Abstract
We have developed a RecA-mediated simple, rapid and scalable method for identifying novel alternatively spliced full-length cDNA candidates. This method is based on the principle that RecA proteins allow to carry radioisotope-labeled probe DNAs to their homologous sequences, resulting in forming triplexes. The resulting complex is easily detected by mobility difference on electrophoresis. We applied this exon profiling method to four selected mouse genes as a feasibility study. To design probes for detection, the information on known exonic regions was extracted from public database, RefSeq. Concerning the potentially transcribed novel exonic regions, RNA mapping experiment using Affymetrix tiling array was performed. As a result, we were able to identify alternative splice variants of Thioredoxin domain containing 5, Interleukin1β, Interleukin 1 family 6 and glutamine-rich hypothetical protein. In addition, full-length sequencing demonstrated that our method could profile exon structures with >90% accuracy. This reliable method can allow us to screen novel splice variants from a huge number of cDNA clone set effectively.
INTRODUCTION
It is known that most mammalian genes are subjected to alternatively splicing, probably to maximize the diversity of proteins from the unexpectedly small number of genes encoded by the genome (1–4). Many alternative splicing transcripts have been reported to play critical roles in living cells, e.g. in disease cascades (5,6). Conventionally, pairwise comparison of full-length cDNAs (7–12) and expressed sequence tags (ESTs) have been used to identify alternative transcripts. As a tool for expression profiling analysis, DNA microarray was innovated (13) and has been a widely used tool in finding responsible genes for various diseases (14). Recently, exon junction arrays (15) and oligonucleotide tiling arrays (10,16–19) have also been employed to study alternatively spliced transcripts. More recently, the newly developedexon arrays promise to identify alternative exons genome-wide. Although these methods are powerful tools to help us to identify individual alternative exons, it is still difficult to determine the entire exon structure including the information on exon boundaries, which is indispensable for functional analysis. From this aspect, collecting physical cDNAs is still the only solution for acquiring the whole structure of transcripts.
With the aim to describe the whole picture of transcriptome, several cDNA projects have been conducted intensively. They, however, emphasize on identifying novel genes, and most of collected cDNA clones were clustered based on their EST information and stored without being fully sequenced. In the case of our RIKEN full-length mouse cDNA project and FANTOM activities (20,21), a few millions of full-length enriched cDNA clones have been collected, but only ∼100 000 representative cDNA clones were fully sequenced and functionally annotated. This fact implies that current cDNA projects unveiled just one tiny part of the transcriptome. In addition, recent analyses showed that >40% of genes were subjected to alternative splicing, producing various protein isoforms (12,22,23). Taken together, a huge number of splice variants are potentially stored among clustered cDNA clones.
To shed light on potential alternative splice variants within the huge cDNA clone set, the most accurate and reliable way is to determine the entire sequence of all transcripts by sequencing. However, it is time-consuming and cost-consuming procedure and far from the reality. At present, $100 000 and $1000 genome projects are ongoing to develop next-generation type sequencers. Some pioneering sequencers are getting available (24), but these sequencers usually take shotgun-like sequencing strategy and would not be suitable for full-length cDNA sequencing since it requires the correspondence of each clone to each sequence data. Therefore, if a novel and simple method for exon profiling is available, it would be useful to screen alternatively spliced transcripts from the huge cDNA clone set.
To this end, we have developed a simple RecA-mediated exon profiling method to investigate exon structures without sequencing. In this method, the sequence-specificity of homologous recombinase, RecA protein, was utilized (25–27). Radioisotope-labeled ‘probe’ oligonucleotides werefirst carried to their homologous sequences by RecA protein in a sequence-specific manner, resulting in pairing at the same sequences between the single-stranded (ss) and double-stranded (ds) DNA molecules. Although there is no stable triple-stranded DNA formation since RecA filaments within the strands are already exchanged (28), we call the paired DNA complexes as ‘triplex’. The triplexes were detectedby mobility difference on electrophoresis. Full-length sequencing demonstrated that our method was able to detect exonic regions with high accuracy. Moreover, several novel splice variants were successfully identified with this method. Here we report the principle of the simple and reliable RecA-mediated method for exon profiling.
MATERIALS AND METHODS
The sequence information on fully sequenced cDNAs in this study has been deposited to DDBJ with the library name, RMEP. DDBJ accession nos = AK224911-AK225023.
