Methylotrophy is defined as the ability to “grow at the expense of reduced carbon compounds containing one or more carbon atoms but containing no carbon-carbon bonds” (3). It is an intriguing example of microbial metabolic agility, with the use of a class of chemicals disregarded by the majority of organisms. Even though the ability to grow methylotrophically was first discovered in the early 1900s (cited in reference 3), it was not until the 1960s to 1970s that an understanding of the biochemical nature of this capability started to emerge. Fascination with methylotrophy in those years was fueled by the commercial interest in single-cell protein production, and as a result, the specific details of the biochemistry of methylotrophy began to be revealed. Enzymes for the primary oxidation of C1 substrates such as methanol dehydrogenase and methylamine dehydrogenase were characterized, and distinct modes of C1 assimilation, such as the ribulose monophosphate cycle and the serine cycle were discovered. The biochemical processes involved in methylotrophy that were known by 1982 are described in detail in the now classic book Biochemistry of Methylotrophs by Christopher Anthony (3). In the 20 years following the publication of Biochemistry of Methylotrophs, a few additional methylotrophy biochemical pathways have been discovered, such as the pathway for C1 transfer linked to methanopterin and methanofuran, which solved the long-standing mystery of formaldehyde oxidation in many methylotrophs (15, 53), and novel pathways for primary C1 oxidation, such as the pathways for degradation of chlorinated methanes and methanesulfonic acid (21, 50).
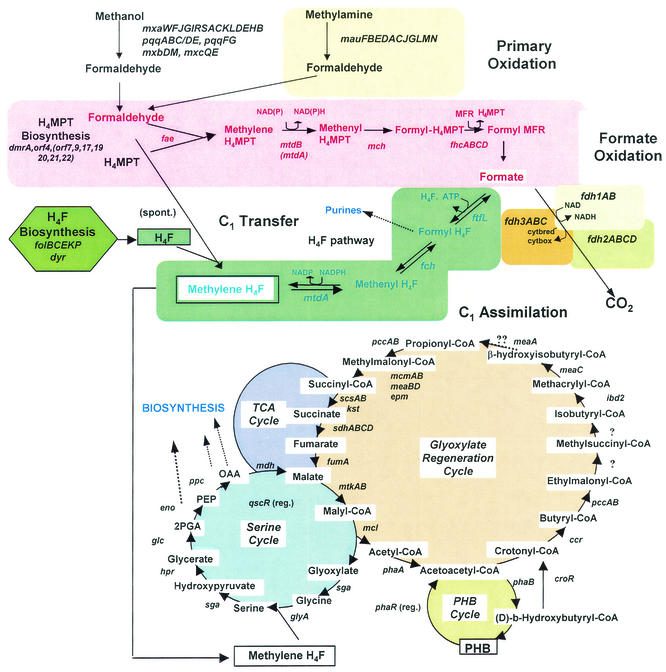
The knowledge concerning the biochemistry and physiology of methylotrophic organisms accumulated over the past three decades suggests a new framework for understanding methylotrophy as a novel metabolic mode. In this framework, methylotrophy is envisioned as a set of specific metabolic functional modules, with different combinations of such modules being present in different methylotrophs (Fig. 1 for the methylotrophic metabolic modules in Methylobacterium extorquens AM1). However, until recently, a number of important details of these modules were missing, and so the picture remained incomplete. The availability of two unfinished genome sequences for the important model organisms M. extorquens AM1 (http://www.integratedgenomics.com/genomereleases.html#list6)and Methylococcus capsulatus Bath (http://tigrblast.tigr.org/ufmg/) is transforming our understanding of methylotrophy. Annotation of these two genomes combined with functional analysis will delineate the set of genes and functions that is both sufficient and necessary to define a methylotroph. Expanding genomic analyses to include other groups of methylotrophs will in turn provide clues to the origins of methylotrophy and the evolution of various methylotrophic pathways. In this publication, we summarize the existing knowledge of the genes involved in methylotrophic pathways in M. extorquens AM1, analyze its yet unfinished genome with respect to location and clustering of methylotrophy genes, and present a comprehensive list of methylotrophy genes and enzymes known at this time in M. extorquens AM1 (Table 1).
FIG. 1.
Methylotrophy metabolic modules in M. extorquens AM1. Known genes are in italic. For simplicity, redox reactions in the assimilatory pathways are not indicated. For details, refer to the references given in Table 1.
TABLE 1.
