Abstract
Human individuals differ from one another at only ∼0.1% of nucleotide positions, but these single nucleotide differences account for most heritable phenotypic variation. Large-scale efforts to discover and genotype human variation have been limited to common polymorphisms. However, these efforts overlook rare nucleotide changes that may contribute to phenotypic diversity and genetic disorders, including cancer. Thus, there is an increasing need for high-throughput methods to robustly detect rare nucleotide differences. Toward this end, we have adapted the mismatch discovery method known as Ecotilling for the discovery of human single nucleotide polymorphisms. To increase throughput and reduce costs, we developed a universal primer strategy and implemented algorithms for automated band detection. Ecotilling was validated by screening 90 human DNA samples for nucleotide changes in 5 gene targets and by comparing results to public resequencing data. To increase throughput for discovery of rare alleles, we pooled samples 8-fold and found Ecotilling to be efficient relative to resequencing, with a false negative rate of 5% and a false discovery rate of 4%. We identified 28 new rare alleles, including some that are predicted to damage protein function. The detection of rare damaging mutations has implications for models of human disease.
INTRODUCTION
Nucleotide variation is a major source of heritable phenotypic change. Recent advances in the discovery and cataloguing of single nucleotide polymorphisms (SNPs) promise to broaden our understanding of phenotypic differences in human populations (1,2). It is hoped that knowledge of the nucleotide variation throughout a genome will allow for more effective drug development and treatment of diseases (3). Most nucleotide variation is in the form of SNPs, which are considered common if they are found in the population at a frequency >5%. Those that fall below this threshold are classified as rare, although on a cumulative basis, rare SNPs account for a large fraction of polymorphisms in a population (1). There has been some debate as to the usefulness of either common or rare alleles for the characterization and identification of the causative agent(s) of disease [e.g. (4–8)]. Although the debate continues, it is clear that rare variants can contribute to complex disease (9–12), and their potential importance has initiated a call for studies directed specifically towards the discovery of rare variants (13). In contrast, current efforts to catalogue human nucleotide diversity have largely focused on the identification of common variants. This focus may in part be driven by available technologies, especially Sanger dideoxy sequencing, which is suitable for the discovery of common nucleotide differences but becomes increasingly inefficient and costly as the frequency of a variant falls below ∼5%. This means that alternative technologies are needed to detect rare variants in a high-throughput and cost-effective manner.
There is also an urgent need for technologies to reduce the cost of discovering new mutations, such as those that occur in cancer. The National Institutes of Health has announced recently a $100 million pilot project, The Cancer Genome Atlas, which aims to use large-scale genomic sequencing to discover mutations that are important in different cancers. For this project to succeed, it will be essential to greatly increase the efficiency and accuracy of detection of rare mutations because the density of somatic mutations in several cancers has been estimated to be on the order of ∼1/1 000 000 bp (14–16). The majority of new mutations are likely to be heterozygous; thus, the high levels of quality and redundancy needed to achieve sufficient detection accuracy for this project makes it potentially far more expensive than standard genomic sequencing performed by high-throughput facilities. Therefore, a pre-screening method for discovering rare mutations will allow sequencing centers to reduce the number of sequence reads many-fold, with approximately proportional cost-savings.
Here we describe the application of a practical method that has the potential of addressing these needs. Previously, we developed TILLING (Targeting Induced Local Lesions IN Genomes) for high-throughput and low-cost discovery of chemically induced mutations in genomes (17,18). We later modified the method for the discovery and genotyping of natural nucleotide diversity, termed ‘Ecotilling’, because the method was first used to study diversity in accessions of Arabidopsis thaliana known as ecotypes (19). TILLING and Ecotilling are based on a common set of methods. Target fragments of ∼1.5 kb are amplified by PCR with gene-specific oligonucleotide primers that are 5′-end-labeled with fluorescent IRDye 700 or IRDye 800 dyes. After amplification, samples are denatured and then annealed to form heteroduplexes between strands of DNA harboring nucleotide polymorphisms. Heteroduplexes are digested using a single-strand specific nuclease and then size-fractionated by denaturing PAGE (20).
In this study, we have applied the Ecotilling method to the screening of human DNA samples. We screened 90 samples from the human polymorphism discovery resource (PDR) panel (21) for nucleotide changes in five target gene fragments that were subjected previously to resequencing as part of a large-scale effort. After blindly scoring the Ecotilling data, we compared the results to the public resequencing data and to sequencing data collected in our own laboratory. From this, we estimate error rates that are low relative to those achieved by resequencing technologies. To increase the efficiency of discovery of rare alleles and to further reduce errors, we applied an 8-fold pooling and 2D arraying strategy that resulted in the discovery of many rare SNPs, some of which are predicted to damage the encoded protein. We also developed a universal primer strategy that dramatically reduces Ecotilling costs. Finally, we developed a set of algorithms for automated detection and scoring of SNPs and incorporated these into the GelBuddy interactive automated band-mapping program (22).
MATERIALS AND METHODS
Sample arraying and Ecotilling
DNA samples from the human PDR were obtained from Dr Stephen Tapscott in preparation for a survey of SNPs in the neuroD2 gene (23). For unpooled Ecotilling, the first 96 samples from the PDR (samples PD0001–PD0096) were arrayed into individual wells of a 96-well plate and sample concentrations were adjusted to 0.875 ng/μl in TE [10 mM Tris–HCl and 1 mM EDTA, pH 7.4]. Before pooling samples, the concentration of each DNA was verified on a 1.5% agarose gel using lambda DNA as a concentration reference (Invitrogen, Carlsbad, CA, USA). Samples were arrayed in an 8 × 8 grid and equal volumes of samples within a row and within a column were combined to create the 8-fold pool (Figure 2A). Samples PD0001 to PD0384 were used to make pools. Gene targets were selected from the National Institute of Environmental Health Sciences (NIEHS) SNPs finished genes directory (http://egp.gs.washington.edu/directory.html). Primers (Supplementary Table 1) were selected using the CODDLe and Primer3 programs as described previously (24). Forward and reverse primers, both unlabeled and 5′-end-labeled with either IRD700 dye (forward) or IRD800 dye (reverse) were ordered from MWG Biotech. PCR amplification, nuclease digestion of heteroduplexes and polyacrylamide gel analysis were performed as described previously (20) with the following exceptions: 10 μl PCRs were performed with 4.4 ng of genomic DNA and 1 unit of celery juice extract was used per reaction in place of CEL 1 nuclease (25).
Universal priming
Dye-labeled universal primers complementary to bacteriophage T3 (5′-IRD700-ATTAACCCTCACTAAAGGG-3′) and T7 (5′-IRD800-AATACGACTCACTATAGGG-3′) promoters were used, because tests showed no amplification products when used in PCRs with human DNA (data not shown). Gene-specific primer sequences were synthesized 3′ of T3 (forward) or T7 (reverse) sequences (Supplementary Table 1). Nucleotides from the 3′ ends of standard Tm = 70°C gene-specific primers were excluded from the primer design to generate a gene-specific primer sequence of Tm = ∼60°C. PCR was performed in a final volume of 10 μl. Reaction mixes were as described previously (20) with the following exceptions: 60 nmol of each unlabeled gene primer, 300 nmol of each universal IRD-labeled primer and 175 ng of genomic DNA were used per reaction. Samples were amplified using the following parameters: 95°C for 2 min; loop 1 for 8 cycles 94°C for 20 s, 58°C for 30 s, reduce temperature 1°C per cycle, ramp to 72°C at 0.5°C/s, 72°C for 1 min); loop 2 for 45 cycles (94°C for 20 s, 50°C for 30 s, ramp to 72°C at 0.5°C/s, 72°C for 1 min); 72°C for 5 min; 99°C for 10 min; loop 3 for 70 cycles (70°C for 20 s, reduce temperature 0.3°C per cycle); hold at 8°C. Nuclease digestion and polyacrylamide gel analysis were performed similar to the method using IRD-labeled gene-specific primers.