Optimization of RecA-mediated exon profiling conditions
Oligonucleotides and target DNA
Oligonucleotides with identical and non-identical sequences to pUC19 (‘Target’ DNA) were designed from 10mer to 50mer with the increment of 10 bases. The pUC19 was used as a model to optimize the detection conditions. The oligonucleotides for detection are defined as probes in this study. After designing probes, a BLAST search (29) was performed to investigate whether unintended homologous sequences were present in target DNA sequence. To avoid cross hybridization, the most specific probes to the target sequences were carefully selected. The probe sequences are listed in Supplementary Table S1. The pUC19 DNA was purified with Mini Prep kit according to the manufacture's protocol (Qiagen). Oligonucleotides were purchased from Operon Biotechnology, Inc.
Labeling of oligonucleotides
Oligonucleotides were labeled at the 5′-terminus with T4 polynucleotide kinase (TaKaRa-Bio) in the presence of [γ-32P]ATP (3000 Ci/mmol; GE Healthcare Bio-Sciences Corp.) by incubating for 30 min at 37°C. To terminate the labeling reaction and to purify, 50 µl of resultant mixture was, then, mixed with 1 µl of 0.5 M EDTA, 1 µl of 10% SDS, and 1 µl of 20 mg/ml Proteinase K and incubated for 30 min at 45°C followed by phenol-chloroform extraction. Then, the purified labeled DNA solution was applied to Microspin S-400 HR columns (GE Healthcare Bio-Sciences Corp.) to remove excess of [γ-32P]ATP.
The RecA-mediated triplex formation analysis
The probe DNAs were diluted to ∼1 pmol/l and the triplex formation reaction was performed in 20 µl of the solution containing 25 mM Tris–acetate, 5 mM Magnesium acetate, 1 mM DTT and 0.1 mM ATPγS (Roche Diagnostics), 3 µg of Escherichia coli RecA protein (New England Biolabs) and 50 ng of pUC19 plasmid. The reaction mixture was incubated for 1 h at 37°C. The reaction was terminated by placing on ice and adding 6× loading dye, which is composed of 10 mM Tris–HCl (pH 7.6), 0.03% bromophenol blue, 0.03% xylene cyanol FF, 60% glycerol and 60 mM EDTA; and then it was followed by 1.2% agarose gel electrophoresis in 1× TAE buffer. The gel was dried with a gel drier (Bio-Rad Laboratories), and exposed to an IP sheet (Fuji Photo Film Co., Ltd) for 3–5 h. The exposed sheet was analyzed by BAS2500 (Fuji Photo Film Co., Ltd).
The RecA-mediated exon profiling analysis using RIKEN clone sets
Selection of drug-related genes from the RIKEN Clone Bank
The RIKEN clone sets were used for this analysis. Four human genes, which were related to commercially available drugs, were randomly selected. With the sequences of these four drug-related genes, the mouse orthologs to these genes were searched with HomoloGene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=homologene). The RIKEN cDNA clones corresponding to these mouse orthologs were computationally identified using their gene locus information. When necessary, Gene Ontology information was used to select the RIKEN cDNA clones that included similar functional domains to drug-related genes. The E.coli carrying the selected cDNA sequences in plasmids were cultivated, and the plasmids were purified with Qiagen Mini preparation kit (Qiagen) for further analyses.
RNA mapping experiment with tiling arrays
To obtain the information on novel exon candidates, we used the GeneChip Mouse Tiling Array ver. 1.0 (Affymetrix). The RNAs, which were used for cDNA library construction in the RIKEN mouse encyclopedia project, were utilized for microarray-based RNA mapping. A total of 66 different RNA samples with the same amount were mixed and 10 µg of the mixed RNA were used for hybridization. The tissue/cell names of the mixed 66 RNAs are provided in Supplementary Table S2. The first-strand cDNA was synthesized with random primers using SuperScript II (Invitrogen). The resulting cDNAs were then used as template for the second-strand cDNA synthesis under conditions described in the manufacture's protocol. After the second-strand synthesis, the remaining RNAs were degraded using a combination of RNaseA/T1 cocktail (Ambion) and RNaseH (Ambion). The dscDNAs were then purified with PCR purification kit (Qiagen, Inc.) and fragmented with DNase I, and end-labeled with biotin as described previously (10). One set of whole genomic tiling array comprises 16 separate chips. For each chip, ∼4 µg of the end-labeled cDNA samples were applied. The labeled DNAs were hybridized to the array chips for 16–18 h at 45°C. The array chips were washed as per the manufacture's protocol, and scanned using a GeneChip Scanner 3000 7G (Affymetrix, Inc). The hybridization experiments were carried out in duplicate.