Methylotrophy genes in M. extorquens AM1
| Gene | Function | Cluster | Essential for C1 growtha | Metabolic moduleb | Previous name | Accession no. | Source or reference |
|---|---|---|---|---|---|---|---|
| Primary oxidation | |||||||
| mxaW | Unknown | 1 | − | MOX | moxW | M31108 | 41 |
| mxaF | Methanol dehydrogenase large subunit | 1 | + | MOX | moxF | M31108 | 1, 2, 26 |
| mxaJ | Unknown | 1 | + | MOX | moxJ | M31108 | 1, 2, 26 |
| mxaG | Cytochrome c1 | 1 | + | MOX | moxG | X07856 | 1, 2, 26, 30 |
| mxaI | Methanol dehydrogenase small subunit | 1 | + | MOX | moxI | X15792 | 1, 26, 30, 31 |
| mxaR | Unknown | 1 | + | MOX | moxR | Y07864 | 1, 26, 44 |
| mxaS | Unknown | 1 | + | MOX | moxS | L41608 | 1, 26, 44 |
| mxaA | Essential for Ca2+ insertion into MDH | 1 | + | MOX | moxA | L41608 | 1, 26, 29 |
| mxaC | Essential for Ca2+ insertion into MDH | 1 | + | MOX | moxC | L41608 | 1, 26, 29, 44 |
| mxaK | Essential for Ca2+ insertion into MDH | 1 | + | MOX | moxK | L41608 | 1, 26, 29 |
| mxaL | Essential for Ca2+ insertion into MDH | 1 | + | MOX | moxL | L41608 | 1, 26, 29 |
| mxaD | Essential for Ca2+ insertion into MDH | 1 | − | MOX | L41608 | 1, 26, 29, 44 | |
| mxaE | Unknown | 1 | + | MOX | AF017434 | 41 | |
| mxaH | Unknown | 1 | NM | MOX | AF017434 | 41 | |
| mxaB | Transcriptional regulator | 1 | + | MOX | moxB | AF017434 | 41 |
| mxbD | Sensor kinase | 2 | + | MOX | moxD | AF032114 | 42 |
| mxbM | Transcriptional regulator | 2 | + | MOX | moxM | AF032114 | 42 |
| pqqA | PQQ synthesis | 2 | − | PQQ-S | pqqD, moxT | L25889 | 26, 45, 46 |
| pqqB | PQQ synthesis | 2 | + | PQQ-S | pqqG, moxO | U72662 | 26, 45 |
| pqqC/D | PQQ synthesis | 2 | + | PQQ-S | pqqB, pqqC, moxV, moxT | L25889/U72662 | 26, 45 |
| pqqE | PQQ synthesis | 2 | + | PQQ-S | pqqA, moxC | U72662 | 26, 45 |
| mxcQ | Sensor kinase | 3 | + | MOX | moxQ | 26, 42 | |
| mxcE | Transcriptional regulator | 3 | + | MOX | moxE | 26, 42 | |
| pqqF | PQQ synthesis | 4 | + | PQQ-S | pqqE, moxH | L43135 | 26, 43 |
| pqqG | PQQ synthesis | 4 | + | PQQ-S | pqqF, moxU | L43135 | 26, 43 |
| mauF | Unknown | 5 | + | MAU | L26406 | 5 | |
| mauB | Methylamine dehydrogenase large subunit | 5 | + | MAU | L26406 | 5 | |
| mauE | Essential for small subunit maturation | 5 | + | MAU | L26406 | 5 | |
| mauD | Essential for small subunit maturation | 5 | + | MAU | L26406 | 5 | |
| mauA | Methylamine dehydrogenase small subunit | 5 | + | MAU | L26406 | 5 | |
| mauC | Amicyanin | 5 | + | MAU | L26406 | 5 | |
| mauJ | Unknown | 5 | − | MAU | L26406 | 5 | |
| mauG | Unknown | 5 | + | MAU | L26406 | 5 | |
| mauI | Unknown | 5 | + | MAU | L26406 | 5 | |
| mauM | Unknown | 5 | − | MAU | L26406 | 5 | |
| mauN | Unknown | 5 | − | MAU | L26406 | 5 | |
| Formate oxidation | |||||||
| fdh1A | Tungsten-dependent formate dehydrogenase, α subunit | 6 | − | FOX1 | AF489516 | 25 | |
| fdh1B | Tungsten-dependent formate dehydrogenase, β subunit | 6 | − | FOX1 | AF489516 | 25 | |
| fdh2A | Molybdenum-dependent formate dehydrogenase, α subunit | 7 | − | FOX2 | AY183757 | Unpublished | |
| fdh2B | Molybdenum-dependent formate dehydrogenase, β subunit | 7 | − | FOX2 | AY183757 | Unpublished | |
| fdh2C | Molybdenum-dependent formate dehydrogenase, γ subunit | 7 | − | FOX2 | AY183757 | Unpublished | |
| fdh2D | Molybdenum-dependent formate dehydrogenase, δ subunit | 7 | − | FOX2 | AY183757 | Unpublished | |
| fdh3A | Cytochrome-linked formate dehydrogenase, α subunit | 8 | − | FOX3 | AY181035 | Unpublished | |
| fdh3B | Cytochrome-linked formate dehydrogenase, β subunit | 8 | − | FOX3 | AY181035 | Unpublished | |
| fdh3C | Cytochrome-linked formate dehydrogenase, γ subunit | 8 | − | FOX3 | AY181035 | Unpublished | |
| C1 transfer | |||||||
| dmrA | Dihydromethanopterin reductase | 9 | + | H4MPT-T | AY093431 | 28 | |
| fae | Formaldehyde activating enzyme | 2 | + | H4MPT-T | orf18 | AF032114 | 52 |
| mtdB | Methylene-H4MPT dehydrogenase | 2 | + | H4MPT-T | orfX | AF032114 | 15, 18 |
| mch | Methenyl-H4MPT cyclohydrolase | 2 | + | H4MPT-T | orfZ | AF032114 | 15, 36 |
| fhcA | Formyltransferase/hydrolase complex, α subunit | 2 | + | H4MPT-T | orf1 | AF032114 | 15, 34, 35 |
| fhcB | Formyltransferase/hydrolase complex, β subunit | 2 | + | H4MPT-T | orf2 | AF032114 | 15, 34, 35 |
| fhcC | Formyltransferase/hydrolase complex, γ subunit | 2 | + | H4MPT-T | orf3 | AF032114 | 15, 34, 35 |
| fhcD | Formyltransferase/hydrolase complex, δ subunit | 2 | + | H4MPT-T | ffsA | AF032114 | 15, 34, 35 |
