Abstract
The heterogeneous nuclear ribonucleoprotein (hnRNP) F belongs to the hnRNP H family involved in the regulation of alternative splicing and polyadenylation and specifically recognizes poly(G) sequences (G-tracts). In particular, hnRNP F binds a G-tract of the Bcl-x RNA and regulates its alternative splicing, leading to two isoforms, Bcl-xS and Bcl-xL, with antagonist functions. In order to gain insight into G-tract recognition by hnRNP H members, we initiated an NMR study of human hnRNP F. We present the solution structure of the three quasi RNA recognition motifs (qRRMs) of hnRNP F and identify the residues that are important for the interaction with the Bcl-x RNA by NMR chemical shift perturbation and mutagenesis experiments. The three qRRMs exhibit the canonical βαββαβ RRM fold but additional secondary structure elements are present in the two N-terminal qRRMs of hnRNP F. We show that qRRM1 and qRRM2 but not qRRM3 are responsible for G-tract recognition and that the residues of qRRM1 and qRRM2 involved in G-tract interaction are not on the β-sheet surface as observed for the classical RRM but are part of a short β-hairpin and two adjacent loops. These regions define a novel interaction surface for RNA recognition by RRMs.
INTRODUCTION
The heterogeneous nuclear ribonucleoprotein (hnRNP) H family consists of four highly homologous proteins, namely hnRNP H, hnRNP H′, hnRNP F and hnRNP 2H9. Members of the hnRNP H family specifically recognize poly(G) sequences (G-tracts) that are abundant in both DNA and RNA. These G-tracts are known to form a specific structure, the G-quadruplex, that consists of four guanine bases arranged in a square planar conformation and stabilized by hydrogen bonds [reviewed in (1,2)]. In DNA, these structures are mainly located in telomeres and also in some promoter regions. In RNA, G-tracts are frequent splicing recognition elements found both in introns and exons and are crucial for 5′ splice site recognition (3–5). They are also abundant downstream of mammalian polyadenylation signals (6). hnRNP F and H, through binding to these G-tracts, are responsible for the regulation of polyadenylation (6,7) and the splicing regulation of numerous pre-mRNAs such as the Bcl-x (8), the rat β-tropomyosin (9), the Rous sarcoma virus NRS (10), the HIV type 1 tat (11), the HIV-1 tev (12), the HIV-1 p17gag instability (13) and the c-src (14) pre-mRNAs. Furthermore, mutations in G-tract splice sites correlate with many diseases [reviewed in (15)] and in some cases, such as the neurofibromatosis type 1 disease involving the NF1 gene (16), the congenital hypothyroidism involving the TSH-beta subunit gene (17), or the cystic fibrosis through the CFTR gene (18), these mutations directly affect (disrupt or enhance) the binding of hnRNP H/F to the pre-mRNA (18,19).
Bcl-x is a member of the Bcl-2 family of apoptotic genes and plays an important role during development by regulating apoptosis of damaged or aged cells. Bcl-x naturally exists in two isoforms, Bcl-xL (233 amino acids) and Bcl-xS (170 amino acids). These two isoforms result from alternative splicing, Bcl-xS containing a truncated exon 2 as compared to Bcl-xL (20). The effect of these isoforms is antagonistic; Bcl-xL is anti-apoptotic while Bcl-xS is pro-apoptotic. It was shown that in a number of cancer cells, the Bcl-xL isoform is overexpressed, which decreases apoptosis and therefore increases the risk of metastases (21,22). The ratio of Bcl-xS and Bcl-xL in cells depends on two cis-acting elements flanking the 5′ splice site of Bcl-xS (23) and hnRNP H and F promote the production of the Bcl-xS isoform by recognizing one of these elements that contains three consecutive G-tracts. Mutations of these G-tracts abolish hnRNP F/H binding and thereby the production of the Bcl-xS isoform (8).
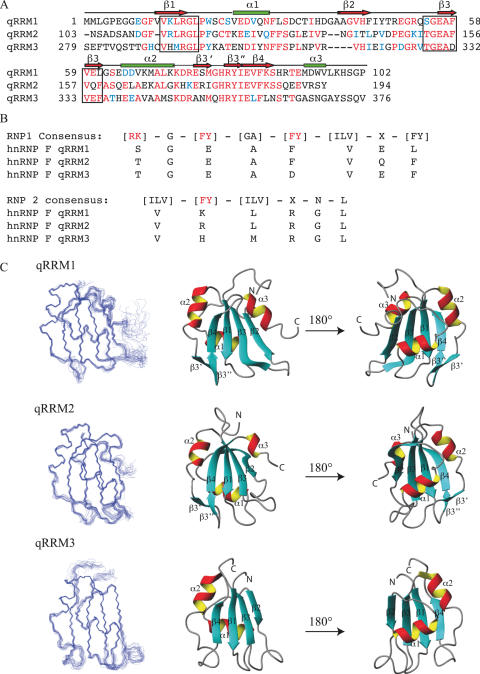
HnRNP H family members contain two (2H9) or three (H, H′ and F) quasi RNA recognition motifs (qRRMs) and one or two glycine rich auxiliary domains and are highly similar in sequence (hnRNP F and H share 78% sequence identity) (24–26). Human hnRNP F is a 45 kDa protein, which consists of two N-terminal qRRMs (residues 1–102 and 111–194) followed by a glycine-rich motif (residues 195–276) and a third qRRM (residues 277–366) (Figure 1A). These domains were denoted qRRMs because of the ability of these proteins to bind RNA and the small resemblance between the qRRM and the classical RRM motif. However, two conserved sequences found in all RRMs (RNP 1 and 2, located in the β-strands 3 and 1, respectively) that contain positively charged and aromatic residues involved in RNA binding are poorly conserved in hnRNP H family members (Figure 1B) (24).
Figure 1.
Overview of the structures of qRRM1, qRRM2 and qRRM3 of human hnRNP F. (A) Sequence alignment of the three qRRMs of hnRNP F. Identical residues are colored red and homologous residues are colored blue. Residues corresponding to RNP1 and RNP2 sequences are boxed. (B) Comparison of the consensus RNP1 and RNP2 sequence of RRMs with corresponding residues of hnRNP F qRRMs. Consensus residues important for RNA binding are colored red. (C) Overlay of the 20 final structures and ribbon representation of the lowest energy structure of qRRM1, qRRM2 and qRRM3. Figures were generated with MOLMOL (51).
In order to gain insight into G-tract recognition by hnRNP H members, we initiated an NMR study of human hnRNP F. We have determined the solution structure of the isolated qRRMs of hnRNP F and identified the residues that are important for the interaction with the Bcl-x G-tract RNA by NMR chemical shift perturbation and mutagenesis experiments. We show that the structures of the three qRRMs adopt a classical RRM fold and that the two N-terminal qRRMs are responsible for RNA binding. Our data also show that the mode of RNA recognition by hnRNP F differs from classical RNA recognition by RRMs as the β-sheet surface of the qRRM is not engaged in RNA binding.