Gel image and sequence analysis
Ecotilling gels were analyzed using the program GelBuddy in manual mode (22). Analysis was performed without knowledge of sequencing data. Nucleotide changes identified by Ecotilling were verified by sequencing using an ABI 3730 sequencer and Sequencher analysis software as described previously (19). Public sequence data were obtained from http://egp.gs.washington.edu and manually compared to lab-derived sequence and Ecotilling data.
Automated signal detection
The image processing algorithm takes as input a pair of 8-bit or 16-bit grayscale images, the length ltotal of the full PCR product, a set of lane tracks and de-smiling curves constructed as in Ref. (22), with de-smiling curves at 200 bp, 700 bp, and full-length (ltotal), a peak detection threshold (Tpeak) and a pairing threshold (Tpair). The output is a list of scored peak pairs [spair, (s700, l700),(s800, l800)], each corresponding to two image bands and a single putative cleavage product.
The algorithm first constructs an electropherogram for each channel i of each lane j by summing a 9-pixel wide horizontal window centered on the lane track. The region of interest of each lane (bounded below by 100 bp and above by ltotal − 75 bp) is resampled to the calibration standard of the leftmost lane and scaled to mean value 1, resulting in a set of input signals sij(y) in which co-migrating bands appear at the same y-coordinate in each lane and the effect of gross differences in signal intensity have been reduced.
For each image channel i, a background signal bi(y) approximating the signal that would result from a negative control sample in which no cleavage fragments are present is constructed by calculating for each vertical coordinate value y the top value of the bottom quintile of the source signal sij(y) among all lanes. A decorrelation algorithm based on a simplified version of robust principal component analysis (26) is then employed to find deviations of sij(y) from bi(y), resulting in a conditioned foreground signal gij(y) in which cleavage fragment peaks are enhanced (Supplementary Data 2).
A threshold function tij(y) is determined by computing the mean of the second quartile of all the values in a 96-sample sliding window centered at y with the addition of a small constant (0.002) to force tij(y) > 0. The algorithm considers potential peaks above this threshold. For each interval P such that gij(y) > tij (y) for all y ∈ P, the peak score s = 26 · max({gij(y)}y∈P) is calculated. The peak score and inferred fragment length of each peak with s ≥ Tpeak is recorded.
Sporadic mispriming products and other artifacts result in bands that appear at the same location in both channels. To prevent these bands from being mistaken for cleavage products, pairs of peaks appearing at approximately the same location in both channels are removed whenever the score of both peaks exceeds 4Tpeak. Peaks corresponding to lane markers (200 bp PCR products added to every eighth lane) are also removed at this stage.
The final step identifies pairs of peaks corresponding to single cleavage products with inferred fragment lengths summing to approximately ltotal. For every possible pair of peaks [(s700, l700),(s800, l800)] in a given lane, the distance penalty
and the pair score
are calculated, where dmax = 100 is the maximum allowed difference between ltotal and the summed fragment lengths. If spair > Tpair, the item [spair, (s700, l700), (s800,l800)] is added to a list of scored peak pairs for the current lane. This list is subsequently sorted in descending order of spair, and each 700 nm peak is assigned a complementary 800 nm peak and a pair score according to its first appearance in the list.
Analysis of unpooled gel images was performed using thresholds Tpeak = 100 and Tpair = 200. The decorrelation algorithm is described in Supplementary Data 2.
RESULTS
Ecotilling of human DNA samples
In a typical Ecotilling assay, a ∼1.5 kilobase target region is PCR-amplified using gene-specific primers. The forward primer is 5′-end-labeled with the fluorescent dye IRDye 700, and the reverse primer with the IRDye 800 dye. After amplification, samples are denatured and annealed to form heteroduplexes between DNA strands. Mismatched regions of the heteroduplexes are digested with CEL I endonuclease (25,27). Samples are then size-fractionated by PAGE using a LI-COR DNA analyzer in 96-lane format, producing a pair of IRDye 700 and IRDye 800 images (Figure 1). For any duplexed molecule containing a nucleotide difference, the molecular weight of the cleaved product in the IRDye 700 channel image plus the molecular weight of the cleaved product in the IRDye 800 channel image will equal the molecular weight of the full-length PCR product. We have shown previously that only a fraction of the DNA duplexes are cleaved at each site, allowing for the detection of multiple polymorphisms per reaction, and that all types of nucleotide changes and small insertions/deletions are detected (19).
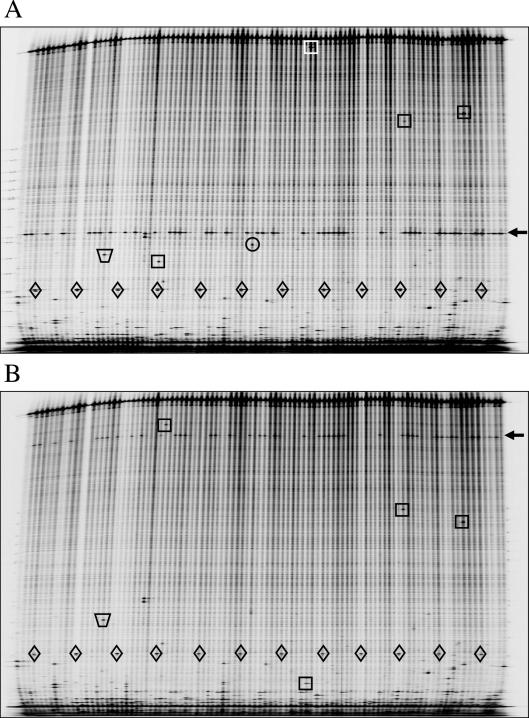
Figure 1.
SNP discovery in individual human DNA samples by Ecotilling. LI-COR gel analyzer images from the (A) IRDye 700 channel and (B) IRDye 800 channel are shown for a 1489 bp region of the DCLRE1A gene. Each lane contains a sample from a unique individual. Rare heterozygous polymorphisms are boxed, and a common SNP is marked by an arrow. Cleavage of polymorphisms with crude celery extract produces two fragments, one fluorescing in the IRDye 700 channel and its complement in the IRD 800 channel. Complementary fragments are marked in each channel image. Rare SNPs on this gel are found in only 1/90 individuals. Diamonds mark a 200-bp marker that marks every eighth lane beginning with lane 4. The trapezoid marks a band from mispriming. There are several such bands on this gel image, and none are scored as true polymorphisms because they lack a complementary fragment of the appropriate molecular weight in the other IRDye channel image. The band marked with a circle was scored as a low quality putative polymorphism. No appropriately sized fragment is found in this lane in (B), and thus the band represents a false positive error that could have been avoided.