| orf4 | H4MPT biosynthesis | 2 | + | H4MPT-T | AF032114 | 15, 38 | |
| orf5 | Unknown | 2 | − | H4MPT-T | AF032114 | 15 | |
| orf7 | Cofactor biosynthesis | 2 | + | H4MPT-T | AF032114 | 15 | |
| orf9 | Cofactor biosynthesis | 2 | + | H4MPT-T | AF032114 | 15 | |
| orf17 | Cofactor biosynthesis | 2 | + | H4MPT-T | AF032114 | 15 | |
| orf19 | Cofactor biosynthesis | 2 | + | H4MPT-T | AY117134 | Unpublished | |
| orf20 | Cofactor biosynthesis | 2 | + | H4MPT-T | AY117134 | Unpublished | |
| orf21 | Cofactor biosynthesis | 2 | + | H4MPT-T | AY117134 | Unpublished | |
| orf22 | Cofactor biosynthesis | 2 | + | H4MPT-T | AY117134 | Unpublished | |
| orfY | Unknown | 2 | − | H4MPT-T | AF032114 | 15 | |
| mtdA | Methylene-H4MPT/Methylene-H4F dehydrogenase | 2 | + | H4F-T/H4MPT-T | L27235 | 9, 16, 17, 51 | |
| fch | Methenyl-H4F cyclohydrolase | 2 | + | H4F-T | orf4 | L33465 | 36 |
| ftfL | Formyl-H4F ligase | 10 | + | H4F-T | 28 | ||
| folK | 2-Amino-4-hydroxy-6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase | 2 | E | H4F-S | folA | U72662 | 13 |
| folB | Dihydroneopterin aldolase | 2 | E | H4F-S | folB | U72662 | 13 |
| folP | Dihydropteroate synthase | 2 | E | H4F-S | folC | U72662 | 13 |
| folE | GTP cyclohydrolase | 11 | E | H4F-S | AY183759 | Unpublished | |
| folC | Dihydrofolate synthase | 12 | E | H4F-S | AY183760 | Unpublished | |
| dyr | Dihydrofolate reductase | 13 | NM | H4F-S | dfrA | AY093432 | 28 |
| C1 assimilation | |||||||
| sga | Serine glyoxylate aminotransferase | 2 | + | QSC | sgaA | L27235 | 9 |
| hpr | Hydroxupyruvate reductase | 2 | + | QSC | hprA | L27235 | 7, 8, 9 |
| ppc | PEP carboxylase | 2 | + | QSC | ppcA | U72662 | 4 |
| mtkA | Malate thiokinase, α subunit | 2 | + | QSC/GRC | L33465 | 10 | |
| mtkB | Malate thiokinase, β subunit | 2 | + | QSC/GRC | L33465 | 10 | |
| mcl | Malyl-CoA lyase | 2 | + | QSC/GRC | mclA | U72662 | 4, 13 |
| glyA | Serine hydroxymethyltrasferase | 14 | + | QSC | L33463 | 11 | |
| gck | Glycerate kinase | 15 | + | QSC | gckA | U87316 | 14 |
| eno | Enolase | 16 | E | QSC/GNG | AY181039 | Unpublished | |
| mdh | Malate dehydrogenase | 17 | E | QSC/TCAC | AY181040 | Unpublished | |
| qscR | Serine cycle transcriptional regulator | 18 | + | QSC | AF516903 | 20 | |
| phaA | β-ketothiolase | 19 | + | GRC/PHBC | AF287907 | 22, 23, 24 | |
| phaB | Acetoacetyl-CoA reductase | 19 | + | GRC/PHBC | AF287907 | 22, 23, 24 | |
| phaR | Regulator of PHB synthesis and acetyl-CoA flux | 19 | + | GRC/PHBC | orf3 | AF287907 | 23, 24 |
| croR | Crotonase | 20 | + | GRC | AF416777 | 22 | |
| pccA | Propionyl-CoA carboxylase, α subunit | 21 | + | GRC | AY181038 | 22 | |
| pccB | Propionyl-CoA carboxylase, β subunit | 22 | + | GRC | pccA | L48340 | 12, 22 |
| crr | Crotonyl-CoA reductase | 22 | + | GRC | adhA | L48340 | 12, 22 |
| meaA | A mutase | 22 | + | GRC | U28335 | 12, 22, 40 | |
| meaB | Essential for methylmalonyl-CoA mutase reaction | 22 | + | GRC | AF416776 | 22 | |
| meaC | Unknown | 23 | + | GRC | 28 | ||
| meaD | Unknown | 24 | + | GRC | 28 | ||
| ibd2 | Isobutyryl-CoA dehydrogenase | 25 | + | GRC | AY054980 | 22 | |
| mcmA | Methylmalonyl-CoA mutase, α subunit | 26 | + | GRC | AY181037 | 22 | |
| mcmB | Methylmalonyl-CoA mutase, β subunit | 27 | + | GRC | 22 | ||
| epm | Epimerase | 28 | + | GRC | AY183758 | Unpublished | |
| sdhA | Succinate dehydrogenase flavoprotein subunit | 29 | EG | GRC/TCAC | AY181036 | 48, 49 | |
| sdhB | Succinate dehydrogenase iron-sulfur binding subunit | 29 | NM | GRC/TCAC | AY181036 | 48, 49 | |
| sdhC | Succinate dehydrogenase cytochrome B556 subunit | 29 | NM | GRC/TCAC | AY181036 | 48, 49 | |
| sdhD | Succinate dehydrogenase membrane anchor subunit | 29 | NM | GRC/TCAC | AY181036 | 48, 49 | |
| fumA | Fumarase | 13 | EG | GRC/TCAC | AF497854 | 49 |
+, mutation impairs growth on a C1 compound; −, mutation does not impair growth on a C1 compound; NM, no mutant available: involvement in methylotrophy implicated by regulation, location, or phenotype of mutant in other subunit(s); E, essential for both methylotrophic and heterotrophic growth; EG, growth on C1 compounds rescued by glyoxylate.
Metabolic module designations: MOX, methanol oxidation; PQQ-S, PQQ biosynthesis; MAU, methylamine utilization; H4F-T, H4F-linked C1 transfer; H4F-S, H4F biosynthesis; H4MPT-TS, H4MPT-linked C1 transfer and H4MPT biosynthesis; FOX1, FOX2, and FOX3, three nonhomologous formate dehydrogenases; QSC, (Quayle) serine cycle; PHBC, PHB cycle; TCAC, TCA cycle.