MATERIALS AND METHODS
Cloning, expression and purification of hnRNP F subdomains
The DNA sequences encoding the first (residues 1–102), second (residues 103–194), third (residues 277–381) and the first two qRRMs (residues 1–194) of human hnRNP F (Swiss-Prot entry P52597) were cloned into the Xho1/BamH1 site of the Pet28b(+) vector containing an N-terminal hexa-histidine tag and overexpressed in BL21 (DE3) codon plus strains (Stratagene). qRRMs were uniformly labeled by overexpression in M9 minimum medium, containing 15NH4Cl and/or 13C-glucose as the sole nitrogen and carbon source. Cells were grown at 37°C to OD600 ∼0.6 and induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. Cells were harvested 2 h after induction and centrifuged. Cell pellets were resuspended in lysis buffer (50 mM Na2HPO4, 1 M NaCl and 10 mM Imidazole, pH 8), and lysed by two passages through a cell cracker (Avestin Inc.). Cell lysates were centrifuged 30 min at 20 000 g. His-tagged proteins were purified using Ni-NTA affinity column, dialyzed against NMR Buffer (25 mM NaH2PO4, 50 mM NaCl and 10 mM β-mercaptoethanol, pH 6.2) and concentrated to ∼0.4 mM (qRRM1 and qRRM1–2) and 2 mM (qRRM2 and qRRM3).
Mutagenesis experiments were carried out using the Quickchange Kit (Stratagene) following manufacturer's instructions.
RNA oligonucleotides were purchased from Dharmacon Research, deprotected according to manufacturer's instructions, desalted using a G-15 size exclusion column (Amersham), lyophilized and resuspended in NMR buffer.
NMR measurement
All NMR experiments were carried out at 313K using Bruker DRX-500 MHz equipped with a cryoprobe, DRX-600 MHz and Avance-900 MHz spectrometers. Data were processed using XWINNMR (Bruker) and analyzed with Sparky (http://www.cgl.ucsf.edu/home/sparky/). Sequence-specific backbone assignments were achieved using 2D (15N-1H)-HSQC, 2D (13C-1H)-HSQC, 3D HNCA, 3D HNCACB, 3D HN(CO)CA and 3D CBCA(CO)NH experiments [for a review see (27)]. 1H and 13C side chain assignments were performed using 3D H(C)CH-TOCSY, 3D (H)CCH-TOCSY, 3D NOESY-(15N-1H)-HSQC and 3D NOESY-(13C-1H)-HSQC. NH2 resonances of Asparagine and Glutamine were identified using 3D NOESY-(15N-1H)-HSQC. Aromatic proton assignments were performed using 2D-(1H-1H)-TOCSY and 2D-(1H-1H)-NOESY in 100% D2O. All NOESY spectra were recorded with a mixing time of 150 ms, the 3D TOCSY spectra with a mixing time of 23 ms and the 2D TOCSY with a mixing time of 50 ms.
Relaxation measurements were performed on a BRUKER DRX-600 (heteronuclear NOE) and DRX-500 (T1 and T2) equipped with a cryoprobe (1H frequency of 500.13 MHz,), using a 15N labeled qRRM1–2 sample with a concentration of ∼0.4 mM. The (15N-1H) heteronuclear NOE was recorded in an interleaved fashion, recording alternatively one increment for the reference and one for the NOE spectrum. A relaxation delay of 2 s and a 1H presaturation delay of 3 s were used in the NOE experiment while a 5 s relaxation delay was used in the reference experiment. 15N T1 relaxation times were derived from seven spectra with different values for the relaxation delay: 10.01, 75.16, 205.48, 355.84, 506.20, 756.80 and 1007.40 ms and an interscan delay of 3 s. Similarly, 15N T2 relaxation times were derived from seven CPMG experiments with different values for the relaxation delay: 12.40, 24.81, 43.42, 62.04, 80.66, 105.46 and 124.08 ms and an interscan delay of 3 s. T1 and T2 values were extracted using a curve-fitting subroutine included in the program Sparky. Overall correlation times (τc) were estimated from the average T1/T2 ratio of the rigid amide resonances (1H-15N NOE > 0.65) that have non-overlapping peaks in the HSQC spectrum.
Structure calculation
The automated peak picking, NOE assignment and structure calculation of the three individual qRRMs were performed using the AtnosCandid software (28,29). For each qRRM, peak picking and NOE assignments were performed using the two 3D NOESY (15N- and 13C- edited) spectra and the 2D homonuclear NOE spectrum recorded in D2O. Additionally, H-bond constraints were added based on hydrogen–deuterium exchange experiments on the amide protons. Seven iterations were performed and 100 independent structures were calculated at each iteration steps.
The 20 structures with the lowest target function were refined using the SANDER module of AMBER 7.0 (30) using the simulated annealing protocol described previously (31). The 20 final structures were analyzed with PROCHECK (32).
Data Bank accession numbers
Chemical shifts of qRRM1–2 were deposited previously to the BioMagResBank with entry number 6745 (33). Coordinates of qRRM1, qRRM2 and qRRM3 have been deposited to the Protein Data Bank with accession numbers 2HGL, 2HGM and 2HGN, respectively.
RESULTS
Structure of the three qRRMs of hnRNP F
The three qRRMs of hnRNP F were studied separately by NMR. qRRM1, qRRM2 and qRRM3 comprise residues 1–102, 103–194 and 277–381, respectively. Additionally, a construct containing both N-terminal qRRMs (qRRM1–2, residues 1–194) was also studied. We reported previously the resonance assignment of qRRM1–2 (33). Similarly, resonance assignment of qRRM3 was obtained using classical methods (Materials and Methods). For each qRRM, automated NOE peak picking and assignment were performed using the software AtnosCandid (28,29) based on (15N-1H)-NOESY-HSQC spectrum, (13C-1H)-NOESY-HSQC spectrum and 2D NOE spectrum recorded in D2O. A total of 1548, 1931 and 1818 NOE were extracted for qRRM1, qRRM2 and qRRM3, respectively (Table 1). Additionally, 26 (qRRM1), 22 (qRRM2) and 24 (qRRM3) hydrogen bond restraints derived from slowly exchanging amide protons in presence of D2O were used in the structure calculation. The 20 structures with the lowest target function in the last iteration were refined in implicit solvent.
Table 1.
Structural Statistics of the 20 best structures of qRRM1, qRRM2 and qRRM3
| qRRM1 | qRRM2 | qRRM3 | |
|---|---|---|---|
| Number of experimental restraints | |||
| Intra-residues NOEs | 389 | 383 | 281 |
| Sequential NOEs | 409 | 477 | 542 |
| Medium-range NOEs | 237 | 328 | 312 |
| Long-range NOEs | 513 | 743 | 683 |
| Total number of NOEs | 1548 | 1931 | 1818 |
| Hydrogen bonds | 26 | 22 | 24 |
| Root mean square deviation (r.m.s.d.) from the mean structurea (Å) | |||
| All backbone atoms | 0.82 ± 0.18 | 0.54 ± 0.11 | 0.60 ± 0.14 |
| All heavy atoms | 1.66 ± 0.29 | 1.23 ± 0.18 | 1.23 ± 0.20 |
| Secondary structure backbone atoms | 0.48 ± 0.09 | 0.45 ± 0.09 | 0.30 ± 0.07 |
| Secondary structure heavy atoms | 1.10 ± 0.14 | 1.21 ± 0.20 | 1.00 ± 0.17 |
| r.m.s.d. from idealized covalent geometry | |||
| Bond length (Å) | 0.0109 ± 0.0002 | 0.0112 ± 0.0001 | 0.0111 ± 0.0002 |
| Bond angle (°) | 2.73 ± 0.03 | 2.65 ± 0.04 | 2.91 ± 0.03 |
| Energies (kcal.mol−1) | |||
| Average AMBER | −3484 ± 24 | −3173 ± 15 | −2749 ± 16 |
| Average constraint violations | 130 ± 8 | 127 ± 8 | 107 ± 8 |
| Number of NOE violationsb | 10 | 9 | 6 |
| Maximum NOE violation (Å) | 0.53 | 0.71 | 0.40 |
| Ramachandran analysis (%) | |||
| Most favored regions | 73.3 | 69.3 | 63.8 |
| Additional allowed regions | 25.0 | 25.1 | 29.4 |
| Generously allowed regions | 1.0 | 4.0 | 4.5 |
| Disallowed regions | 0.7 | 1.7 | 2.3 |
aAll atoms comprise residues 11–98, 111–192 and 289–362 for qRRM1, qRRM2 and qRRM3, respectively. Secondary structure comprise residues 12–17, 24–30, 43–47, 55–61, 64–73, 76–78, 81–89 and 92–97 for qRRM1, residues 112–117, 124–130, 140–143, 153–159, 163–171, 174–176, 179–187 and 188–192 for qRRM2, and residues 289–294, 302–308, 315–318, 330–335, 338–345 and 357–361 for qRRM3.