To determine the accuracy of the Ecotilling method using human samples, we screened the PDR panel in gene regions that were scanned previously by resequencing as part of the NIEHS SNPs program. Five targets were selected from the directory of finished genes on the NIEHS SNPs program website (http://egp.gs.washington.edu/) (Table 1). For each target, we screened the 90 PDR samples that overlapped with the NIEHS dataset, plus 6 additional samples, in a 96-well format. Figure 1 shows gel images representing one of the targets. Ecotilling gel images provide information on the presence or absence of a nucleotide change in a particular sample and the approximate location of the change. In previous work, we have shown that the resolution achievable using this system is a few nucleotides. To verify that changes identified by Ecotilling correspond to changes identified by resequencing, we confirmed at least one sample of each allele type by sequencing, and we compared alleles identified by Ecotilling to those identified in the NIEHS SNPs project. As samples were screened individually, only heterozygous changes were detected. SNPs within 100 bp of the ends of the fragment were not included, because SNPs are more difficult to detect in low molecular weight regions of the gel where misprimed failure products typically migrate (28).
Table 1.
Comparison of alleles identified by Ecotilling with NIEHS SNPs
| GenBank ID | Target name | Start position | Window size (bp) | Alleles | ||
|---|---|---|---|---|---|---|
| Ecotilling/NIEHS | New by Ecotilling | Total by Ecotilling | ||||
| AY607842 | DCLRE1A | 005669 | 1289 | 3/3 | 2 | 5 |
| AY337516 | GAD1 | 043917 | 1299 | 6/6 | 0 | 6 |
| AY632118 | HK2 | 040539 | 1297a | 3/4 | 4 | 7 |
| AY800271 | NAT1 | 054558 | 1027 | 6/6 | 0 | 6 |
| AY504960 | TNFRSF5 | 003810 | 1298 | 6/6b | 1 | 8 |
| Total | 24/25 | 7 | 32 | |||
aData are not reported between positions 41 090 and 41 160 of HK2 because of difficulties in sequencing and Ecotilling caused by low nucleotide complexity and the presence of heterozygous indels.
bOne allele reported by NIEHS was not validated by resequencing the corresponding sample screened by our lab.
Using Ecotilling, we detected 24 of the 25 SNP alleles previously identified by NIEHS for the 5 primer pairs screened (Table 1). In addition, we discovered 7 new alleles, which we then verified by sequencing. Each new SNP is represented in only 1 of 90 individuals, and thus the 7 newly discovered alleles are considered rare.
To determine the accuracy of the Ecotilling method, we chose to compare the 90 test samples based on individual NIEHS-determined genotype (Table 2). As with SNP comparison, a small percentage of genotypes not detected in the NIEHS dataset were sequence-verified in our laboratory and thus are counted as true positives. By genotype comparison, we calculate a false discovery rate of 4% (7/163) and a false negative rate of 5% (9/171). Examination of gel images revealed that no data were collected for five of the nine false negatives due to unscorable gel lanes, one was missed because of human error, leaving four false negatives (2%) attributable to the Ecotilling method.
Table 2.
Comparison of polymorphisms identified by sequencing, Ecotilling with individual samples and Ecotilling with pooled samples
| Target name | Allele position | Base change | No. of SNPs by sequencing | No. of SNPs by Ecotilling | No. of SNPs by pooled Ecotillingc | Effectd | SIFTe score | PARSESNPf score | |
|---|---|---|---|---|---|---|---|---|---|
| NIEHSa | Newb | ||||||||
| DCLRE1A | 005855 | C→T | 1 | 1 | 1 | P287L | 1.00 | 15.2 | |
| 005944 | C→G | 38 | 38 | — | H317D | 1.00 | −0.1 | ||
| 006371 | C→T | 1 | 1 | 0 | P459L | 0.01+ | 12.2 | ||
| 006419 | G→A | 1 | 1 | 1 | G475E | 0.18 | 3.6 | ||
| 006939 | G→A | 1 | 1 | 1 | A648= | ||||
| GAD1 | 043953 | C→G | 28 | 26g | — | Intron | |||
| 044207 | G→A | 6 | 5h | (10)i | R532Q | 0.37 | 8.6 | ||
| 044526 | A→G | 2 | 2 | 2 | Intron | ||||
| 044582 | T→C | 1 | 1 | 1 | Intron | ||||
| 044940 | A→G | 7 | 6h | (11)i | Intron | ||||
| 044971 | G→A | 1 | 1 | 0 | Intron | ||||
| HK2 | 040543 | C→T | 1 | 1 | 0 | Intron | |||
| 040750 | G→A | 1 | 1 | 1 | Intron | ||||
| 040966 | T→C | 1 | 1 | 1 | Intron | ||||
| 041056 | A→C | 1 | 1 | 1 | Intron | ||||
| 041233 | G→C | 1 | 1 | 1 | V204= | ||||
| 041606 | G→A | 10 | 9h | — | Intron | ||||
| 041696 | T→C | 31 | 30 | — | D251= | ||||
| 041763 | C→T | 1 | 0 | 0j | R274C | 0.00 | 14.7 | ||
| NAT1 | 054792 | A→T | 3 | 3 | 3 | utr | |||
| 054796 | A→T | 6 | 5h | 4 | utr | ||||
| 055194 | C→T | 1 | 1 | 1 | V121= | ||||
| 055276 | G→A | 3 | 3 | 3 | V149I | 1.00 | −3.8 | ||
| 055290 | G→A | 3 | 3 | 0 | T153= | ||||
| 055471 | T→G | 3 | 3 | 3 | S214A | 0.40 | 1.6 | ||
| TNFRSF5 | 004013 | C→T | 1 | 1 | 0 | Intron | |||
| 004356 | T→C | 1 | 1 | 1 | Intron | ||||
| 004439 | C→T | 9 | 9 | (10)i | Intron | ||||
| 004641 | A→G | 1 | 1 | 1 | Intron | ||||
| 004694 | C→T | 1 | 1 | 1 | Intron | ||||
| 004695 | G→A | 3 | 3 | 2 | Intron | ||||
| 004764 | A→C | 1k | 0 | 0 | |||||
| 004952 | C→T | 2 | 2 | 2 | S124L | 0.32 | 0.0 | ||
| Total | 171 | 163 | 170 | ||||||
aAlleles sequenced by the NIEHS SNPs program.
bAlleles sequenced by STP to confirm TILLING results.
cIn some cases, the frequency of the polymorphism is sufficiently high that genotypes cannot be assigned to individuals in 8× pools (indicated by —). To calculate the total number of SNPs detected in these pools, we used the number of SNPs detected by NIEHS.
dSynonymous (=) and non-synonymous changes are shown, where the amino acid residue number is based on the exon–intron model for the TILLed fragment. Utr = 5′ or 3′ untranslated.
eA non-synonymous SNP is predicted to be damaging to the encoded protein if the SIFT score is <0.05 (in boldface). Low-confidence predictions are indicated (+).
fA non-synonymous SNP is predicted to be damaging to the encoded protein if the PARSESNP score is >10 (in boldface).
gNo data collected in two individuals.
hNo data collected in one individual.
iHomozygous SNPs are discovered in pools. The SNP frequency is too high to assign genotypes in 8× pools. The number in parenthesis indicates the number of individuals with the SNP determined by sequencing. We used this number of SNPs to calculate the total number of SNPs detected in pools.
jThis polymorphism was overlooked when screening blind. Upon comparison with the known sequence, it was determined that the allele was clear on the gel and overlooked because of human error.
kResequencing of the individual identified by NIEHS showed that this SNP is not present in the corresponding sample screened by our group. This SNP is not counted when calculating false negative errors.