FROM GENETICS TO GENOMICS
M. extorquens AM1 is the most-well-studied methylotroph to date. In the 1960s and 1970s, this organism served as a model to characterize the reactions of the serine cycle for C1 assimilation, and methanol- and methylamine dehydrogenases from M. extorquens AM1 were among the first primary C1 oxidation enzymes characterized (3). Genetic studies of M. extorquens AM1 began with the isolation and characterization of chemically induced C1-negative mutants (3). Later, mutants with defects in methanol oxidation were isolated via a specific allyl alcohol selection (32, 33). The availability of these C1-negative mutants allowed identification and isolation of the corresponding DNA regions encoding key methylotrophy genes, and it became evident that some of these genes are clustered together (2, 4, 5, 6, 8, 9, 11). As sequencing became a routine technique, these clusters were analyzed and expanded via chromosomal walking (5, 12, 15, 27). Concurrently, directed mutagenesis techniques were developed and applied to the analysis of the genes present in these methylotrophy islands (5, 8-15). By the end of 1990s, classical genetic approaches in combination with chromosomal walking and directed mutagenesis had resulted in characterization of about 70 genes involved in methylotrophy in M. extorquens AM1, and these were localized to eight regions on the chromosome (27). One methylotrophy island spans about 60 kb and contains a number of tightly linked genes enabling different methylotrophy metabolic modules: most of the reactions of the serine cycle, most of the formaldehyde oxidation reactions, and some functions involved in methanol oxidation (27). Most of the methanol oxidation genes were found in a different location on the chromosome, where they formed a large operon (1). All of the genes enabling methylamine oxidation were found in one location and tightly linked (5). However, some important methylotrophy genes were not parts of methylotrophy islands (i.e., gck and glyA) (11, 14), suggesting that further expansion of the existing methylotrophy clusters via chromosomal walking promised limited opportunity for finding new methylotrophy genes. Meanwhile, a number of essential methylotrophy genes were still missing from the picture. These included (i) two of the serine cycle genes assumed to also participate in multicarbon metabolism, encoding malate dehydrogenase and enolase; (ii) formate dehydrogenase genes; and (iii) genes for the novel glyoxylate regeneration pathway involving propionyl coenzme A (CoA) as an intermediate (12). In addition, no regulators were known for assimilatory C1 metabolism. In order to locate and study the missing methylotrophy genes in M. extorquens AM1, a whole-genome sequencing project was initiated in 1998, in collaboration with the Human Genome Sequencing Center at University of Washington and later Integrated Genomics, Inc. (Chicago, Ill.). At the time of this writing, a gapped sequence of the M. extorquens AM1 genome (6.5× coverage) is available (http://www.integratedgenomics.com/genomereleases.html#list6). Data mining began at the very early stages of the sequencing project, via BLAST analysis and key word searches against the partial genome database. At these early stages, many genes of interest were isolated and sequenced from a cosmid library of M. extorquens AM1. At the later stages when analysis of larger fragments became possible, genes of interest were PCR amplified from the chromosome and subjected to mutation analysis. A random (transposon-induced) mutagenesis approach was also employed, and sites of insertions resulting in a C1-negative phenotype were identified (28). The whole-genome-based gene-mining approach outlined above has resulted in identification of about 30 new genes involved in methylotrophy. The major outcomes of the whole-genome-analysis approach have been twofold: (i) filling in existing metabolic gaps in our knowledge of methylotrophy in serine cycle facultative methylotrophs and (ii) providing, for the first time, comprehensive knowledge on the suite of necessary genes as well as the suite of sufficient genes for enabling methylotrophy in a given organism. Our data at this time suggest that we have accounted for the majority of methylotrophy genes in M. extorquens AM1. Only a few genes still remain unidentified: namely genes involved in the yet unsolved reactions of the glyoxylate regeneration cycle and a few regulatory genes participating in C1 oxidation, C1 assimilation, or both. Below, the current information about each of the methylotrophic metabolic modules is presented in more detail.
PRIMARY C1 OXIDATION
M. extorquens AM1 possesses two primary oxidation metabolic modules for methylotrophy, which function in the oxidation of methanol and methylamine, respectively. All 11 of the known methylamine utilization genes (mauFBEDACJGIMN)—i.e., the genes for the catalytic subunits for methylamine dehydrogenase, the specific electron acceptor amicyanin, and the specific cofactor (TTQ) biosynthesis enzymes—are located in a single 8.4-kb gene cluster with all genes transcribed in the same direction (5), except the regulatory gene or genes, which remain unknown.
The genes enabling methanol oxidation in M. extorquens AM1 are found in three different locations on the chromosome. One 12.5-kb cluster (cluster 1) contains 14 genes (mxaFJGIRSACKLDEHB), all transcribed in the same direction. These genes encode the structural polypeptides of methanol dehydrogenase, the specific cytochrome c that accepts electrons from methanol dehydrogenase, the proteins essential for calcium insertion into the apoprotein, one regulatory protein, and a few proteins whose functions are still unknown (1, 2, 27, 29-31, 41, 44). One gene (mxaW) is located immediately upstream of this cluster, transcribed in the opposite direction by a methanol-inducible promoter, but its function is unknown (41). One pair of genes involved in transcriptional regulation of MeDH (mxbMD) are a part of the 60-kb methylotrophy island (cluster 2) (42), and another pair (mxcQE) are located elsewhere on the chromosome. The six genes for PQQ biosynthesis are located in two different clusters: one of them (pqqABC/DE) is located in the large methylotrophy island immediately downstream of mxbMD (45), while another cluster (pqqFG) is not linked to other methylotrophy genes (cluster 2) (43). In this work, we view PQQ biosynthesis as a separate metabolic module, because it is a cofactor of other dehydrogenases in M. extorquens AM1, based on genomic predictions.
C1 TRANSFER BETWEEN FORMALDEHYDE AND FORMATE
Two distinct metabolic modules operate in M. extorquens AM1 for transferring C1 units between the oxidation levels of formaldehyde and formate, both linked to folate cofactors. One module uses tetrahydrofolate (H4F) as a cofactor. The enzymes converting methylene-H4F to formyl-H4F, methylene-H4F dehydrogenase, and methenyl-H4F cyclohydrolase are encoded by genes (mtdA and fch) (9, 36, 51) unique to methylotrophs, while in most known bacteria, both reactions are performed by a bifunctional enzyme encoded by folD. While mtdA and fch are cotranscribed and are a part of the serine cycle gene cluster (9, 20, 36), the gene encoding formyl-H4F ligase (ftfL) is located elsewhere on the M. extorquens AM1 chromosome, and the encoded polypeptide shows high similarity to known formyl-H4F ligases (28). mtdA and fch are regulated coordinately with the serine cycle enzymes, suggesting a potential link between the H4F-linked C1 transfer module in M. extorquens AM1 and C1 assimilation (20).