Construction of transcription map for novel exonic region
The signal intensity at each probe was computed using the Affymetrix software, TAS. A Wilcoxon signed-rank testwas applied to the hybridization intensities (background-subtracted intensity, PM–MM, where PM and MM indicate intensities detected by a pair of 25mers matching perfectly and onebase mismatching to the genome, respectively) measured at probes located within ±50 bp of every probe location. The pseudo median generated by the Wilcoxon test was assigned as an estimate of the signal intensity to the probe position. The signal intensity was given on log2 scale as shown in Figure 3.
The graphs from these processed data were generated by INTEGRATED GENOME BROWSER software (Affymetrix, Inc.). The transfrags, genomic regions where a significant level of expression was detected, were identified by joining positive probes (signal intensity >7.4) separated by less than a certain distance (maxgap = 100 bp) and selecting regions with an extension of ≥30 bp (minrun).
Probe design and exon profiling analysis
Oligonucleotide probes were designed according to the following work order: (i) Extraction of known and candidate exonic regions for the drug-related genes from RefSeq and transfrag data (transcriptional fragments data from tiling arrays). (ii) Designing of all possible combinations of 30mer oligonucleotide sequence probes within each extracted exonic region. (iii) Comparison of each 30mer oligonucleotide sequence with the cloning vector sequence by BLAST. The most specific oligonucleotide sequences for detection were chosen. All oligonucleotides were purchased from Operon Biotechnology, and the probes for RIKEN clone sets were prepared as described above. Exon profiling for RIKEN clone sets was performed as described above, except that 96-well plates were used (Abgene, Inc.).
Full-length cDNA sequence determination
All cDNA clones clustered within the Txndc5 locus and all alternative splice variant candidates in IL-1β, IL-1F6 and glutamine-rich hypothetical protein (GR) were fully sequenced by using an ABI3700 capillary sequencer (Applied Biosystems) and Big Dye terminator sequencing kit (Applied Biosystems). For sequencing, primers were designed for every 300 bp on the sequence of fully sequenced representative clones. Their sequences were base-called by Phred (30) and assembled by Phrap (31).
Further characterization of newly identified splice variants. The sequences of splice variant candidates were compared to sequences in genetic databases; GeneBank, the Mouse Genome Database and TIGR databases. The comparisonwas performed using the nucleotide-to-nucleotide sequence local alignment algorithm, BLASTN. In addition, deduced amino acid sequences for longest ORF were computationally determined. Finally, the splice variant candidates were analyzed with the SCOP (32), Pfam (33) and InterPro (34,35) programs for identifying potential functional domains. SOSUI program (36) was also applied for transmembrane protein prediction.
RESULTS
Principle of RecA-mediated exon profiling method
In this study, radioisotope-labeled ss oligonucleotides were defined as probes, and dsDNAs containing their homologous sequences were called as targets. The principle of our simple exon profiling method utilizing RecA protein is described in Figure 1. At the first step, radioisotope-labeled oligonucleotide probes were incubated with RecA protein and Adenosine 5′-O-(3-thio)triphosphate (ATPγS), which stabilizes the interaction of a short ss oligonucleotide with dsDNA (37,38). The resulting complex was further incubated with dsDNA to pair with its homologous sequence. And then, the paired complex, ‘triplex’ formation was evaluated by mobility difference on agarose electrophoresis and by autoradiography.
Figure 1.
Principle of RecA-mediated exon profiling method. Oligonucleotides were designed for exon candidate regions. (a) Incubation of the labeled oligonucletides (probes) with RecA, ATPγS and Mg2+ for 5 min at 37°C. (b) Incubation for 1 h at 37°C after adding dsDNA (e.g. pUC19) to the reaction mixture. (c) Electrophoresis and autoradiography for detection. The shifted band with slower migration represents triplex formation, indicating the presence of the sequence corresponding to the probe (arrow I). The bands for the probes without forming the triplex are represented as faster migration (arrow II).