Discovery of rare single-nucleotide differences in pooled samples
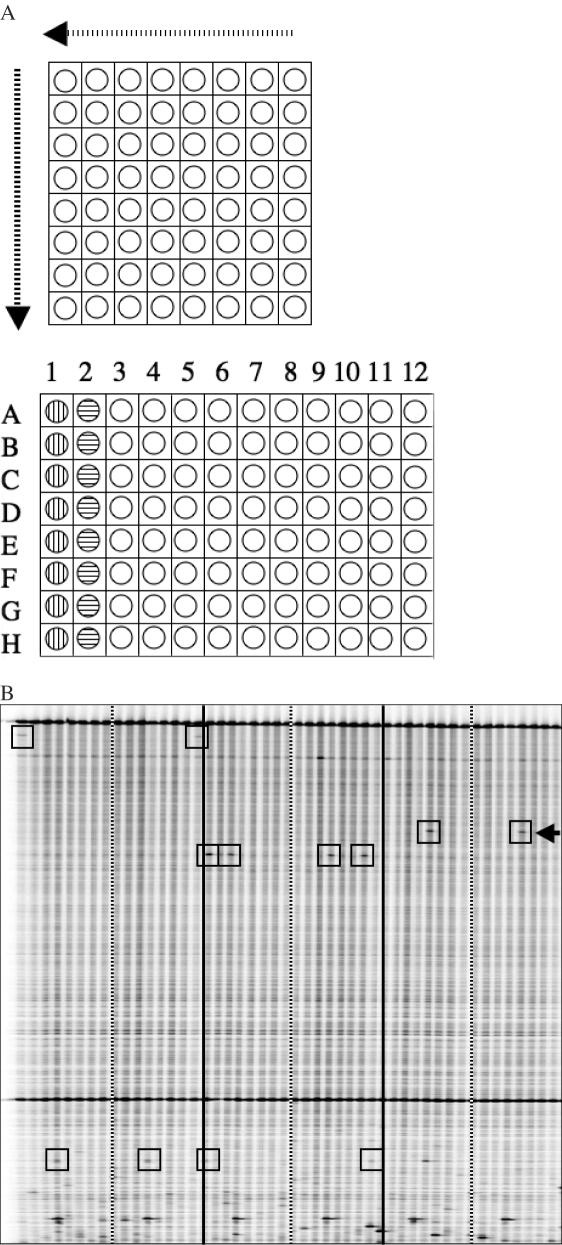
TILLING technology is ideally suited for the discovery of rare nucleotide differences and has been used successfully to identify induced point mutations in a variety of organisms from Arabidopsis to zebrafish (24,29–35). Having established the accuracy of Ecotilling with human DNA samples, we next sought to develop a pooled Ecotilling platform designed specifically to discover rare nucleotide differences in the human genome. To do this, we used a pooling and 2D arraying strategy. Samples are pooled 8-fold such that each single sample is arrayed into two pools, with only one individual common between the two pools (Figure 2A). A nucleotide polymorphism in an individual results in a band in two lanes of the gel. The lane numbers provide coordinates to decipher the unique polymorphic individual in the pool. Thus, in a single assay, the 2D strategy provides both an independent confirmation of the nucleotide change and the identity of the unique sample. We pooled eight individuals together, based on evidence for robust detection of heterozygous mutations in 8-fold DNA pools (1 in 16) (28). Figure 2B shows a typical example. With 8-fold pooling, most SNPs with a frequency of <5% will be present in separate pools allowing for unambiguous identification of the individual harboring the nucleotide change.
Figure 2.
Ecotilling of pooled samples to discover rare nucleotide changes. (A) Schematic diagram of sample pooling and arraying. A 2D arraying strategy is used whereby 64 unique samples are first arranged in an 8 × 8 grid (upper panel), pooled by row, and deposited into a 96-well screening plate (vertical striped wells, lower panel). Samples are then pooled by column and deposited in the adjacent column of the 96-well plate (horizontal striped wells). Each well in the 96-well plate contains eight pooled samples. Per set of 64 samples, an individual sample is present only once in a row pool and only once in a column pool. Samples are robotically loaded onto gels with sample A1 in lane 1, B1 in lane 2, A2 in lane 9 and so on. A true nucleotide change present in one of the first eight lanes must be present again in one of lanes 9 through 15. The exact lane numbers provide the coordinates to determine the individual harboring the nucleotide change. A total of 384 unique samples can be assayed per gel run. (B) Example of a pooled Ecotilling image (IRDye 700 shown). The first 48 of 96 lanes are shown from this run screening for polymorphisms in the DCLRE1A gene. Individuals screened in Figure 1 lanes 1–64 are rescreened in pooled lanes 1–16. Lanes to the left of the striped bars are row pools, and to the right are the corresponding column pools from a set of 64 samples. Solid black lines separate sets of 64 unique samples. Rare polymorphism are boxed. The arrow indicates a rare nucleotide change that was not found in the first 96 individuals screened (First two sets of lanes and Figure 1).
Using 8-fold pooling, we performed screens with the five test targets listed in Table 1. Gels were blindly scored for polymorphisms and then compared to results from Ecotilling unpooled samples and from resequencing (Table 2). This comparison implied a false discovery rate of 2% (4/170) and a false negative rate of 7% [12/(171 heterozygous SNPs + 9 homozygous SNPs)]. With this strategy, we have successfully screened 384 unique DNA samples in a single gel run and have discovered 21 additional rare alleles (Table 3). Some of these alleles are likely to be damaging to the protein. One is a nonsense mutation in the middle of the coding region, and seven are predicted to be damaging to the protein using either SIFT or PARSESNP, which are web-based tools that predict damaging non-synonymous SNPs (36,37). All eight of these predicted deleterious SNPs were heterozygous and were discovered at a frequency of <0.5% (3/768) in the 384 samples screened. In contrast, none of the five common non-synonymous SNPs were predicted to be damaging by either program, consistent with evidence that there are very few damaging common polymorphisms in the human genome (38).
Table 3.
Additional rare alleles discovered by 2D Ecotillinga
| Target name | Allele position | Base change | Discovered in | Effectb | SIFTc score | PARSESNPd score |
|---|---|---|---|---|---|---|
| DCLRE1A | 005797 | G→A | P0253 | D268N | 0.03+ | 9.8 |
| 006494 | C→A | P0263 | T500N | 0.05+ | — | |
| 006497 | A→G | P0165 | N501S | 0.43 | — | |
| GAD1 | 044479 | A→G | P0324 | Intron | ||
| 044724 | C→G | P0357 | Intron | |||
| HK2 | 040806 | G→C | P0117 | Intron | ||
| 041438 | C→A | P0263 | Intron | |||
| 041527 | A→G | P0168 | Intron | |||
| NAT1 | 054682 | A→T | P0193 | Non-coding | ||
| 054852 | T→G | P0111,P0334 | L7= | |||
| 055021 | C→T | P0108 | R64W | 0.00 | 10.6 | |
| 055291 | G→T | P0213 | E154* | Stop codon | Stop codon | |
| 055484 | C→A | P0249 | T218N | 0.02 | 13.7 | |
| 055608 | T→C | P0111,P0334 | S259= | |||
| 055608 | T→G | P0097 | S259R | 0.31 | 6.9 | |
| TNFRSF5 | 004313 | G→C | P0288 | Intron | ||
| 004362 | T→C | P0335 | Intron | |||
| 004501 | G→A | P0363 | T57= | |||
| 004629 | A→G | P0092 | Intron | |||
| 004850 | G→A | P0141 | R90Q | 0.48 | 4.2 | |
| 005004 | A→G | P0218,P0332 | Intron |
aNot found in individuals P0001 to P0090, which were scrutinized by both NIEHS SNPs and Ecotilling (Table 2).