In this study, we place the genes for H4F biosynthesis into a separate metabolic module, because H4F is involved not only methylotrophy functions, but also in general metabolism during growth on multicarbon compounds. fol genes (involved in folate synthesis) are therefore essential, and no null mutants can be isolated in these genes (13). Three fol genes (folKBP) are linked together, transcribed in the same direction, and are a part of a methylotrophy island (cluster 2); one gene (dyr) is loosely linked to the gene for fumarase (fumA; cluster 13); and two other genes (folC and folE) are not linked to other fol or methylotrophy genes.
The H4MPT-linked metabolic module that involves “archaeal-like” genes and enzymes appears to be the pathway responsible for the majority of formaldehyde oxidation (15). Seventeen of the genes involved in this module (fhcCDAB-orf4-mtdB-orfY-mch-orf5-orf7-fae-orf17-orf9-[3 non-“archaeal” genes]-orf19-orf20-orf21-orf22) are clustered together in a 20-kb region on the chromosome, located at one end of the largest methylotrophy island (cluster 2). Some of these genes are in the opposite orientation with respect to others, and therefore the region must be composed of a number of transcriptional units. One gene involved in this module, dmrA, encoding putative dihydromethanopterin reductase, was found in a different location on the chromosome, and was not linked to any other known methylotrophy genes (28). One other gene has been identified as a putative methanopterin biosynthesis gene, orf4, encoding the first enzyme in the methanopterin biosynthesis pathway, β-ribofuranosylaminobenzene 5′-phosphate synthase (40). dmrA and orf4 remain, at this point, the only genes proposed to participate in archaeal cofactor (H4MPT) biosynthesis in M. extorquens AM1. It is likely, however, that many of the archaeal-like genes of yet unknown function in the “archaeal-like” gene cluster are involved in biosynthesis of H4MPT or methanofuran or are involved in regulation of these biosynthetic pathways. As such, we have assigned these genes to the H4MPT-linked C1 transfer metabolic module.
FORMATE OXIDATION
Until recently, the formate oxidation step was believed to be essential for energy generation during methylotrophic growth (3). However, no randomly generated mutants were available with lesions in formate oxidation. The whole-genome approach has revealed the presence of three gene clusters unlinked to each other, encoding three nonhomologous formate dehydrogenases in M. extorquens AM1 designated fdh1AB, fdh2ABCD, and fdh3ABC (25; L. Chistoserdova, M. Laukel, J. A. Vorholt, and M. E. Lidstrom, unpublished observations). In each case, the genes are transcribed in the same orientation with respect to each other. Mutation analysis has shown that the formate oxidation step is not essential for energy generation during growth on methanol or methylamine. It is essential, however, for growth on formate, but any of the three formate oxidation modules can fulfill this energy-generating function (Chistoserdova et al., unpublished).
SERINE CYCLE
The serine cycle is the pathway for formaldehyde assimilation during methylotrophic growth of M. extorquens AM1. Even though the role of the pathway, the net production of one C3 molecule (phosphoglycerate) from two molecules of formaldehyde and one molecule of CO2, is uniquely methylotrophic, little is unique about the 11 genes involved in this module. The first enzyme in the pathway, serine hydroxymethyltransferase (GlyA) is a traditional enzyme found in most known organisms, where it functions in supplying C1 units in the form of methylene-H4F for biosynthetic pathways, for instance, purine biosynthesis. Mutants in this enzyme are normally deficient in the biosynthesis of purines (37, 47). GlyA in M. extorquens AM1, however, is specialized to methylotrophy and is not required for growth on multicarbon compounds (11), so an alternative source of C1 units must exist for purine biosynthesis. Homologs of other enzymes of the serine cycle are also found in nonmethylotrophic bacteria. It therefore seems that the functionality of the serine cycle must be determined by subtle substrate specificity adjustments for the enzymes involved and by common regulation. The glyA gene is not linked to other serine cycle genes. However, six of the serine cycle genes are clustered at one end of the large methylotrophy island (the end opposite to that containing the H4MPT module genes), together with two of the H4F-linked C1 transfer module genes (mtdA and fch), and these are transcribed in two units, sga-hpr-mtdA-fch and mtkA-mtkB-ppc-mcl (20). The serine cycle enzymes encoded by this gene cluster are serine-glyoxylate aminotransferase (sga), hydroxypyruvate reductase (hpr), the two subunits of malate thiokinase (mtkAB), an acetyl-CoA-independent phosphoenolpyruvate (PEP) carboxylase (ppc), and malyl-CoA lyase (mcl). Another serine cycle gene, gck, encoding glycerate kinase, is not linked to other serine cycle genes. Most of the serine cycle genes are regulated coordinately, but so far, only one regulator is known, QscR (20), the gene for which has been discovered via random mutagenesis (28). QscR is a LysR-type regulator with high identity to CbbR, a regulator of autotrophy and photosynthesis in other bacteria (39). The location of qscR on the chromosome, adjacent to the fructose-1,6-bisphosphatase and phosphoribulokinase genes, might be indicative of a relatively recent acquisition from an autotrophic bacterium. While most of the reactions of the serine cycle are enabled by genes specific to the methylotrophic mode of metabolism, and mutants with mutations in these genes grow normally on multicarbon compounds, two enzymes in the pathway are directly borrowed from other metabolic modules and therefore have dual functions. The genome of M. extorquens AM1 contains only one gene for malate dehydrogenase (mdh), and therefore the same enzyme functions in the serine cycle during growth on C1 compounds and in the tricarboxylic acid (TCA) cycle during growth on multicarbon compounds. Likewise, only one gene is found for enolase (eno); therefore, one and the same enzyme functions in both the serine cycle and gluconeogenesis. Mutant analysis has confirmed that both genes, mdh and eno, are essential, and no null mutants can be obtained on either C1 or multicarbon substrates (L. Chistoserdova and M. E. Lidstrom, unpublished observations).