bNOE violations >0.3 Å and present in >50% of the structures.
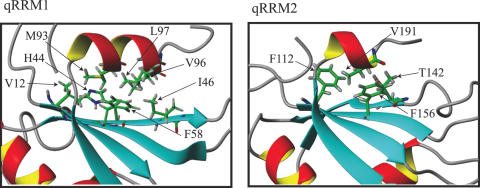
The three qRRM structures display a compact β1α1β2β3α2β4 fold resulting in a 4-stranded antiparallel β-sheet and two α-helices packed against the β-sheet (Figure 1C). This fold is very similar to the classical RRM fold. The core structure consists of residues 11–98 (qRRM1), 111–192 (qRRM2) and 289–362 (qRRM3). The first β-strand comprises residues 12–17, 112–117 and 289–294, for qRRM1, qRRM2 and qRRM3, respectively. It is followed by the first α-helix (residues 24–30, 124–130 and 302–308), the second (residues 43–47, 140–143 and 315–318) and third β-strands (residues 55–61, 153–159 and 330–335), the second α-helix (residues 64–73, 163–171 and 338–345) and the last β-strand (residues 84–89, 182–187 and 357–361). Furthermore, the structures of qRRM1 and qRRM2 are characterized by an additional β-hairpin (β3′ and β3′) located between α2 and β4 (residues 76–78 and 81–83 for qRRM1, and 174–176 and 179–181 for qRRM2) and a C-terminal α-helix (α3) (residues 92–97 and 188–192 for qRRM1 and qRRM2, respectively) that lies on the β-sheet surface (Figure 1C). This additional C-terminal α-helix forms a small hydrophobic core involving residues of this helix and of the β-sheet, mainly β1 and β3 (Figure 2). Especially hydrophobic residues of the C-terminal α-helix (M93, V96 and L97 for qRRM1, and V191 for qRRM2) are in contact with hydrophobic and aromatic residues of the β-sheet (V12, H44, I46 and F58 for qRRM1, and F112, T142 and F156 for qRRM2).
Figure 2.
The hydrophobic cluster formed between the C-terminal α-helix and the β-sheet of qRRM1 and qRRM2. In qRRM1, M93, V96 and L97 of the C-terminal α-helix interact with V12, H44, I46 and F58 of the β-sheet. In qRRM2, V191 of the C-terminal α-helix interact with F112, T142 and F156 of the β-sheet. Figures were generated with MOLMOL (51).
Dynamics of qRRM1 and qRRM2
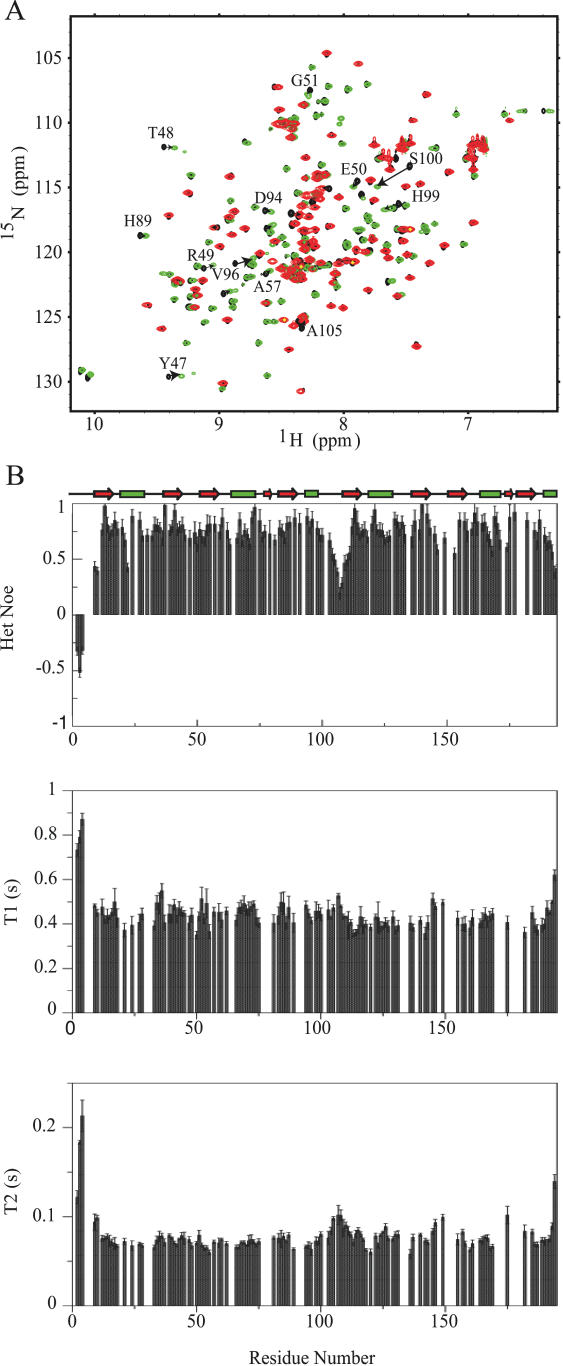
It was reported previously that two consecutive RRMs can interact with each other (34,35). We therefore studied a longer construct containing the two N-terminal qRRMs (qRRM1–2, residues 1–194) by NMR. The HSQC spectrum of qRRM1–2 is very similar to the HSQC spectra of qRRM1 and qRRM2 indicating that the structures of the individual domains are very similar in the context of qRRM1–2. Some small differences, however, can be observed for some residues located, as expected, at the C-terminus of qRRM1 and the N-terminus of qRRM2, and also in the loop connecting β2 and β3 of the first qRRM (Y47 to S54) suggesting that the two qRRMs might interact with each other (Figure 3A). Careful analysis of the NOESY spectra of qRRM1–2, qRRM1 and qRRM2, however, did not provide a clear evidence for the presence of inter-domain NOEs. Similarly, structure calculation of qRRM1–2 generated the proper RRM fold for qRRM1 and qRRM2 but the two domains were completely independent (data not shown). To further investigate whether these two domains are independent in solution or adopt a fixed relative orientation, we measured NMR relaxation experiments and performed a dynamic study of qRRM1–2. (1H-15N)-NOE, T1 and T2 were measured (Figure 3B). Except for the 10 first residues that are highly flexible [negative (1H-15N)-NOE values], the 2 qRRMs possess a rigid core [(1H-15N)-NOE values higher than 0.7]. The linker region (residues 100–110), however, shows low (1H-15N)-NOE values (between 0.19 and 0.5), which indicates that the linker between the two qRRMs is flexible and suggests that the two qRRMs tumble independently in solution. As a confirmation, we calculated the T1/T2 ratio and estimated the overall correlation time (τc) of qRRM1–2 and also of qRRM1 and qRRM2 in the context of qRRM1–2 (Table 2). The estimated overall correlation time for qRRM1–2 is 8.3 ± 0.6 ns, which is too short for a compact domain of 21.6 kDa. This value is in agreement with two globular domains of ∼10 kDa that tumble independently (Discussion).
