (Nityanand Maddodi, Kumar M. R. Bhat and Vijayasaradhi Setaluri, unpublished data), indicating a tight regulation of this gene during melanoma progression.
In summary, our findings on the regulation of neuronal MAP2 promoter gene in melanoma cells are relevant to understanding molecular mechanisms of melanoma progression. A more detailed understanding of the regulation of MAP2 gene transcription in melanoma cells might allow us to design and develop strategies that can activate this gene as a novel therapeutic approach for malignant melanoma.
Acknowledgments
We thank Dr Kageyama for providing pCI-Hes1 construct and anti-Hes1 antibody, Dr Jan for providing pcDNA-NeuroD1-FLAG construct, and Dr Suh-Kim for providing NeuroD antibody. We also thank Ms. Smita Ramprakash, Ms. Namratha Sangha and Dr Pichardo for valuable technical help, and Drs Rajendra Kedlaya and Akihiro Ikeda for comments and suggestions. Funding to pay the Open Access publication charges for this article was provided by the Department of Dermatology, University of Wisconsin, Madison, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fraichard A., Chassande O., Bilbaut G., Dehay C., Savatier P., Samarut J. In vitro differentiation of embryonic stem cells into glial cells and functional neurons. J. Cell. Sci. 1995;108:3181–3188. doi: 10.1242/jcs.108.10.3181. [DOI] [PubMed] [Google Scholar]
- 2.Megiorni F., Mora B., Indovina P., Mazzilli M.C. Expression of neuronal markers during NTera2/cloneD1 differentiation by cell aggregation method. Neurosci. Lett. 2005;373:105–109. doi: 10.1016/j.neulet.2004.09.070. [DOI] [PubMed] [Google Scholar]
- 3.Caceres A., Mautino J., Kosik K.S. Suppression of MAP2 in cultured cerebellar macroneurons inhibits minor neurite formation. Neuron. 1992;9:607–618. doi: 10.1016/0896-6273(92)90025-9. [DOI] [PubMed] [Google Scholar]
- 4.Dehmelt L., Smart F.M., Ozer R.S., Halpain S. The role of microtubule-associated protein 2c in the reorganization of microtubules and lamellipodia during neurite initiation. J. Neurosci. 2003;23:9479–9490. doi: 10.1523/JNEUROSCI.23-29-09479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada A., Teng J., Takei Y., Oguchi K., Hirokawa N. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J. Cell Biol. 2002;158:541–549. doi: 10.1083/jcb.200110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn J., Meissner C., Oehmichen M. Microtubule-associated protein 2 (MAP2)—a promising approach to diagnosis of forensic types of hypoxia–ischemia. Acta Neuropathol. 2005;110:579–586. doi: 10.1007/s00401-005-1090-9. [DOI] [PubMed] [Google Scholar]
- 7.Blumcke I., Muller S., Buslei R., Riederer B.M., Wiestler O.D. Microtubule-associated protein-2 immunoreactivity: a useful tool in the differential diagnosis of low-grade neuroepithelial tumors. Acta Neuropathol. 2004;108:89–96. doi: 10.1007/s00401-004-0873-8. [DOI] [PubMed] [Google Scholar]
- 8.Lopes M.B.S., Altermatt H.J., Scheithauer B.W., Shepherd C.W., VandenBerg S.R. Immunohistochemical characterization of subependymal giant cell astrocytomas. Acta Neuropathol. 1996;91:368–375. doi: 10.1007/s004010050438. [DOI] [PubMed] [Google Scholar]
- 9.Leung M.F., Sokoloski J.A., Sartorelli A.C. Changes in microtubules, microtubule-associated proteins, and intermediate filaments during the differentiation of HL-60 leukemia cells. Cancer Res. 1992;52:949–954. [PubMed] [Google Scholar]