GLYOXYLATE REGENERATION CYCLE
The glyoxylate regeneration cycle (GRC) in serine cycle methylotrophs not containing isocitrate lyase has remained a mystery for three decades. The pregenomic efforts resulted in identification of four genes loosely linked on the chromosome (cluster 22); two genes encoding polypeptides of unknown function, MeaA (12, 40) and MeaB (23); and the genes encoding crotonyl-CoA reductase and propionyl-CoA carboxylase (crr, pccB) (12, 23). The genomic approach combined with gene-specific, as well as random mutagenesis and metabolite analysis, has resulted in the identification of most of the reactions of the GRC (23, 24). The pathway involves an elaborate series of reactions proceeding via the CoA derivatives of C3, C4, and C5-carboxylic acids and involves two carboxylation and two decarboxylation reactions and at least two mutase reactions (23). A few pieces of the puzzle are still missing: i.e., the two enzymes participating in conversion of ethylmalonyl-CoA into isobutyryl-CoA (a putative mutase and a putative decarboxylase) are yet to be identified, and the substrate for MeaA, a putative mutase, remains unknown, as do the other enzymes involved in the conversion of β-hydroxyisobutyryl-CoA into propionyl-CoA (Fig. 1). Mutant analysis shows that early steps of the GRC overlap with the pathway for poly-β-hydroxbutyrate (PHB) biosynthesis (24), and the late steps overlap with late steps of the TCA cycle and the serine cycle (23). A total of 12 genes are known that are specific to this pathway (croR, crr, pccAB, ibd2, meaABCD, mcmAB, and epm), 3 that overlap with PHB biosynthesis (phaABR), 5 that overlap with the TCA cycle (sdhABCD, fumA), and 3 that overlap with the serine cycle (mtkAB, mcl). With the exception of the four genes that are part of cluster 22 and the sdh and the pha genes, the genes for the GRC are not linked to each other or to other known methylotrophy genes. Why such an elaborate pathway is employed by many serine cycle methylotrophs instead of the classic glyoxylate shunt remains unknown. However, it may in part reflect the need for low carbon flux through the initial steps of the TCA cycle during methylotrophic growth. The GRC is viewed here as a separate metabolic module, as opposed to a part of the serine cycle due to its role not only in C1, but also in C2 metabolism in M. extorquens AM1 and possibly in other bacteria. The presence of homologs for the GRC genes in other α-proteobacterial genomes points towards this pathway being widespread in non-methylotrophs, and at least in Streptomyces, the pathway has been shown to be involved in C2 metabolism (19).
PHB CYCLE
Serine cycle methylotrophs accumulate PHB as a reserve material. Metabolism of PHB in M. extorquens AM1 is intimately interlinked with its C1 metabolism. The first two reactions of the PHB cycle (catalyzed by PhaA and PhaB) are also the first reactions of the GRC (23, 24). Besides the catalytic steps, there are common regulatory mechanisms controlling both PHB production and C1 metabolism. These are not completely understood at this point, but one such common regulator, PhaR, seems to be involved in directing flows of acetyl-CoA between C1 assimilation and PHB accumulation (23).
TCA CYCLE
During growth on multicarbon compounds, the TCA cycle plays its classical role in carbon and energy metabolism in M. extorquens AM1 (3). In contrast, it is not involved in energy generation during growth on C1 compounds. Instead, α-ketoglutarate dehydrogenase is repressed, and the incomplete cycle plays a strictly assimilatory role (3, 48, 49). However, many of the TCA cycle enzymes are involved in C1 assimilation. Besides malate dehydrogenase, a series of the TCA cycle reactions converting succinyl-CoA into malate form a part of the GRC. Genes encoding two enzyme systems capable of converting succinyl-CoA into succinate have been identified in the genome of M. extorquens AM1: for succinyl-CoA synthase (the genes scsA and scsB are linked to mdh) and keto acid succinyl-CoA transferase (kst). In addition, cell extracts contain a succinyl-CoA hydrolase activity, but the gene responsible for this activity is unknown (Chistoserdova and Lidstrom, unpublished). Null mutations in scsB and kst caused no effect on growth of M. extorquens AM1 on C1 or multicarbon compounds (Chistoserdova and Lidstrom, unpublished). This result points toward either succinyl-CoA hydrolase being the essential enzyme for this conversion or the three systems being degenerate for this function. The genes for succinate dehydrogenase (SDH) are all linked on the chromosome (cluster 29), and SDH null mutants cannot be obtained either on methanol or on succinate (48, 49). However, they can be obtained on methanol supplemented with glyoxylate (N. Korotkova and M. E. Lidstrom, unpublished observations). This growth condition circumvents the GRC, confirming the hypothesis that SDH is involved in glyoxylate regeneration during growth on C1 compounds. Two genes showing homology to fumarases (fumA and fumB) are present in the genome. One of the potential fumarase genes (fumA) has been subjected to mutagenesis, and, as in the case of SDH, null mutants could only be obtained on methanol supplemented with glyoxylate (Korotkova and Lidstrom, unpublished), confirming the dual function of this enzyme in C1 and multicarbon metabolism. The role of the second homolog of fumarase remains unknown.
CONCLUSIONS
We have presented here the genome-based analysis of methylotrophic metabolism in one model facultative methylotroph, M. extorquens AM1. This analysis presents a comprehensive picture of the complex genetic and biochemical makeup of methylotrophy in a given organism. A little over 100 genes participate in C1 metabolism in M. extorquens AM1, and these belong to a few specialized metabolic modules. Some genes involved in these modules are located in “methylotrophy islands,” while others are scattered around the chromosome and are present as singular entities. While some genes are specialized in methylotrophic metabolism, others are shared with nonmethylotrophic pathways. Such a complex framework of methylotrophy in M. extorquens AM1 may reflect a complex and nonlinear history of this metabolic capability. Because a nearly complete set of genes involved in the different steps of methylotrophic metabolism in M. extorquens AM1 is now defined, the stage is now set for studies directed at understanding how this complex network of genes and enzymes is coordinated and what are the important mechanisms for switching between C1 and multicarbon metabolic modes in this organism.