Figure 3.
Relaxation studies of hnRNP F qRRM1–2. (A) HSQC spectrum of qRRM1–2 (black) overlaid with HSQC spectra of qRRM1 (green) and qRRM2 (red). (B) Heteronuclear NOE values (top panel), T1 relaxation rates (middle panel) and T2 relaxation rates from CPMG experiments (bottom panel). The secondary structure elements are also displayed.
Table 2.
T1, T2, apparent correlation time (App τc) and molecular weight (MW) of qRRM1–2
| qRRM1–2 | QRRM1–2 (qRRM1) | qRRM1–2 (qRRM2) | |
|---|---|---|---|
| T1 (s) | 0.43 ± 0.04 | 0.44 ± 0.04 | 0.41 ± 0.03 |
| T2 (s) | 0.074 ± 0.007 | 0.072 ± 0.007 | 0.076 ± 0.009 |
| T1/T2 | 5.84 ± 0.75 | 6.24 ± 0.57 | 5.30 ± 0.60 |
| App τc (ns) | 8.3 ± 0.6 | 8.6 ± 0.4 | 7.8 ± 0.5 |
| MW (kDa) | 21.4 | 11.4 | 10.2 |
NMR studies on hnRNP F binding to G-tract RNA
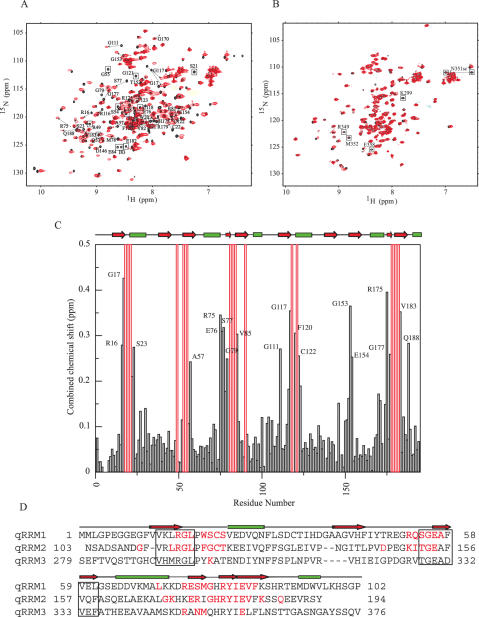
We used NMR titration experiments to test the ability of each qRRM of hnRNP F to bind G-tract RNA. For this purpose, different RNAs containing one or more G-tracts were used. First, we studied complex formation between a construct containing the two N-terminal qRRMs (qRRM1–2) and CGAUGGGAA (the underlined sequence corresponds to the G-tract, which is the minimum RNA sequence recognized by hnRNP F), which is the HIV-1 p17gag instability (INS) sequence that promotes export of unspliced HIV-1 transcripts (13). NMR studies of the free RNA show the presence of protected imino protons indicating that intramolecular or intermolecular hydrogen bonds are formed in our NMR conditions. Therefore, the free RNA is not unstructured but adopts a compact conformation that is most likely a G-quadruplex. Our titration experiments show that both qRRM1 and qRRM2 domains are binding RNA, and that two molecules of RNA bind one molecule of qRRM1–2, suggesting that each qRRM binds one G-tract (data not shown). Imino protons that were present in the free RNA are not observed in complex indicating that the RNA unfolds upon binding. Moreover, the complex is in fast exchange on the NMR time scale, which is indicative of a low-binding affinity. We then tested the ability of qRRM1–2 to bind a longer RNA containing two consecutive G-tracts and chose a sequence corresponding to the two first G-tracts of the Bcl-x RNA, CGGGAUGGGGUA (Figure 4A). In this case, the complex is in intermediate exchange (peaks disappear during the titration and reappear when a 1:1 complex is formed), which is indicative of a higher binding affinity. As observed with the previous RNA, the free Bcl-x RNA forms a compact structure that is disrupted upon qRRM1–2 binding. To ascertain that these two qRRMs can recognize RNA independently from each other, we titrated the p17gag INS sequence with qRRM1 and qRRM2 and observed the same perturbations as in the context of qRRM1–2, demonstrating that qRRM1 and qRRM2 of hnRNP F can bind and unfold G-tract RNA irrespective of the presence of the other qRRM. We then tested the ability of hnRNP F qRRM3 to bind RNA and chose the third G-tract sequence of the Bcl-x RNA, CUGGGGU (Figure 4B). In this case, no changes are observed in the HSQC spectra, which signify that qRRM3 of hnRNP F does not—or very weakly—bind G-tract RNA when isolated. As a consequence, imino protons of the RNA are still observed in the presence of qRRM3. We can therefore conclude that the two N-terminal qRRMs of hnRNP F are primarily responsible for G-tract RNA recognition.
Figure 4.
MR chemical shift perturbation experiments of qRRM1–2 and qRRM3 with Bcl-x G-tract RNAs. (A) HSQC of free qRRM1–2 (black) overlaid with the HSQC of qRRM1–2 in complex with the Bcl-x G-tract RNA, CGGGAUGGGGUA, in a 1:1 ratio (red). Peaks corresponding to residues showing large chemical shift changes upon RNA binding are labeled and the shifts are indicated. Boxed peaks correspond to peaks for which no assignment could be derived in the bound form. (B) HSQC of free qRRM3 (black) overlaid with the HSQC of qRRM3 in complex with CUGGGGU in a 1:1 ratio (red). Boxed peaks tend to disappear during RNA titration. (C) Combined chemical shift perturbations (Δδ=[(δHN)2 + (δN/6.51)2]1/2) of qRRM1–2 upon binding with Bcl-x G-tract RNA as a function of qRRM1–2 amino acid sequence. Red bars correspond to residues for which no assignments could be derived in complex with RNA. (D) Sequence alignment of qRRM1, qRRM2 and qRRM3 of human hnRNP F. Residues showing a significant chemical shift perturbation (>0.1) or that disappear upon RNA binding are colored red. Residues corresponding to RNP1 and RNP2 sequences are boxed.