- 10.Veitia R., David S., Barbier P., Vantard M., Gounon P., Bissery M.C., Fellous A. Proteolysis of microtubule associated protein 2 and sensitivity of pancreatic tumours to docetaxel. Br. J. Cancer. 2000;83:544–549. doi: 10.1054/bjoc.2000.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang D., Hallman J., Sangha N., Kute T.E., Hammarback J.A., White W.L., Setaluri V. Expression of microtubule-associated protein 2 in benign and malignant melanocytes: implications for differentiation and progression of cutaneous melanoma. Am. J. Pathol. 2001;158:2107–2115. doi: 10.1016/S0002-9440(10)64682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soltani M.H., Pichardo R., Song Z., Sangha N., Camacho F., Satyamoorthy K., Sangueza O.P., Setaluri V. Microtubule-associated protein 2, a marker of neuronal differentiation, induces mitotic defects, inhibits growth of melanoma cells, and predicts metastatic potential of cutaneous melanoma. Am. J. Pathol. 2005;166:1841–1850. doi: 10.1016/S0002-9440(10)62493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoek K., Rimm D.L., Williams K.R., Zhao H., Ariyan S., Lin A., Kluger H.M., Berger A.J., Cheng E., Trombetta E.S., et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 14.Balint K., Xiao M., Pinnix C.C., Soma A., Veres I., Juhasz I., Brown E.J., Capobianco A.J., Herlyn M., Liu Z.-J. Activation of Notch1 signaling is required for {beta}-catenin-mediated human primary melanoma progression. J. Clin. Invest. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z.-J., Xiao M., Balint K., Smalley K.S.M., Brafford P., Qiu R., Pinnix C.C., Li X., Herlyn M. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 2006;66:4182–4190. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- 16.Fang D., Tsuji Y., Setaluri V. Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF. Nucleic Acids Res. 2002;30:3096–3106. doi: 10.1093/nar/gkf424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer I., Richter-Landsberg C., Safaei R. Regulation of microtubule associated protein 2 (MAP2) expression by nerve growth factor in PC12 cells. Exp. Cell Res. 1991;194:195–201. doi: 10.1016/0014-4827(91)90354-w. [DOI] [PubMed] [Google Scholar]
- 18.Bai S., Ghoshal K., Datta J., Majumder S., Yoon S.O., Jacob S.T. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol. Cell. Biol. 2005;25:751–766. doi: 10.1128/MCB.25.2.751-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Ito T., Udaka N., Yazawa T., Okudela K., Hayashi H., Sudo T., Guillemot F., Kageyama R., Kitamura H. Basic helix–loop–helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127:3913–3921. doi: 10.1242/dev.127.18.3913. [DOI] [PubMed] [Google Scholar]
- 20.Kim J.-W., Seghers V., Cho J.-H., Kang Y., Kim S., Ryu Y., Baek K., Aguilar-Bryan L., Lee Y.-D., Bryan J., et al. Transactivation of the mouse sulfonylurea receptor I gene by BETA2/NeuroD. Mol. Endocrinol. 2002;16:1097–1107. doi: 10.1210/mend.16.5.0934. [DOI] [PubMed] [Google Scholar]
- 21.Shashikant C., Bieberich C., Belting H., Wang J., Borbely M., Ruddle F. Regulation of Hoxc-8 during mouse embryonic development: identification and characterization of critical elements involved in early neural tube expression. Development. 1995;121:4339–4347. doi: 10.1242/dev.121.12.4339. [DOI] [PubMed] [Google Scholar]
- 22.Norton A.J., Jordan S., Yeomans P. Brief, high-temperature heat denaturation (pressure cooking): a simple and effective method of antigen retrieval for routinely processed tissues. J. Pathol. 1994;173:371–379. doi: 10.1002/path.1711730413. [DOI] [PubMed] [Google Scholar]