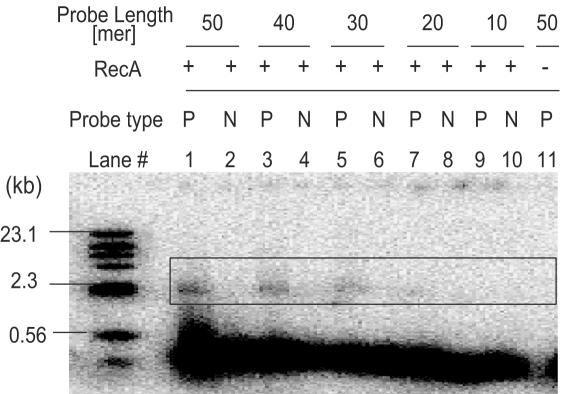
Examination of probe length in RecA-mediated exon profiling method
Our computational analysis using the FANTOM cDNA clone set showed that out of 126 322 internal exons ∼93% of the exons were longer than 50 bp and 99.3% of the exons were longer than 30 bp (S. Kondo, unpublished data), indicating that the detection for exons shorter than 50 bp is required. To determine the minimum probe length in our method, various probes in length were examined using pUC19 as a target dsDNA.
Each probe DNA labeled with 32P at the 5′ end was incubated with RecA, ATPγS and pUC19 DNA, as described in Materials and Methods. The resultant products were electrophoresed, and signals were detected by autoradiography. As shown in Figure 2, the RecA-mediated triplex formation corresponding to the presence of probe sequence was detected with mobility difference on electrophoresis. Although the signals derived from triplexes were detected down to 10mer probes, the signals using 20mer and 10mer probes were faint and seemed not to be reliable (Figure 2, lanes 7 and 9). As for non-homologous probes, no mobility change was detected (Figure 2, lanes 2, 4, 6, 8 and 10), indicating that triplex formation with pUC19 DNA occurred in a sequence-specific manner. In addition, it was also verified that the triplex formation took place in a RecA protein-dependent manner (Figure 2, lane 11). Altogether, we decided to utilize 30mer probes for further analyses, and it means that our method is able to detect >99% of internal exons theoretically.
Figure 2.
Examination of probe length with RecA-mediated exon profiling method. Two sets of 50, 40, 30, 20 and 10mer probes were prepared; identical (P) and non-identical (N) to pUC19 DNA. Identical (P) probes with 50, 40, 30, 20 and 10mer probes were examined in lanes 1, 3, 5, 7 and 9, respectively. Non-identical (N) probes with 50, 40, 30, 20 and 10mer were examined in lanes 2, 4, 6, 8 and 10, respectively. In lane 11, the reaction without RecA protein was examined. Open box indicates the position of the band corresponding to triplex. The DNA size marker (λHind III digests) is shown at the left with the approximate molecular sizes in kb. The sequences for all the probes are listed in Supplementary Table S1.
Selection of gene loci for evaluation
To evaluate the RecA-mediated exon profiling method, we next selected four drug-related gene loci for exon profiling from the RIKEN clone bank: Thioredoxin domain containing 5 (Txndc5), Interleukin1 beta (IL-1β), Interleukin 1 family 6 (IL-1F6) and glutamine-rich hypothetical protein. Through our FANTOM activity, all collected cDNA clones were clustered with their EST information on their genomic loci, intending to encompass all overlapping transcripts derived from the same strand of a single gene locus (21). These clusters are called as transcriptional units (TUs) (21). All cDNA clones clustered within the four selected TUs were computationally identified. As shown in Table 1, a total of 139 clones were clustered into the four loci, and the Txndc5, IL-1β, IL-1F6 and glutamine-rich hypothetical protein loci contained 95, 14, 17 and 13 clones, respectively (Table 1 and Supplementary Tables S7–S10).
Table 1.