In order to identify the minimum RNA sequence recognized by qRRMs, we tested the binding of qRRM2 with GGG and GG RNAs by NMR titration experiments. Complex formation with GGG is very similar to what is observed with CGAUGGGGAA while GG is not recognized (data not shown). This indicates that three consecutive guanosines are necessary and sufficient for hnRNP F binding. We also tested the binding of qRRM2 with another RNA sequence, the UGCAUG Fox RNA-binding site, that does not contain a G-tract (31). As expected, hnRNP F qRRM2 does not bind this sequence showing that hnRNP F recognition to G-tract RNA is specific (data not shown).
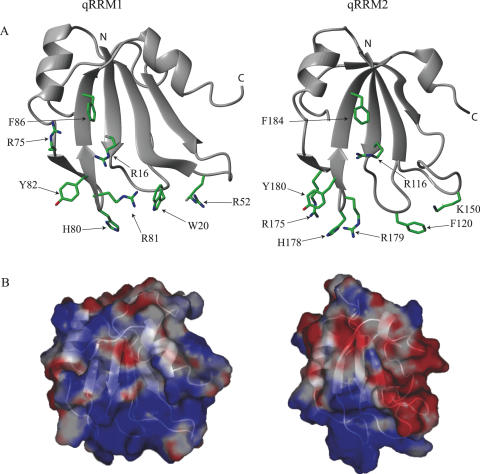
Since the complex between qRRM1–2 and Bcl-x G-tract RNA has the highest affinity, we used triple resonance experiments to assign the chemical shifts of qRRM1–2 in complex with Bcl-x RNA. Figure 4C shows the combined chemical shift differences between the free and bound qRRM1–2 amide proton and nitrogen atoms. For each qRRM, the same regions are affected by RNA binding. These are the loop connecting β1 and α2 (residues 16–23 and 115–123 for qRRM1 and qRRM2, respectively), the loop between β2 and β3 (residues 52–57 and 150–154) and the small β-hairpin between α2 and β4 (residues 75–86 and 174–184) (Figure 4C and D).Residues that align with the classical RNP1 and 2 sequences (13–18 and 113–118 for RNP2 and 54–61 and 152–159 for RNP1) do not display a significant chemical shift perturbation upon RNA binding. Resonances corresponding to residues forming the C-terminal α-helix are also not significantly perturbed upon complex formation. Furthermore, NOEs between residues of the C-terminal α-helix and the β-sheet are still present when qRRM is bound to the RNA, indicating that in complex, the hydrophobic cluster between the C-terminal α-helix and the β-sheet is conserved (Supplementary Data 1).Mapping the perturbed residues on the structure of qRRM1 and qRRM2 shows that these two domains bind the RNA through their β-hairpin and the two loops connecting β1−α1 and β2−β3 (Figure 5A). These three regions are rich in aromatic and positively charged residues making a suitable RNA-binding platform (Figure 5B).
Figure 5.
Residues of qRRM1 and qRRM2 showing a large chemical shift perturbation are clustered in the β-hairpin, the β1−α1 loop and the β2−β3 loop. (A) Ribbon representation of qRRM1 and qRRM2. Aromatic and positively charged side chains showing a significant chemical shift perturbation are displayed and labeled. Figures were generated with MOLMOL (51). (B) Surface representation of qRRM1 and qRRM2 colored according to electrostatic potential (red and blue indicate negative and positive charges, respectively). Figures were generated with PYMOL (http://www.pymol.org).
Mutagenesis of hnRNP F qRRM1 and qRRM2 aromatic residues
To test the importance of the aromatic residues located in the β-hairpin and the β1−α1 loop for G-tract recognition, we performed single mutations of F120, H178 and Y180 of qRRM2 to Alanine. These aromatic residues are at the surface of the protein, solvent exposed in the free structure and display a significant chemical shift perturbation upon RNA binding (Figures 4 and 5). As a control, we also mutated F156 to alanine. F156 is located in β3 and corresponds to a very important residue of the RNP1 sequence for RNA binding in classical RRM. In the case of qRRM2, however, this residue is participating in the hydrophobic cluster involving the β-sheet and the C-terminal α-helix (Figure 2). HSQC spectrum of this mutant shows that the F156A mutation drastically affects the fold of qRRM2 since most of the peaks disappear (Supplementary Data 2) most probably due to unfolding or aggregation. This suggests that this phenylalanine is important for proper folding or solubility of qRRM2 most likely due to its interaction with the C-terminal α-helix. The three other mutations (F120A, H178A and Y180A), however, do not affect the fold of qRRM2, which is consistent with the observation that these residues have their side chains exposed to the solvent in the structure of the free qRRM2 and therefore do not participate in the fold of the domain (Supplementary Data 2). We then analyzed the binding ability of the F120A, H178A and Y180A mutants to the CGAUGGGAA RNA sequence by NMR titration experiments. These mutations do not disrupt RNA binding (Supplementary Data 3) but differences with the wild type are observed. In the case of H178A, chemical shift perturbations are similar to those observed for the wild type indicating that this residue does not play a major role in G-tract recognition. F120A and Y180A, however, show a different binding pattern. When F120, located in the β1−α1 loop, is mutated to Ala, peaks corresponding to the three regions of the RNA interface (the β-hairpin, the β1−α1 loop and the β2-β3 loop) display smaller chemical shift perturbations than the wild type, indicative of a lower affinity for RNA. When Y180, located in the β-hairpin, is mutated to Ala, only peaks corresponding to residues in the β-hairpin and β2−α3 loops display a smaller perturbation, while peaks corresponding to the β2−β3 loop are not affected. We then produced the F120A/Y180A double mutant. In this case, the double mutation does not disrupt the fold of the qRRM (Supplementary Data 2) and completely abrogates G-tract binding (Supplementary Data 3). Therefore, we can conclude that, in contrast to classical RRMs, the non-canonical F120 and Y180 residues are crucial for RNA binding.
DISCUSSION
The three qRRMs of hnRNP F adopt the canonical RRM fold but qRRM1 and qRRM2 display extra secondary structure elements
The canonical fold of the RRM is well known. To date, >40 structures of this domain free or in complex with RNA have been solved [for a review see (34)]. It consists of ∼90 amino acids and is composed of a 4-stranded anti-parallel β-sheet and two α-helices packing on one surface of the β-sheet. In some cases, extra secondary structure elements could also be observed in addition to the canonical RRM fold, in particular a small β-hairpin located between α-helix 2 and β-strand 4. Sequence conservation between various RRMs is low except for two well-defined regions, RNP 1 and 2, of ∼7–8 amino acids located in the β-strands 1 and 3 that are responsible for RNA binding. hnRNP H family members are able to bind RNA, more precisely poly(G) sequences (G-tract), and three domains of the proteins were identified as possible RNA-binding domains. These domains are ∼90 amino acids and display a small resemblance to the classical RRM motif. The two RNP sequences, however, are not conserved (Figure 1B) and these domains were therefore termed qRRMs (24).
We solved the structures of the three qRRMs of hnRNP F using NMR spectroscopy. The structures are well defined and consist of the canonical β1α1β2β3α2β4 RRM fold (Figure 1C). For qRRM1 and qRRM2, two additional secondary structure elements are present. A small β-hairpin is formed between α2 and β4 and, more strikingly, the C-terminal part adopts an α-helical conformation and interacts with the β-sheet surface through hydrophobic and aromatic residues (Figure 2). Interestingly, residues of the β-sheet that interact with this C-terminal α-helix correspond to aromatic residues (F58 in qRRM1, and F112 and F156, in qRRM2) that are often essential for RNA interactions in classical RRM–RNA complexes. These residues are buried by the C-terminal α-helix and not solvent exposed as observed in most structures of free RRMs. The presence of a C-terminal α-helix packing against the β-sheet surface is only observed in a few RRM structures, such as the C-terminal RRM of LA protein (36), the N-terminal RRM of U1A (37), the N-terminal RRM of CstF-64 (38) and the p14 spliceosomal protein (39).