bSynonymous (=), non-synonymous and stop codon (*) changes are indicated, where the amino acid residue number is based on the exon–intron model for the TILLed fragment.
cA non-synonymous SNP is predicted to be damaging to the encoded protein if the SIFT score is <0.05 (in boldface). Low-confidence predictions are indicated as (+). SIFT analysis with default settings used the full-protein sequence as query of SWISS-PROT 48.7 + TREMBL 31.7.
dA non-synonymous SNP is predicted to be damaging to the encoded protein if the PARSESNP score is >10 (in boldface). PARSESNP (http://proweb.org/parsesnp) used default alignments.
Universal priming for Ecotilling
The 5′ IRDye labeled primers represent the largest material cost in the Ecotilling assay. A single set of primers is purchased for each target, and thus the cost per sample is reduced as the test population size increases. For TILLING assays, we typically screen thousands of pooled individuals, so the cost per sample is minor (∼$0.04 per sample). However, Ecotilling typically involves screening of much smaller populations, which requires a substantial investment in IRDye-labeled custom primers (∼$0.32 per sample for screening 384 samples pooled in a single 96-well plate). Because the IRDye-labeled primers are ∼10-fold more expensive than unlabeled primers, using a universal IRDye-labeled primer with unlabeled custom primers can dramatically reduce primer cost.
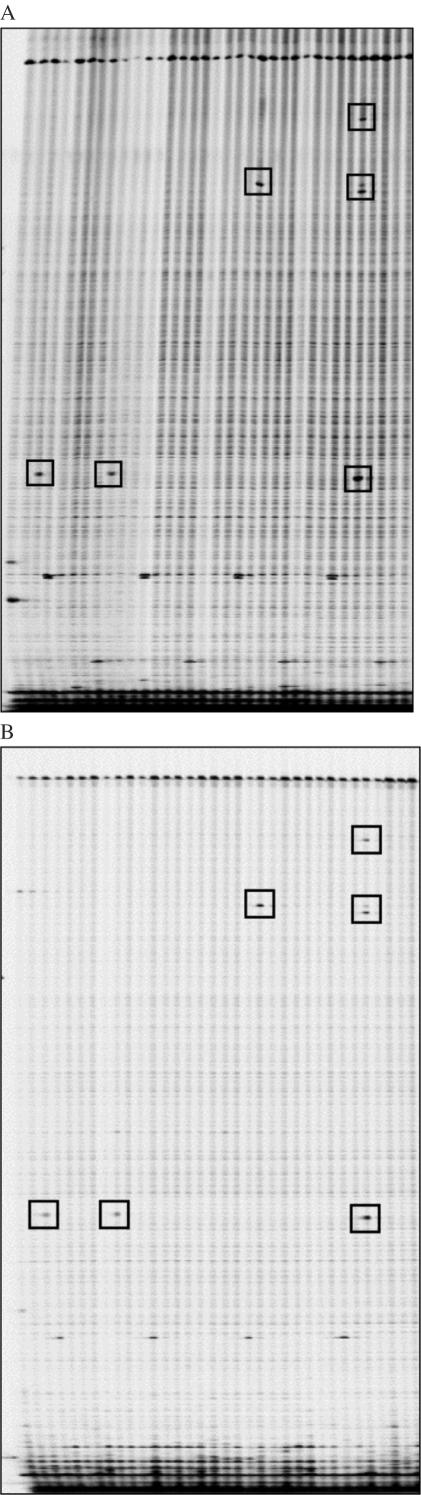
A two-step universal priming method was described previously for TILLING zebrafish (33). A first round of PCR amplification was performed on individual samples with an unlabeled custom primer, followed by sample dilution and a second round of amplification using a universal primer hybridizable to the 5′ ends of each custom primer. Following this, samples were pooled and digested with CEL I. The large number of separate PCRs and the complexity required to apply this strategy to 8-fold two-dimensionally pooled samples makes it prohibitive for Ecotilling. Therefore, we developed a single-step method. Unlabeled gene-specific primers are designed with either the T3 (forward) or the T7 (reverse) promoter sequence added to the 5′ end of the primer. In the PCR mixture, these primers are added together with universal IRDye 700-labeled T3 and IRDye 800-labeled T7 primers, and a single cycling program is run. All other Ecotilling steps are performed as when using 5′-end-labeled primers. Gel data quality using this universal primer strategy is comparable with that obtained using gene-specific 5′-end-labeled primers (Figure 3). We have also successfully applied this method to Arabidopsis TILLING assays, with slight adjustments to the ratio of labeled to unlabeled primers (data not shown). By using universal primers, the primer cost for screening 384 pooled samples is reduced ∼40-fold ($0.0085 per sample).
Figure 3.
Comparison of Ecotilling results using gene specific 5′-end-labeled primers (A) and universal 5′-end-labeled primers (B). Unadjusted IRDye 700 images displayed. Results from 34 individual samples are shown. Polymorphisms identified in a 1227 bp region of the NAT1 gene are boxed.
Automated band detection
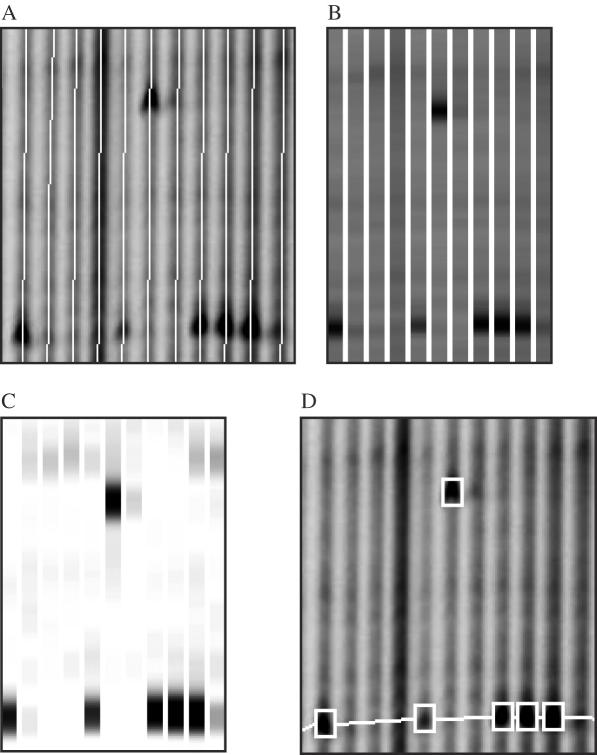
Data handling and analysis can be major determinants in the overall throughput of any production-scale genomics operation. We introduced previously the GelBuddy software package for analysis of TILLING and Ecotilling gel images on PC and Macintosh computers (22). GelBuddy automatically calls the lanes and calibrates fragment lengths based on background information. When GelBuddy is run in its manual mode, the user scores mutations or polymorphisms by moving the cursor over the relevant bands on the gel image and clicking the mouse. GelBuddy matches complementary 5′ (IRDye700)-labeled and 3′ (IRDye800)-labeled fragments, groups co-migrating fragments, and identifies samples of the same genotype. Lane number, mobility and grouping statistics for each selected fragment are automatically deposited in a database managed by the Perl program Squint (17,19). For the present study, we added new functions to GelBuddy to visualize and manipulate genotype information, allowing the user to easily copy a set of scored bands from one lane to another (or to a range of adjacent lanes) in cases where visual inspection reveals lanes containing multiple co-migrating cleavage products. To expedite accurate scoring of TILLING and Ecotilling images, we developed algorithms for automated detection of cleavage fragment bands and incorporated these algorithms in designing the ‘GelBrain’ option for GelBuddy. To detect and assign scores to candidate bands, GelBuddy constructs an estimated common background pattern for each LI-COR image channel and searches each lane for deviations from this pattern, assigning a score to each candidate band. The list of candidate bands for each lane is then searched to generate scores for pairs of bands corresponding to complementary cleavage fragments, based on the score of a single IRDye 700 band, a single IRDye 800 band, and the difference between the sum of the predicted length of each fragment and the length of the full-length PCR product (Figure 4).