REFERENCES
- 1.Amaratunga, K., P. M. Goodwin, C. D. O'Connor, and C. Anthony. 1997. The methanol oxidation genes mxaFJGIR(S)ACKLD in Methylobacterium extorquens. FEMS Microbiol. Lett. 146:31-38. (Erratum, 150:175-177.). [DOI] [PubMed]
- 2.Anderson, D. J., C. J. Morris, D. N. Nunn, C. Anthony, and M. E. Lidstrom. 1990. Nucleotide sequence of the Methylobacterium extorquens AM1 moxF and moxJ genes involved in methanol oxidation. Gene 90:173-176. [DOI] [PubMed] [Google Scholar]
- 3.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 4.Arps, P. J., G. F. Fulton, E. C. Minnich, and M. E. Lidstrom. 1993. Genetics of serine pathway enzymes in Methylobacterium extorquens AM1: phosphoenolpyruvate carboxylase and malyl coenzyme A lyase. J. Bacteriol. 175:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistoserdov, A. Y., L. V. Chistoserdova, W. S. McIntire, and M. E. Lidstrom. 1994. Genetic organization of the mau cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J. Bacteriol. 176:4052-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chistoserdova, L. V. 1996. Metabolism of formaldehyde in M. extorquens AM1. Molecular genetic analysis and mutant characterization, p. 16-24. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Chistoserdova, L. V., and M. E. Lidstrom. 1991. Purification and characterization of hydroxypyruvate reductase from the facultative methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 173:7228-7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chistoserdova, L. V., and M. E. Lidstrom. 1992. Cloning, mutagenesis, and physiological effect of a hydroxypyruvate reductase gene from Methylobacterium extorquens AM1. J. Bacteriol. 174:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chistoserdova, L. V., and M. E. Lidstrom. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification of sgaA and mtdA and sequences of sgaA, hprA, and mtdA. J. Bacteriol. 176:1957-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chistoserdova, L. V., and M. E. Lidstrom. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification, sequence, and mutation of three new genes involved in C1 assimilation, orf4, mtkA, and mtkB. J. Bacteriol. 176:7398-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chistoserdova, L. V., and M. E. Lidstrom. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: cloning, sequence, mutation, and physiological effect of glyA, the gene for serine hydroxymethyltransferase. J. Bacteriol. 176:6759-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chistoserdova, L. V., and M. E. Lidstrom. 1996. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA into glyoxylate in the isocitrate-lyase-negative methylotroph, Methylobacterium extorquens AM1. Microbiology 142:1459-1468. [DOI] [PubMed] [Google Scholar]
- 13.Chistoserdova, L. V., and M. E. Lidstrom. 1997. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology 143:1729-1736. [DOI] [PubMed] [Google Scholar]
- 14.Chistoserdova, L. V., and M. E. Lidstrom. 1997. Identification and mutation of a gene required for glycerate kinase activity from a facultative methylotroph, Methylobacterium extorquens AM1. J. Bacteriol. 179:4946-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chistoserdova, L. V., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 16.Ermler, U., C. H. Hagemeier, A. Roth, U. Demmer, W. Grabarse, E. Warkentin, and J. A. Vorholt. 2002. Structure of methylene-H4MPT dehydrogenase from Methylobacterium extorquens AM1. Structure 10:11271.137. [DOI] [PubMed]
- 17.Hagemeier, C. H., S. Bartoschek, C. Griesinger, R. K. Thauer, and J. A. Vorholt. 2001. Re-face stereospecificity of NADP dependent methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 as determined by NMR spectroscopy. FEBS Lett. 494:95-98. [DOI] [PubMed] [Google Scholar]
- 18.Hagemeier, C. H., L. Chistoserdova, M. E. Lidstrom, R. K. Thauer, and J. A. Vorholt. 2000. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur. J. Biochem. 267:3762-3769. [DOI] [PubMed] [Google Scholar]
- 19.Han, L., and K. A. Reynolds. 1997. A novel alternative anaplerotic pathway to the glyoxylate cycle in streptomycetes. J. Bacteriol. 179:5157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalyuzhnaya, M., and M. E. Lidstrom. 2003. QscR, a LysR-type transcriptional regulator and CbbR homolog, is involved in regulation of the serine cycle genes in Methylobacterium extorquens AM1. J. Bacteriol. 185:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, D. P., and J. C. Murrell. 1999. Microbial metabolism of methanesulfonic acid. Arch. Microbiol. 172:341-348. [DOI] [PubMed] [Google Scholar]
- 22.Korotkova, N., L. Chistoserdova, V. Kuksa, and M. E. Lidstrom. 2002. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 184:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korotkova, N., L. Chistoserdova, and M. E. Lidstrom. 2002. Poly-β-hydroxybutyrate biosynthesis in the facultative methylotroph Methylobacterium extorquens AM1: identification and mutation of gap11, gap20, and phaR. J. Bacteriol. 184:6174-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korotkova, N., and M. E. Lidstrom. 2001. A connection between poly-β-hydroxybutyrate biosynthesis and growth on C1 and C2 compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 183:1038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laukel, M., L. Chistoserdova, M. E. Lidstrom, and J. A. Vorholt. 2003. The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: purification and properties. Eur. J. Biochem. 270:325-333. [DOI] [PubMed] [Google Scholar]
- 26.Lidstrom, M. E., C. Anthony, F. Biville, F. Gasser, P. Goodwin, R. S. Hanson, and N. Harms. 1994. New unified nomenclature for genes involved in the oxidation of methanol in Gram-negative bacteria. FEMS Microbiol. Lett. 117:103-106. [DOI] [PubMed] [Google Scholar]
- 27.Lidstrom, M. E., L. Chistoserdova, S. Stoliar, and A. L. Springer. 1998. Genetics and regulation of C1 metabolism in methylotrophs, p. 121-134. In E. Vijgenboom and G. Canters (ed.), NATO Symposium on Electron Transfer Chains.