An analysis of RRM structures solved to date shows that two consecutive RRMs that are separated by a short linker (10–20 residues) can interact with each other to form a compact fold. This RRM–RRM interaction is often induced by the presence of RNA but can also occur in the absence of RNA (34,35). Since qRRM1 and qRRM2 of hnRNP F are separated by a short linker (residues 100–110), it is possible that these two domains interact with each other. We therefore studied a construct containing the two N-terminal qRRMs (qRRM1–2) by NMR. Relaxation measurements and dynamical studies clearly show that the linker between the two domains is flexible (Figure 3B). Furthermore, estimation of the overall correlation time for each qRRM strongly suggests that these two domains tumble independently in our conditions. The estimated overall correlation time for qRRM1–2 is 8.3 ± 0.6 ns, which is too short for a compact domain of 21.6 kDa, while using peaks corresponding to the first qRRM or the second qRRM only, estimations of the overall correlation times are 8.6 ± 0.4 and 7.8 ± 0.5 ns, respectively (Table 2). These values are in agreement with a globular domain of ∼10 kDa. We compared our relaxation analysis with those of RRM3 and RRM4 of the protein PTB. These two domains are interdependent in solution and show a large inter-domain interface involving 27 residues (35). NMR relaxation measurements and dynamical studies were performed on the wild-type PTB34, as well as on a mutant protein with a disrupted interface. For the wild-type construct, (1H-15N)-NOE values of the linker were higher than 0.68 and the estimated overall correlation time was 10.4 ns, while for the mutant, the estimated overall correlation time was 8.0 and 6.9 ns for RRM3 and RRM4, respectively (35). We can therefore conclude that, in our NMR conditions, qRRM1 and qRRM2 of hnRNP F are independent in solution.
HnRNP F qRRM1 and qRRM2 but not qRRM3 are responsible for G-tract recognition and two consecutive G-tracts are necessary for high-affinity binding
HnRNP F is a key regulator of splicing events and specifically recognizes RNAs containing poly(G) sequences (G-tracts) (3). It is, however, not clear which part of the protein is responsible for RNA recognition, although the three qRRMs are good candidates. Furthermore, it was postulated previously that the bases flanking the G-tract are important for RNA recognition (11,13). Jacquenet et al. (11) defined the minimum RNA sequence recognized by the hnRNP H family as UGGG, while Caputi and Zahler (13) defined the minimum RNA motif as GGGA. In order to identify which part of the protein is involved in RNA recognition and also what is the minimum RNA sequence recognized by hnRNP F, we performed NMR chemical shift perturbation experiments and tested the binding ability of each qRRM with different G-tract RNAs.
Our data clearly indicate that qRRM1 and qRRM2 are responsible for G-tract recognition (Figure 4). qRRM1–2 is able to bind single G-tracts in a one to two ratio indicating that each qRRM binds one G-tract. Furthermore, we observe that longer RNAs containing two consecutive G-tracts separated by a short linker bind qRRM1–2 with a considerably higher affinity, as was observed previously (40). Our results also show that the minimum RNA sequence recognized by qRRM1 and qRRM2 is GGG, which indicate that three consecutive guanosines are important for hnRNP F binding while the bases flanking the G-tract are not. We could not detect binding of qRRM3 with G-tract RNA. These results are striking since the three qRRMs of hnRNP F display a high sequence similarity (Figure 1A). The structure of qRRM3, however, differs slightly from the two N-terminal qRRM structures. qRRM3 adopts the canonical RRM fold and does not exhibit additional secondary structure elements like the small β-hairpin and the C-terminal α-helix. Since in qRRM1 and qRRM2, the β-hairpin is involved in G-tract recognition (see below), its absence in the qRRM3 structure might prevent RNA binding, although most of the residues that seem to be involved in RNA binding by qRRM1 and qRRM2 are conserved in qRRM3. It should, however, be noticed that, although no clear interaction between qRRM3 and G-tract could be observed, few peaks tend to disappear during the NMR titration (Figure 4B) and these residues are also located in the β-hairpin region.
qRRM1 and qRRM2 of HnRNP F recognize G-tracts by an unusual binding surface
Based on the numerous structures of RRM-RNA complexes, it is now well established that the RNA recognition by RRM is mediated by amino acids present at the surface of the β-sheet, in particular two sequences located in the central β3 (RNP1) and β1 (RNP2) strands [reviewed in (34)]. These mainly positively charged (R, K) and aromatic residues (F, Y) are crucial for RNA binding through H-bond formation and base stacking. In the case of hnRNP F qRRMs, these RNP sequences are not canonical, especially a positively charged residue of RNP1 is changed to a Serine (S54 in qRRM1) or a Threonine (T152 and T328 in qRRM2 and 3) and two highly conserved aromatic residues are changed to a Glutamate in all qRRMs (E56, E154 and E330) and to a positively charged residues in qRRM1 (K14) and in qRRM2 (R114) (Figure 1B). Furthermore, the structures of qRRM1 and qRRM2 of hnRNP F show that a C-terminal α-helix that is unusual for the RRM fold packs against the β-sheet, forming a hydrophobic core with residues of RNP1 and RNP2, therefore masking the classical RNA-binding site (Figure 2).
We identified the residues of qRRM1–2 that are important for Bcl-x G-tract binding using NMR chemical shift perturbation experiments. Strikingly, the residues that show the highest perturbations are not found in the β-sheet, but are located in the extra β-hairpin and in the β1−α1 and β2−β3 loops. Furthermore, the C-terminal α-helix that interacts with the canonical RNP1 and RNP2 residues is still present in the complex (Supplementary Data 1), which provides strong evidence that hnRNP F qRRMs bind RNA in a way distinct from the canonical RRM. It was reported previously that in some RRMs, a C-terminal α-helix is present in the free form and makes hydrophobic interactions with the β-sheet. However, these RRMs either do not bind RNA (LA C-terminal domain) or when they do, the β-sheet surface is the primary binding surface (U1A N-terminal RRM, CstF-64). In the case of U1A, the C-terminal α-helix rotates away from the β-sheet, and repositions in a way that the β-sheet surface binds to the RNA (41). For CstF-64, no structure in complex with RNA is available but NMR titration and relaxation studies show that the C-terminal α-helix unfold upon RNA binding and that the β-sheet RNP1 and RNP2 residues are responsible for RNA binding (38,42).