Figure 4.
Automated band recognition using the GelBrain feature of GelBuddy. There are four main features of automated band detection. Lanes are automatically defined (A), a normalized electropherogram is constructed from image data (B), a decorrelation algorithm then detects bands that deviate from background signal (C) and bands are automatically detected and marked on the image (D). Bands are boxed in white. Common bands of the same molecular weight are linked by a horizontal connector. Discovery of both a rare SNP (upper box) and common SNPs (lower linked boxes) are shown. Data shown were extracted from the DCLRE1A IRDye 800 image (Figure 1B). When complete, a user can make manual modifications to the automatically marked up gel.
To compare the performance of automated band-calling with that of manual band-calling, we used GelBuddy to construct lane paths and calibration curves for each image pair as described by Zerr and Henikoff (22), and executed the automated band detection function. We applied this algorithm to images generated by Ecotilling of unpooled DNA samples and compared its output to heterozygosity inferred from NIEHS data and sequence verification data generated by our own lab (Table 4). Excluding Hexokinase 2 (HK2), this approach resulted in a 10% (12/117) false discovery rate and 16% (20/125) false negative rate, which compares to 5 and 4% scored manually. GelBrain failed on HK2, calling only 1 of 46 sequenced SNPs. Gel data from HK2 was highly atypical in that a diffuse band was present in all lanes on the gel (Figure 5). This complicated both manual and automatic image analysis. Examination of the target sequence revealed a 47 nt stretch containing only guanine and adenine residues, with 28 bases comprising a GA dinucleotide repeat in the approximate region of the diffuse band, which made sequence verification difficult. Nevertheless, the automated detection of the large majority of SNPs demonstrates the potential of GelBrain to reduce the considerable manual effort required for processing complex Ecotilling gel images. By superimposing a set of automatically detected bands upon the raw image data, the relatively small number of errors can be easily corrected after a brief visual inspection.
Table 4.
Comparison of GelBrain detection in Ecotilling images to sequence detection
| Target | Allele | Heterozyotesa | GelBrain false positives | |
|---|---|---|---|---|
| No. of SNPs by sequencing | No. of SNPs by GelBrain | |||
| DCLRE1A | 005855 | 1 | 1 | |
| 005944 | 38 | 38 | ||
| 006371 | 1 | 1 | ||
| 006419 | 1 | 1 | ||
| 006939 | 1 | 1 | ||
| 0 | ||||
| Total | 42 | 42 | 0 | |
| GAD1 | 043953 | 28 | 26 | |
| 044207 | 6 | 0 | ||
| 044526 | 2 | 2 | ||
| 044582 | 1 | 1 | ||
| 044940 | 7 | 6 | ||
| 044971 | 1 | 0 | ||
| 6 | ||||
| Total | 45 | 35 | 6 | |
| HK2 | 040543 | 1 | 0 | |
| 040750 | 1 | 0 | ||
| 041056 | 1 | 0 | ||
| 041233 | 1 | 1 | ||
| 041606 | 10 | 0 | ||
| 041696 | 31 | 0 | ||
| 041763 | 1 | 0 | ||
| 5 | ||||
| Total | 46 | 1 | 5 | |
| NAT1 | 054792, 054796b | 9 | 8 | |
| 055194 | 1 | 1 | ||
| 055276 | 3 | 3 | ||
| 055290 | 3 | 2 | ||
| 055471 | 3 | 0 | ||
| 0 | ||||
| Total | 19 | 14 | 0 | |
| TNFRSF5 | 004013 | 1 | 1 | |
| 004356 | 1 | 1 | ||
| 004439 | 9 | 9 | ||
| 004641 | 1 | 0 | ||
| 004694 | 1 | 0 | ||
| 004695 | 3 | 1 | ||
| 004764 | 1 | 0 | ||
| 004952 | 2 | 2 | ||
| 6 | ||||
| Total | 19 | 14 | 6 | |
aSNP heterozygosity was detected by automatic analysis of images generated by Ecotilling of unpooled DNA samples. Heterozygosity was determined by sequencing using NIEHS SNPs and/or Seattle Tilling Project sequence data.
bAutomated analysis did not distinguish between individuals heterozyous for SNPs 054792 and 054796 (spacing 4 bp).
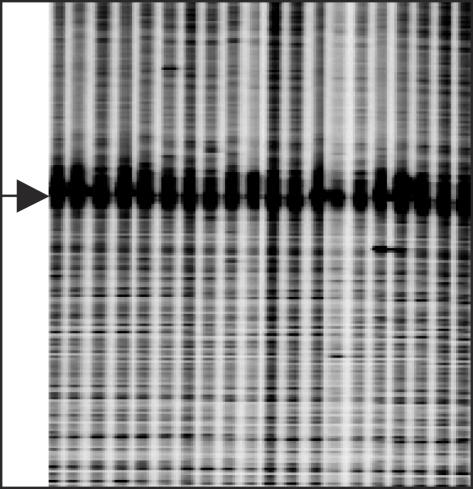
Figure 5.
Partial image of Ecotilling data for target HK2 (IRDye 700 shown). The arrow marks a strong band present in all lanes. Coincident with this band is a 49 bp region of low nucleotide complexity containing only guanine and adenine residues. The band interferes with signal detection in this region of the gel and with sequencing.
DISCUSSION
The ability to rapidly and inexpensively discover nucleotide differences in the human genome promises to be a major step towards determining the basis for phenotypic variation and genetic factors in human disease, including cancer. This promise has led to large-scale resequencing projects, including HapMap and the Human Cancer Genome Atlas. However, current resequencing technology is efficient only for common polymorphisms, and becomes increasingly inefficient as the frequency of an allele decreases. As a result, current SNP databases are heavily biased in favor of common SNPs.
By applying Ecotilling of 5 human genes to a sample of 384 individuals, we have discovered 28 new rare SNPs. Unlike the large majority of common SNPs, which are present at such high frequencies that they are unlikely to be deleterious, SNPs that are sufficiently rare that they almost never become homozygous might often be deleterious. Indeed, 8 of the 12 rare non-synonymous SNPs catalogued in our study are predicted by either SIFT or PARSESNP or both to be damaging to the encoded protein, including a protein truncation. This relatively high proportion of rare potentially damaging SNPs provides support for the common disease rare variant hypothesis and emphasizes the importance of discovering rare variants that are challenging for sequencing methods to detect.