- 28.Marx, C. J., B. N. O'Brien, J. Breezee, and M. E. Lidstrom. 2003. Novel methylotrophy genes of Methylobacterium extorquens AM1 identified by using transposon mutagenesis including a putative dihydromethanopterin reductase. J. Bacteriol. 185:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris, C. J., Y. M. Kim, K. E. Perkins, and M. E. Lidstrom. 1995. Identification and nucleotide sequences of mxaA, mxaC, mxaK, mxaL, and mxaD genes from Methylobacterium extorquens AM1. J. Bacteriol. 177:6825-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunn, D. N., and C. Anthony. 1988. The nucleotide sequence and deduced amino acid sequence of the genes for cytochrome cL and a hypothetical second subunit of the methanol dehydrogenase of Methylobacterium extorquens AM1. Nucleic Acids Res. 16:7722.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nunn, D. N., D. Day, and C. Anthony. 1989. The second subunit of methanol dehydrogenase of Methylobacterium extorquens AM1. Biochem. J. 260:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunn, D. N., and M. E. Lidstrom. 1986. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J. Bacteriol. 166:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunn, D. N., and M. E. Lidstrom. 1986. Phenotypic characterization of 10 methanol oxidation mutant classes in Methylobacterium sp. strain AM1. J. Bacteriol. 166:591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomper, B. K., O. Saurel, A. Milon, and J. A. Vorholt. 2002. Generation of formate by the formyltransferase/hydrolase complex (Fhc) from Methylobacterium extorquens AM1. FEBS Lett. 523:133-137. [DOI] [PubMed] [Google Scholar]
- 35.Pomper, B. K., and J. A. Vorholt. 2001. Characterization of the formyltransferase (Ftr) from Methylobacterium extorquens AM1. Eur. J. Biochem. 269:4769-4775. [DOI] [PubMed] [Google Scholar]
- 36.Pomper, B. K., J. A. Vorholt, L. Chistoserdova, M. E. Lidstrom, and R. K. Thauer. 1999. A methenyl tetrahydromethanopterin cyclohydrolase and a methenyl tetrahydrofolate cyclohydrolase in Methylobacterium extorquens AM1. Eur. J. Biochem. 261:475-480. [DOI] [PubMed] [Google Scholar]
- 37.Rossbach, S., and H. Hennecke. 1991. Identification of glyA as a symbiotically essential gene in Bradyrhizobium japonicum. Mol. Microbiol. 5:39-47. [DOI] [PubMed] [Google Scholar]
- 38.Scott, J. W., and M. E. Rasche. 2002. Purification, overproduction, and partial characterization of β-RFAP synthase, a key enzyme in the methanopterin biosynthesis pathway. J. Bacteriol. 184:4442-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 40.Smith, L. M., W. G. Meijer, L. Dijkhuizen, and P. M. Goodwin. 1996. A protein having similarity with methylmalonyl-CoA mutase is required for the assimilation of methanol and ethanol by Methylobacterium extorquens AM1. Microbiology 142:675-684. [DOI] [PubMed] [Google Scholar]
- 41.Springer, A. L., A. J. Auman, and M. E. Lidstrom. 1998. Sequence and characterization of mxaB, a response regulator involved in regulation of methanol oxidation, and of mxaW, a methanol-regulated gene in Methylobacterium extorquens AM1. FEMS Microbiol. Lett. 160:119-124. [DOI] [PubMed] [Google Scholar]
- 42.Springer, A. L., C. J. Morris, and M. E. Lidstrom. 1997. Molecular analysis of mxbD and mxbM, a putative sensor-regulator pair for oxidation of methanol in Methylobacterium extorquens AM1. Microbiology 143:1737-1744. [DOI] [PubMed] [Google Scholar]
- 43.Springer, A. L., R. Ramamoorthi, and M. E. Lidstrom. 1996. Characterization and nucleotide sequence of pqqE and pqqF in Methylobacterium extorquens AM1. J. Bacteriol. 178:2154-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyama, H., C. Anthony, and M. E. Lidstrom. 1998. Construction of insertion and deletion mxa mutants of Methylobacterium extorquens AM1 by electroporation. FEMS Microbiol. Lett. 166:1-7. [DOI] [PubMed] [Google Scholar]
- 45.Toyama, H., L. L. Chistoserdova, and M. E. Lidstrom. 1997. Sequence analysis of pqq genes required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1 and the purification of a biosynthetic intermediate. Microbiology 143:595-602. [DOI] [PubMed] [Google Scholar]
- 46.Toyama, H., and M. E. Lidstrom. 1998. pqqA is not required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1. Microbiology 144:183-191. [DOI] [PubMed] [Google Scholar]
- 47.Umbarger, H. E., M. A. Umbarger, and P. M. L. Siu. 1963. Biosynthesis of serine in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 85:1431-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Dien, S. J., and M. E. Lidstrom. 2002. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C(3) and C(4) metabolism. Biotechnol. Bioeng. 78:296-312. [DOI] [PubMed] [Google Scholar]
- 49.VanDien, S. J., Y. Okubo, M. T. Hough, N. Korotkova, T. Taitano, and M. E. Lidstrom. 2003.. Reconstruction of C3 and C4 metabolism in Methylobacterium extorquens AM1 using transposon mutagenesis. Microbiology 149:601-609. [DOI] [PubMed]
- 50.Vannelli, T., M. Messmer, A. Studer, S. Vuilleumier, and T. Leisinger. 1999. A corrinoid-dependent catabolic pathway for growth of a Methylobacterium strain with chloromethane. Proc. Natl. Acad. Sci. USA 96:4615-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vorholt, J. A., L. Chistoserdova, M. E. Lidstrom, and R. K. Thauer. 1998. The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J. Bacteriol. 180:5351-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vorholt, J. A., L. Chistoserdova, S. M. Stolyar, R. K. Thauer, and M. E. Lidstrom. 1999. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J. Bacteriol. 181:5750-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]