- 23.Jennings B.H., Tyler D.M., Bray S.J. Target specificities of Drosophila enhancer of split basic helix–loop–helix proteins. Mol. Cell. Biol. 1999;19:4600–4610. doi: 10.1128/mcb.19.7.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kageyama R., Ishibashi M., Takebayashi K., Tomita K. bHLH transcription factors and mammalian neuronal differentiation. Int. J. Biochem. Cell Biol. 1997;29:1389–1399. doi: 10.1016/s1357-2725(97)89968-2. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J., Liu C., Koopman W., Mountz J. Characterization of human Fas gene. Exon/intron organization and promoter region. J. Immunol. 1995;154:1239–1245. [PubMed] [Google Scholar]
- 26.Sobocki T., Jayman F., Sobocka M.B., Duchatellier R., Banerjee P. Isolation, sequencing, and functional analysis of the TATA-less human ATPase II promoter. BBA—Gene Struct. Expr. 2005;1728:186–198. doi: 10.1016/j.bbaexp.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Pleasure S., Page C., Lee V. Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J. Neurosci. 1992;12:1802–1815. doi: 10.1523/JNEUROSCI.12-05-01802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed J.A., Finnerty B., Albino A.P. Divergent cellular differentiation pathways during the invasive stage of cutaneous malignant melanoma progression. Am. J. Pathol. 1999;155:549–555. doi: 10.1016/S0002-9440(10)65150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrix M.J.C., Seftor E.A., Hess A.R., Seftor R.E.B. Molecular plasticity of human melanoma cells. Oncogene. 2003;22:3070–3075. doi: 10.1038/sj.onc.1206447. [DOI] [PubMed] [Google Scholar]
- 30.Naya F.J., Stellrecht C.M., Tsai M.J. Tissue-specific regulation of the insulin gene by a novel basic helix–loop–helix transcription factor. Gene Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 31.Ohsako S., Hyer J., Panganiban G., Oliver I., Caudy M. Hairy function as a DNA-binding helix–loop–helix repressor of Drosophila sensory organ formation. Gene. Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 32.Jung C.-G., Kim H.-J., Kawaguchi M., Khanna K.K., Hida H., Asai K., Nishino H., Miura Y. Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development. 2005;132:5137–5145. doi: 10.1242/dev.02098. [DOI] [PubMed] [Google Scholar]
- 33.Dumonteil E., Laser B., Constant I., Philippe J. Differential regulation of the glucagon and insulin I gene promoters by the basic helix–loop–helix transcription factors E47 and BETA2. J. Biol. Chem. 1998;273:19945–19954. doi: 10.1074/jbc.273.32.19945. [DOI] [PubMed] [Google Scholar]
- 34.Kim J.-Y., Chu K., Kim H.-J., Seong H.-A., Park K.-C., Sanyal S., Takeda J., Ha H., Shong M., Tsai M.-J., et al. Orphan nuclear receptor small heterodimer partner, a novel corepressor for a basic helix–loop–helix transcription factor BETA2/NeuroD. Mol. Endocrinol. 2004;18:776–790. doi: 10.1210/me.2003-0311. [DOI] [PubMed] [Google Scholar]
- 35.Marsich E., Vetere A., Di Piazza M., Tell G., Paoletti S. The PAX6 gene is activated by the basic helix–loop–helix transcription factor NeuroD/BETA2. Biochem. J. 2003;376:707–715. doi: 10.1042/BJ20031021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura Y., Sakakibara S.-I., Miyata T., Ogawa M., Shimazaki T., Weiss S., Kageyama R., Okano H. The bHLH gene Hes1 as a repressor of the neuronal commitment of CNS stem cells. J. Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. Two mammalian helix–loop–helix factors structurally related to Drosophila hairy and enhancer of split. Gene Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 38.Ross S.E., Greenberg M.E., Stiles C.D. Basic helix–loop–helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 39.Balint K., Xiao M., Pinnix C.C., Soma A., Veres I., Juhasz I., Brown E.J., Capobianco A.J., Herlyn M., Liu Z.-J. Activation of Notch1 signaling is required for {beta}-catenin-mediated human primary melanoma progression. J. Clin. Invest. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama A., Odajima T., Murakami H., Mori N., Takahashi M. Characterization of two promoters that regulate alternative transcripts in the microtubule-associated protein (MAP) 1A gene. BBA—Gene Struct. Expr. 2001;1518:260–266. doi: 10.1016/s0167-4781(01)00173-7. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama A., Murakami H., Maeyama N., Yamashiro N., Sakakibara A., Mori N., Takahashi M. Role for RFX transcription factors in non-neuronal cell-specific inactivation of the microtubule-associated protein MAP1A promoter. J. Biol. Chem. 2003;278:233–240. doi: 10.1074/jbc.M209574200. [DOI] [PubMed] [Google Scholar]
- 42.Liu D., Fischer I. Two alternative promoters direct neuron-specific expression of the rat microtubule-associated protein 1B gene. J. Neurosci. 1996;16:5026–5036. doi: 10.1523/JNEUROSCI.16-16-05026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu D., Fischer I. Structural analysis of the proximal region of the microtubule-associated protein 1B promoter. J. Neurochem. 1997;69:910–919. doi: 10.1046/j.1471-4159.1997.69030910.x. [DOI] [PubMed] [Google Scholar]
- 44.Gao L., Tucker K.L., Andreadis A. Transcriptional regulation of the mouse microtubule-associated protein tau. BBA—Gene Struct. Expr. 2005;1681:175–181. doi: 10.1016/j.bbaexp.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Breslin M.B., Zhu M., Lan M.S. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J. Biol. Chem. 2003;278:38991–38997. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kageyama R., Sasai Y., Akazawa C., Ishibashi M., Takebayashi K., Shimizu C., Tomita K., Nakanishi S. Regulation of mammalian neural development by helix–loop–helix transcription factors. Crit. Rev. Neurobiol. 1995;9:177–188. [PubMed] [Google Scholar]
- 47.Lee J. Basic helix–loop–helix genes in neural development. Curr. Opin. Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 48.Ghil S.H., Jeon Y.J., Suh-Kim H. Inhibition of BETA2/NeuroD by Id2. Exp. Mol. Med. 2002;34:367–373. doi: 10.1038/emm.2002.52. [DOI] [PubMed] [Google Scholar]
- 49.Ishibashi M., Moriyoshi K., Sasai Y., Shiota K., Nakanishi S., Kageyama R. Persistent expression of helix–loop–helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H., Thiagalingam A., Chopra H., Borges M.W., Feder J.N., Nelkin B.D., Baylin S.B., Ball D.W. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc. Natl Acad. Sci. USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takebayashi K., Sasai Y., Sakai Y., Watanabe T., Nakanishi S., Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix–loop–helix factor HES-1. Negative autoregulation through the multiple N box elements. J. Biol. Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 52.Hartman J., Müller P., Foster J.S., Wimalasena J., Gustafsson J.A., Ström A. HES-1 inhibits 17-estradiol and heregulin-1-mediated upregulation of E2F-1. Oncogene. 2004;23:8826–8833. doi: 10.1038/sj.onc.1208139. [DOI] [PubMed] [Google Scholar]
- 53.Ju B.-G., Solum D., Song E.J., Lee K.-J., Rose D.W., Glass C.K., Rosenfeld M.G. Activating the PARP-1 sensor component of the Groucho/TLE1 corepressor complex mediates a CaMKinase II[delta]-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 54.Nickoloff B.J., Osborne B.A., Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22:6598–6608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 55.Hendrix M.J.C., Seftor R.E.B., Seftor E.A., Gruman L.M., Lee L.M.L., Nickoloff B.J., Miele L., Sheriff D.D., Schatteman G.C. Transendothelial function of human metastatic melanoma cells: role of the microenvironment in cell-fate determination. Cancer Res. 2002;62:665–668. [PubMed] [Google Scholar]