Even more challenging to detect are new mutations in cancer, estimated to occur only once per megabase (15). The high level of redundancy needed to identify rare heterozygous polymorphisms and to minimize false negative errors makes their detection prohibitively expensive. Furthermore, heterogeneity caused by stromal contamination can make sequencing of tumor samples inaccurate or unfeasible. The high cost of finding cancer mutations by Sanger sequencing is illustrated by a study to detect coding region mutations for 518 protein kinases in 25 breast cancers. To obtain high redundancy, an estimated 750 000 initial sequence reads were collected (15). Assuming a cost of $2/sequence read, this is >$1 million just for the raw data traces that were used to discover 76 new mutations in tumors. Because this study was done only once, it is not known whether there were false negatives that escaped detection despite the redundancy. The Human Cancer Genome Atlas is proposed to be enormously larger, with the aim of discovering all mutations relevant to human cancer by screening several hundreds of tumors. There is thus an urgent need for an efficient screening technology that can reliably detect rare mutations and polymorphisms, and avoid the redundant sequencing that is otherwise necessary to minimize errors.
We have shown that the Ecotilling method can be readily adapted to discover nucleotide variation in human DNA samples. Target regions of up to 1.5 kb were screened using either individual or 8-fold pooled samples. When assaying individuals, 96 samples can be screened in a single gel run, leading to the interrogation of ∼125 000 bp for SNP differences. False positives are very few because of the dual end labeling strategy, in which each real nucleotide change results in a band in the IRDye 700 channel and a band in the IRDye 800 channel, whose molecular weights add up to the molecular weight of the full-length PCR product.
Whereas screening unpooled samples provided a starting point for developing human Ecotilling, the use of pooling increases throughput several-fold and facilitates the detection of rare base changes at the expense of rediscovering common SNPs. The presence of common SNPs in nearly all pools creates a common banding pattern that blends into the background banding pattern of the gel (Figure 2B). We do not consider this loss of common SNP information to be a serious drawback. The majority of these have likely been catalogued previously using Sanger dideoxy sequencing for discovery and custom screening methods, such as SNP-chips, for genotyping. It seems unlikely that such a resequencing and genotyping strategy will extend to less common or rare SNPs, because examination of sequence traces for heterozygous changes is challenging: the best trace analysis methods still require redundancy and human scrutiny to achieve a high degree of accuracy (39).
For heterozygous SNPs and mutations with frequencies of <5%, Ecotilling provides a robust high-throughput method. Indeed, the core TILLING technology used in Ecotilling was originally designed for the discovery of rare induced mutations (17,24). In >4 years of running an NSF-funded public TILLING service for the model plant A.thaliana, we have shown the TILLING method to be robust through the delivery of >6000 induced mostly heterozygous mutations (http://tilling.fhcrc.org:9366/arab/status.html) in a mutagenized population with an overall density of one mutation per ∼250 kb. Analysis of Arabidopsis TILLING production data revealed that heterozygous mutations were discovered efficiently in 8-fold pools (28). We have successfully established TILLING services for both maize and Drosophila, using populations with mutation densities as low as one mutation per 500 kb (http://tilling.fhcrc.org:9366/). Therefore, it is likely that even the low levels of heterozygous mutations found in tumors are within the range of robust detection using Ecotilling. Further robustness is provided by the 2D arraying strategy that we have applied to 8-fold pools, allowing us to screen 384 unique samples in a single 96-well run. With 1.5 kb target regions, ∼0.5 Mb were interrogated in a single gel run, and false positives were reduced compared to screening unpooled samples.
An additional strength of Ecotilling is that it provides high throughput at low cost while using standard methods and equipment. Because TILLING and Ecotilling utilize the same methods, equipment and reagents, we can draw upon our experience from our TILLING production services to estimate the efficiency and throughput for a human Ecotilling service. Since the start of the NSF-funded public Arabidopsis service in August 2001, our overall rate of repeat work has dropped from 24 to 14% for the year 2005. Assay failures have been attributable to equipment error, human error, reagent problems and inadequate primer design. Our production facility, with eight LI-COR gel analyzers, two full-time technicians and a part-time helper, can screen up to 80 gel runs for a maximum of ∼40 Mb per week. Thus, 1 week of output from our facility would interrogate about the same amount of DNA as was reported in the Stephens et al. (15) study for screening ∼1.3 Mb in each of the 25 breast cancer tumors.
We can also draw on experience from our current public TILLING facility to estimate the cost of a human Ecotilling project. Currently, we charge a user fee of $1500 per Arabidopsis allelic series, a price that includes sequencing of each mutation discovered and defrays all ongoing costs including labor, maintenance, failures and overhead. If we include a downward adjustment based on using a universal priming protocol and an upward adjustment based on using 2D pooling (unnecessary for Arabidopsis screening), we estimate a production cost of ∼$0.001 per base screened, or $1000/Mb. This is ∼50-fold cheaper than our estimate of just the data collection cost of the Sanger breast cancer study. Our estimated TILLING cost includes labor and all steps, from receipt of primers to an automatically generated mutation report.
Whereas automated SNP detection did not achieve the accuracy of an expert human, it nevertheless reduced the amount of human data processing effort ∼10-fold. Automated sequence trace analysis programs benefit by allowing human interaction (39), and the same is true for GelBuddy. Using the GelBrain feature allows a quick capture of most of the nucleotide changes while reducing a majority of the human labor involved in data analysis. Poor-scoring targets, such as HK2, become obvious candidates for redesigning primers or target regions. Furthermore, our automated SNP-detection program discovered a few SNPs that were overlooked by a human expert (Table 3), suggesting that the GelBrain band-detection algorithms are insensitive to variations that can obscure true signals. Automated Ecotilling error rates are low when compared to rates for automated detection of heterozygotes from sequence traces. For example, Weckx et al. (39) compared automatic SNP detection using novoSNP, PolyPhred and PolyBayes. For a reasonable compromise between sensitivity and selectivity, best performance was achieved by novoSNP, with an accuracy of 61% [=100 − (false discovery + false negative rate)], compared to the 74% accuracy achieved by GelBrain for typical gel images. In another study, PolyPhred version 5 achieved 80% accuracy (40). Thus the accuracy of GelBrain version 1 is similar to that of sequence trace analyzers that have been subject to many improvements over several years in a highly competitive field. Although no INDELs were present in the sequenceable regions of the targets used in our study, we predict a high accuracy of discovery, as INDELs produce more intense bands than SNPs and are more easily detected on Ecotilling gel images (19).