Analysis of the NMR chemical shift perturbation data show that many of the residues that are perturbed upon RNA binding are positively charged or aromatic (R16, W20, R52, R75, H80, R81, Y82 and F86 for qRRM1, and R116, F120, K150, R175, H178, R179, Y180 and F184 for qRRM2) and hence make a favorable binding platform for a negatively charged RNA molecule (Figures 4 and 5). Correspondingly, analysis of the electrostatic potential at the surface of qRRM1 and qRRM2 shows that these regions form a highly positively charged surface (Figure 5B). Mutagenesis experiments of aromatic residues located in this novel RNA-binding regions confirm their importance for RNA recognition. Our data show that F120, located in the β1−α1 loop, and Y180, located in the β-hairpin, of qRRM2 (corresponding to W20 and Y82 of qRRM1) are crucial for hnRNP F to bind G-tracts since mutation of these two residues to alanine completely abolish RNA binding (Supplementary Data 3). The involvement of the β1−α1 loop and the β-hairpin in RNA binding has been described earlier (31,43). In the structure of the Fox-1–RNA complex, the RNA lies on top of the β-sheet as observed for other RRM domains and the β1−α1 is also involved in binding. In particular, mutagenesis of an aromatic residue (located at the same position as W20 and F120 of qRRM1 and qRRM2 of hnRNP F) decreases significantly the affinity of Fox-1 for RNA (31). The structure of the tcUBP1 protein also contains the β-hairpin observed in hnRNP F qRRM1 and qRRM2, and NMR chemical shift perturbation data showed that RNA binding involves the β-sheet surface but also the β-hairpin extending the RNA binding surface (43). In both cases, however, the RNP1 and RNP2 sequences are involved in RNA binding and the β1−α1 or β-hairpin constitute an additional binding area. Our NMR chemical shift perturbation and mutagenesis data therefore define a novel RNA recognition mode involving the β-hairpin, the β1−α1 and the β2−β3 loops but not the β-sheet surface. Classical RRMs are known to bind single-stranded RNAs. The fact that qRRM1 and qRRM2 of hnRNP F recognize G-quadruplex structures and not single-stranded RNAs might explain the novel interaction interface that we observe.
Biological implication for RNA metabolism
Our data demonstrate that qRRM1 and qRRM2 of hnRNP F bind G-tract RNA via a novel mode of recognition, in which the RNP1 and RNP2 sequences are not involved. This is consistent with previous analyses showing that the amino acid composition of hnRNP H family members in these sequences was different from the classical RRM. We observe that residues in the β-hairpin, the β1−α1 and the β2−β3 loops of hnRNP F qRRM1 and qRRM2 are involved in RNA binding. We therefore performed a sequence alignment of hnRNP F from different species and human hnRNP H, H′ proteins. Full-length human hnRNP F, H and H′ share 40% identity (56% homology) but the sequence identity in the qRRM domains is higher (77% identity and 94% homology for the two first qRRMs). Furthermore, residues of human hnRNP F having a significant chemical shift perturbation upon G-tract RNA binding are strictly conserved between the three proteins. Similarly, full-length human, monkey, dog, bull, mouse and rat hnRNP F share 96% sequence identity (99% similarity) and all residues that show a high chemical shift perturbation upon RNA binding are strictly conserved. We also compared human hnRNP F with glorund, a drosophila homolog (44). These two proteins share 50% sequence homology. When considering the first two RRM domains only (residues 1–194 of human hnRNP F), the homology increases to 78% (42% identity) and residues of hnRNP F that are involved in RNA binding are strictly conserved. This is striking since glorund was shown to interact with a stem loop that does not contain G-tracts. The interaction of glorund with G-tracts, however, was not investigated.
G-tracts found both upstream and downstream of introns cooperate to allow intron definition (4,45). Since hnRNP H family members are able to form homodimers (46), it was proposed that a dimer of hnRNP F or H can simultaneously recognize the two upstream and downstream G-tracts, looping out the intron, therefore facilitating intron definition (45). This model is similar to the one proposed previously for the protein PTB in which PTB dimerizes to loop out alternatively spliced exons (47). In our laboratory, however, we reported recently an alternative model in which one monomer of PTB is sufficient to loop out an exon through the unique organization of the RRM3 and RRM4 domains (48). In this case, the two RRMs interact extensively with one another and each RRM bind one polypyrimidine tract. Our data indicate that both qRRM1 and qRRM2 of hnRNP F are able to bind a G-tract independently. We therefore considered whether each qRRM could bind G-tracts located upstream and downstream of the intron looping it out. Our results, however, are very distinct from what was observed for PTB. In contrast to PTB RRM3–4, hnRNP F qRRM1 and qRRM2 are independent in solution. Furthermore, qRRM1–2 is able to bind two consecutive G-tracts separated by a short linker with high affinity, while PTB RRM3–4 can only bind two polypyrimidine tracts separated by at least 15 bases (48). It is therefore unlikely that one molecule of hnRNP F is sufficient for looping out the intron and a dimer of hnRNP F is probably necessary. Further investigation on full-length hnRNP F should clarify which region of the protein is responsible for dimerization.
G-tract RNAs are frequent splicing recognition elements found downstream of splice sites (4,5). They are also present downstream of polyadenylation sites (6). McCullough and Berget (4) analyzed the base composition of small human introns. They observed that many introns contain a high frequency of G triplets near the splice sites, as illustrated with the 129 nt intron 2 of the human α-globin gene that contains seven G-tracts. Six of them are grouped in pairs with the generic sequence GGG(N)2–4GGG and are important for splicing efficiency and exon–intron definition. Similarly, the intron region near the Bcl-xS 5′ splice site contains three consecutive G-tracts (8). Mutation of one G-tract does not have a tremendous effect on splicing while mutation of two of these G-tracts, leaving one G-tract intact, prevents hnRNP F binding and completely abrogates Bcl-xS production (8). In combination with our data, we can expect that two consecutive G-tracts are important for hnRNP F qRRM1–2 recognition and function. Furthermore, an analysis of different introns containing consecutive G-tracts shows that the length of the linker between two G-tracts is variable. Our dynamical studies of hnRNP F qRRM1–2 demonstrate that the linker between the two domains is flexible and that the two qRRMs do not adopt a fixed conformation relative to each other. This flexibility might be important for the adaptation of qRRM1–2 to bind two consecutive G-tracts separated by linkers of variable length.
G-tracts have been shown to form special structures called G-quadruplexes. In RNA, G-tracts are responsible for recruitment of hnRNP H family members (45,49). In our NMR study, we observe that free G-tracts adopt a G-quadruplex compact fold, and this structure is disrupted upon HnRNP F binding. This is similar to what was observed for hnRNP A1 and hnRNP D. These two proteins, that specifically recognize telomeric DNA with the sequence TTAGGG, unfold the G-quadruplex structure stimulating telomerase activity (50). A model for the role of hnRNP F in alternative splicing and polyadenylation could therefore reside in the unfolding of the RNA, allowing other nearby RNA sites, such as 5′ splice sites or polyadenylation signals, to be available for other proteins. In the case of Bcl-x splicing, the G-tracts are located 20 residues downstream of the Bcl-xS 5′ splice site. Formation of a G-quadruplex might therefore prevent the accessibility of this site for the spliceosome leading to production of the Bcl-xL isoform. When hnRNP H members bind to the G-tract, however, they might prevent G-quadruplex formation and therefore make the Bcl-xS 5′ splice site available for the spliceosome.