We conclude that Ecotilling is a fast and highly cost-effective alternative to the current state-of-the-art techniques for human mutation and rare polymorphism discovery. Ecotilling is a scalable technology that is easily adapted for other types of studies. For example, the high sensitivity of the method could be exploited for screening samples from mixed cell origins or for screening polyploid samples, where nucleotide differences are represented in a small fraction of a sample. The establishment of Ecotilling core facilities to serve as a ‘front-end’ for large-scale sequencing operations will most probably lead to other useful applications for human and model organism genomics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Jeff Harford for pointing out the potential of pooled Ecotilling for human SNP detection and encouraging us to perform this study. We thank Christine Codomo for help with data analysis and sequencing, Rob Laport and Margaret Darlow for help with sample arraying, Samson Kwong and Jorja Henikoff for their database work and for modifications to the Perl program ‘Squint’, and Stephen Tapscott and Jennifer Cooper for comments on the manuscript. This work was supported by Grants 0234960 and 0077737 from the National Science Foundation. Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinds D.A., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 3.Guttmacher A.E., Collins F.S. Realizing the promise of genomics in biomedical research. JAMA. 2005;294:1399–1402. doi: 10.1001/jama.294.11.1399. [DOI] [PubMed] [Google Scholar]
- 4.Reich D.E., Lander E.S. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 5.Altshuler D., Clark A.G. Genetics. Harvesting medical information from the human family tree. Science. 2005;307:1052–1053. doi: 10.1126/science.1109682. [DOI] [PubMed] [Google Scholar]
- 6.Lai E., Bowman C., Bansal A., Hughes A., Mosteller M., Roses A.D. Medical applications of haplotype-based SNP maps: learning to walk before we run. Nature Genet. 2002;32:353. doi: 10.1038/ng1102-353. [DOI] [PubMed] [Google Scholar]
- 7.Couzin J. Genomics. New mapping project splits the community. Science. 2002;296:1391–1393. doi: 10.1126/science.296.5572.1391. [DOI] [PubMed] [Google Scholar]
- 8.Pritchard J.K. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen J.C., Kiss R.S., Pertsemlidis A., Marcel Y.L., McPherson R., Hobbs H.H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 10.Fearnhead N.S., Wilding J.L., Winney B., Tonks S., Bartlett S., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc. Natl Acad. Sci. USA. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaukrodger N., Mayosi B.M., Imrie H., Avery P., Baker M., Connell J.M., Watkins H., Farrall M., Keavney B. A rare variant of the leptin gene has large effects on blood pressure and carotid intima-medial thickness: a study of 1428 individuals in 248 families. J. Med. Genet. 2005;42:474–478. doi: 10.1136/jmg.2004.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savas S., Ahmad M.F., Shariff M., Kim D.Y., Ozcelik H. Candidate nsSNPs that can affect the functions and interactions of cell cycle proteins. Proteins. 2005;58:697–705. doi: 10.1002/prot.20367. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs R. Deeper into the genome. Nature. 2005;437:1233–1234. doi: 10.1038/4371233a. [DOI] [PubMed] [Google Scholar]
- 14.Wang T.L., Rago C., Silliman N., Ptak J., Markowitz S., Willson J.K., Parmigiani G., Kinzler K.W., Vogelstein B., Velculescu V.E. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc. Natl Acad. Sci. USA. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens P., Edkins S., Davies H., Greenman C., Cox C., Hunter C., Bignell G., Teague J., Smith R., Stevens C., et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nature Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 16.Bignell G., Smith R., Hunter C., Stephens P., Davies H., Greenman C., Teague J., Butler A., Edkins S., Stevens C., et al. Sequence analysis of the protein kinase gene family in human testicular germ-cell tumors of adolescents and adults. Genes Chromosomes Cancer. 2006;45:42–46. doi: 10.1002/gcc.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colbert T., Till B.J., Tompa R., Reynolds S., Steine M.N., Yeung A.T., McCallum C.M., Comai L., Henikoff S. High-throughput screening for induced point mutations. Plant Physiol. 2001;126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCallum C.M., Comai L., Greene E.A., Henikoff S. Targeted screening for induced mutations. Nat. Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- 19.Comai L., Young K., Till B.J., Reynolds S.H., Greene E.A., Codomo C.A., Enns L.C., Johnson J.E., Burtner C., Odden A.R., et al. Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. Plant J. 2004;37:778–786. doi: 10.1111/j.0960-7412.2003.01999.x. [DOI] [PubMed] [Google Scholar]
- 20.Till B.J., Colbert T., Tompa R., Enns L.C., Codomo C.A., Johnson J.E., Reynolds S.H., Henikoff J.G., Greene E.A., Steine M.N., et al. High-throughput TILLING for functional genomics. Methods Mol. Biol. 2003;236:205–220. doi: 10.1385/1-59259-413-1:205. [DOI] [PubMed] [Google Scholar]
- 21.Collins F.S., Brooks L.D., Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- 22.Zerr T., Henikoff S. Automated band mapping in electrophoretic gel images using background information. Nucleic Acids Res. 2005;33:2806–2812. doi: 10.1093/nar/gki580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C.H., Hansen S., Wang Z., Storm D.R., Tapscott S.J., Olson J.M. The dosage of the neuroD2 transcription factor regulates amygdala development and emotional learning. Proc. Natl Acad. Sci. USA. 2005;102:14877–14882. doi: 10.1073/pnas.0506785102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Till B.J., Reynolds S.H., Greene E.A., Codomo C.A., Enns L.C., Johnson J.E., Burtner C., Odden A.R., Young K., Taylor N.E., et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Till B.J., Burtner C., Comai L., Henikoff S. Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res. 2004;32:2632–2641. doi: 10.1093/nar/gkh599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubert M., Rousseeuw P.J., Verboven S. A fast method for robust principal components with applications to chemometrics. Chemometr. Intell. Lab. Syst. 2002;60:101–111. [Google Scholar]
- 27.Oleykowski C.A., Bronson Mullins C.R., Godwin A.K., Yeung A.T. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 1998;26:4597–4602. doi: 10.1093/nar/26.20.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greene E.A., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics. 2003;164:731–740. doi: 10.1093/genetics/164.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Till B.J., Reynolds S.H., Weil C., Springer N., Burtner C., Young K., Bowers E., Codomo C.A., Enns L.C., Odden A.R., et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004;4:12. doi: 10.1186/1471-2229-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slade A.J., Fuerstenberg S.I., Loeffler D., Steine M.N., Facciotti D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 2005;23:75–81. doi: 10.1038/nbt1043. [DOI] [PubMed] [Google Scholar]
- 31.Perry J.A., Wang T.L., Welham T.J., Gardner S., Pike J.M., Yoshida S., Parniske M. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 2003;131:866–871. doi: 10.1104/pp.102.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler S., Schwabedissen A., Backasch D., Bokel C., Seidel C., Bonisch S., Furthauer M., Kuhrs A., Cobreros L., Brand M., et al. Target-selected mutant screen by TILLING in Drosophila. Genome Res. 2005;15:718–723. doi: 10.1101/gr.3721805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wienholds E., van Eeden F., Kosters M., Mudde J., Plasterk R.H., Cuppen E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–2707. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Draper B.W., McCallum C.M., Stout J.L., Slade A.J., Moens C.B. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- 35.Caldwell D.G., McCallum N., Shaw P., Muehlbauer G.J., Marshall D.F., Waugh R. A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.) Plant J. 2004;40:143–150. doi: 10.1111/j.1365-313X.2004.02190.x. [DOI] [PubMed] [Google Scholar]
- 36.Taylor N.E., Greene E.A. PARSESNP: a tool for the analysis of nucleotide polymorphisms. Nucleic Acids Res. 2003;31:3808–3811. doi: 10.1093/nar/gkg574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng P.C., Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weckx S., Del-Favero J., Rademakers R., Claes L., Cruts M., De Jonghe P., Van Broeckhoven C., De Rijk P. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005;15:436–442. doi: 10.1101/gr.2754005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens M., Sloan J.S., Robertson P.D., Scheet P., Nickerson D.A. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nature Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.