Supplementary Material
Acknowledgments
We would like to thank J. Nikolic and Prof. D. Black for providing the clone of full-length hnRNP F, Dr T. Hermann for his help in setting up AtnosCandid calculations, Dr S. Jayne and S. Auweter for critical reading of the manuscript, and members of the group for helpful discussion. This investigation was supported by postdoctoral fellowships from the Roche Research Foundation for Biology and from the Novartis Research Foundation. Financial support from the Swiss National Science Foundation, the Structural Biology National Center of Competence in Research and the Roche Research Fund for Biology at the Swiss Federal Institute of Technology in Zurich is also acknowledged. The Open Access publication charges for this article has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Davis J.T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 2.Shafer R.H., Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2000;56:209–227. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 3.Swanson M.S., Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCullough A.J., Berget S.M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Zarudnaya M.I., Kolomiets I.M., Potyahaylo A.L., Hovorun D.M. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arhin G.K., Boots M., Bagga P.S., Milcarek C., Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garneau D., Revil T., Fisette J.F., Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 9.Chen C.D., Kobayashi R., Helfman D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogel B.L., McNally M.T. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 2000;275:32371–32378. doi: 10.1074/jbc.M005000200. [DOI] [PubMed] [Google Scholar]
- 11.Jacquenet S., Mereau A., Bilodeau P.S., Damier L., Stoltzfus C.M., Branlant C. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem. 2001;276:40464–40475. doi: 10.1074/jbc.M104070200. [DOI] [PubMed] [Google Scholar]
- 12.Caputi M., Zahler A.M. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caputi M., Zahler A.M. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 14.Chou M.Y., Rooke N., Turck C.W., Black D.L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 16.Baralle M., Baralle D., De Conti L., Mattocks C., Whittaker J., Knezevich A., Ffrench-Constant C., Baralle F.E. Identification of a mutation that perturbs NF1 agene splicing using genomic DNA samples and a minigene assay. J. Med. Genet. 2003;40:220–222. doi: 10.1136/jmg.40.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pohlenz J., Dumitrescu A., Aumann U., Koch G., Melchior R., Prawitt D., Refetoff S. Congenital secondary hypothyroidism caused by exon skipping due to a homozygous donor splice site mutation in the TSHbeta-subunit gene. J. Clin. Endocrinol. Metab. 2002;87:336–339. doi: 10.1210/jcem.87.1.8154. [DOI] [PubMed] [Google Scholar]
- 18.Pagani F., Buratti E., Stuani C., Baralle F.E. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580–26588. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 19.Buratti E., Baralle M., De Conti L., Baralle D., Romano M., Ayala Y.M., Baralle F.E. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHbeta genes. Nucleic Acids Res. 2004;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boise L.H., Gonzalez-Garcia M., Postema C.E., Ding L., Lindsten T., Turka L.A., Mao X., Nunez G., Thompson C.B. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 21.Olopade O.I., Adeyanju M.O., Safa A.R., Hagos F., Mick R., Thompson C.B., Recant W.M. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J. Sci. Am. 1997;3:230–237. [PubMed] [Google Scholar]
- 22.Clarke M.F., Apel I.J., Benedict M.A., Eipers P.G., Sumantran V., Gonzalez-Garcia M., Doedens M., Fukunaga N., Davidson B., Dick J.E., et al. A recombinant bcl-x s adenovirus selectively induces apoptosis in cancer cells but not in normal bone marrow cells. Proc. Natl Acad. Sci. USA. 1995;92:11024–11028. doi: 10.1073/pnas.92.24.11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massiello A., Salas A., Pinkerman R.L., Roddy P., Roesser J.R., Chalfant C.E. Identification of two RNA cis-elements that function to regulate the 5′ splice site selection of Bcl-x pre-mRNA in response to ceramide. J. Biol. Chem. 2004;279:15799–15804. doi: 10.1074/jbc.M313950200. [DOI] [PubMed] [Google Scholar]
- 24.Honore B., Rasmussen H.H., Vorum H., Dejgaard K., Liu X., Gromov P., Madsen P., Gesser B., Tommerup N., Celis J.E. Heterogeneous nuclear ribonucleoproteins H, H′, and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 25.Mahe D., Mahl P., Gattoni R., Fischer N., Mattei M.G., Stevenin J., Fuchs J.P. Cloning of human 2H9 heterogeneous nuclear ribonucleoproteins. Relation with splicing and early heat shock-induced splicing arrest. J. Biol. Chem. 1997;272:1827–1836. doi: 10.1074/jbc.272.3.1827. [DOI] [PubMed] [Google Scholar]
- 26.Matunis M.J., Xing J., Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sattler M., Schleucher J., Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spec. 1999;34:93–158. [Google Scholar]
- 28.Herrmann T., Guntert P., Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 29.Herrmann T., Guntert P., Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 30.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr, Onufriev A., Simmerling C., Wang B., Woods R. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auweter S.D., Fasan R., Reymond L., Underwood J.G., Black D.L., Pitsch S., Allain F.H. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 33.Dominguez C., Allain F.H. Resonance assignments of the two N-terminal RNA recognition motifs (RRM) of the human heterogeneous nuclear ribonucleoprotein F (HnRNP F) J. Biomol. NMR. 2005;33:282. doi: 10.1007/s10858-005-4287-2. [DOI] [PubMed] [Google Scholar]
- 34.Maris C., Dominguez C., Allain F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 35.Vitali F., Henning A., Oberstrass F.C., Hargous Y., Auweter S.D., Erat M., Allain F.H. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. EMBO J. 2006;25:150–162. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacks A., Babon J., Kelly G., Manolaridis I., Cary P.D., Curry S., Conte M.R. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–843. doi: 10.1016/s0969-2126(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 37.Avis J.M., Allain F.H., Howe P.W., Varani G., Nagai K., Neuhaus D. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 1996;257:398–411. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- 38.Perez Canadillas J.M., Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schellenberg M.J., Edwards R.A., Ritchie D.B., Kent O.A., Golas M.M., Stark H., Luhrmann R., Glover J.N., MacMillan A.M. Crystal structure of a core spliceosomal protein interface. Proc. Natl Acad. Sci. USA. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alkan S.A., Martincic K., Milcarek C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem. J. 2006;393:361–371. doi: 10.1042/BJ20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allain F.H., Gubser C.C., Howe P.W., Nagai K., Neuhaus D., Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 42.Deka P., Rajan P.K., Perez-Canadillas J.M., Varani G. Protein and RNA dynamics play key roles in determining the specific recognition of GU-rich polyadenylation regulatory elements by human Cstf-64 protein. J. Mol. Biol. 2005;347:719–733. doi: 10.1016/j.jmb.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 43.Volpon L., D'Orso I., Young C.R., Frasch A.C., Gehring K. NMR structural study of TcUBP1, a single RRM domain protein from Trypanosoma cruzi: contribution of a beta hairpin to RNA binding. Biochemistry. 2005;44:3708–3717. doi: 10.1021/bi047450e. [DOI] [PubMed] [Google Scholar]
- 44.Kalifa Y., Huang T., Rosen L.N., Chatterjee S., Gavis E.R. Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev. Cell. 2006;10:291–301. doi: 10.1016/j.devcel.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Contreras R., Fisette J.F., Nasim F.U., Madden R., Cordeau M., Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamberi C., Izaurralde E., Beisel C., Mattaj I.W. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol. Cell. Biol. 1997;17:2587–2597. doi: 10.1128/mcb.17.5.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner E.J., Garcia-Blanco M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 49.Han K., Yeo G., An P., Burge C.B., Grabowski P.J. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enokizono Y., Konishi Y., Nagata K., Ouhashi K., Uesugi S., Ishikawa F., Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 51.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.