Summary of exon profiling
| Annotated name/Gene ID/symbol | Total clones | Total exons | Novel exon candidate | Total probes | Total assays |
|---|---|---|---|---|---|
| Thioredoxin domain containing 5 (Txndc5)/105245/Txndc5 | 95 | 10 | 3 | 16 | 1520 |
| Interleukin 1 beta (IL-1β)/16176/Il1b | 14 | 7 | 2 | 14 | 196 |
| Interleukin 1 family, member 6/(IL1-F6)/54448/Il1f6 | 17 | 6 | 3 | 14 | 238 |
| glutamine-rich hypothetical protein (GR)/73332/1700041C02Rik | 13 | 22 | 28 | 50 | 650 |
| Total | 139 | 45 | 36 | 94 | 2604 |
Extraction of known exons and candidate exons
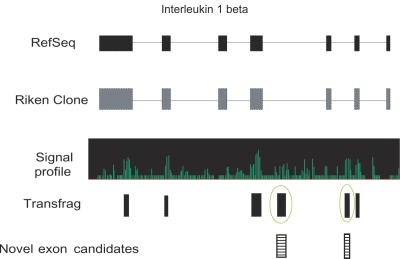
To design probes for exon profiling, all exons were divided into two categories: known exons and candidate exons. The known exons were extracted in silico from RefSeq for the four gene loci. As for the candidate exons, RNA mapping using Affymetrix mouse whole genome tiling array was performed. The RNAs from 66 different tissues/cells were mixed and used for hybridization. We generated the signal intensity using the standard tiling array protocol implemented in the Affymetrix software, TAS (17). The transcription fragments (transfrags) with signal intensity above 7.4 on four different loci were generated as potentially transcribed regions. The transfrags, RefSeq and the RIKEN FANTOM representative clones were aligned on Affymetrix IGB viewer, and the genomic positions of transfrags and those of the known exons from RefSeq and fully sequenced representative clones were compared (Figure 3). The transfrags, which did not overlap with any known exons or which were different in length from the known exons, were considered as novel candidate exons. Number of extracted candidate exons on each gene is summarized in Table 1.
Figure 3.
Selection of novel exon candidates. IL-1β locus is shown as an example. Exon structures of RefSeq, RIKEN full-length cDNA clone and transfrags from tiling array experiments are aligned. Profile of signal intensity by tiling array is shown in the middle. Exons and introns are represented by closed boxes and lines. The transfrags were generated from the array data at signal intensity 7.4. The transfrags which did not overlap with any known exonic regions were considered as novel exon candidates (striped boxes).
Although RNA mapping data using tiling arrays can include false positive transfrags, all transfrag data were used for designing probes because the purpose of this study was to identify alternatively splice variants as many as possible. Within the selected four gene loci, the number of the known exons was 45, and that of novel exon candidates was 36. In this study, one 30mer probe was designed for each 100 bp. If an exon exceeded 100 bp in length, each probe was designed near the ends of exons to improve the efficiency of detection for different exon length. For the four gene loci, 58 probes and 36 probes were designed for known exons and novel candidate exons, respectively.
Evaluation of RecA-mediated exon profiling
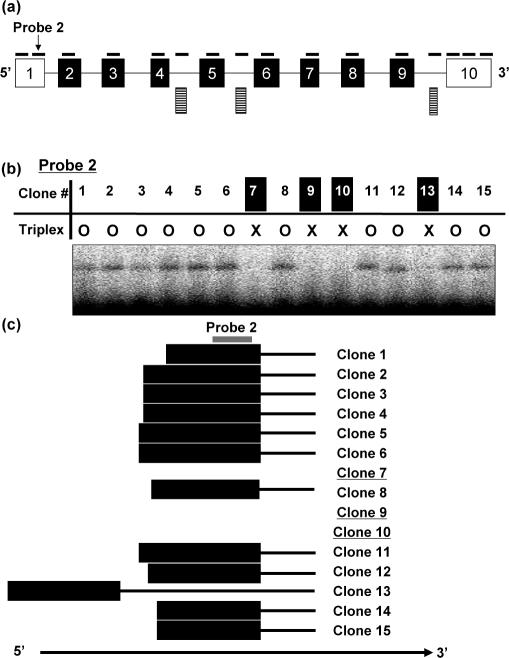
As the first step, we evaluated our RecA-mediated exon profiling method by using the RIKEN cDNA clones clustered within Txndc5 locus. In this locus, 95 RIKEN cDNA clones were entried (Supplementary Table S7). For exon profiling, 13 probes for 10 known exons and 3 probes for 3 novel candidate exons were designed with following the rule described above. The probe sequences are represented in Supplementary Table S3. In total, 1520 assays (=95 cDNA clones × 16 probes) were performed with the RecA-mediated method. One typical result is shown in Figure 4b. To evaluate the method precisely, the full-length sequences of the 95 cDNA clones were determined by sequencing and were compared with the results from our method. Out of 95 cDNA clones, 25 clones were alternative splice variants.
Figure 4.
Exon profiling in the Txndc5 locus. (a) Exon structure of a known clone (GeneID: 105245) in the Txndc5 locus is represented in arbitrary scale. Boxes and lines represent exons and introns, respectively. Closed boxes indicate the region of protein-coding sequence. Solid lines above exonic regions indicate probes. Striped boxes represent novel exon candidates. (b) Typical result of RecA-mediated exon profiling method. The result using probe 2 designed on the first exon is represented. In the table, circles and crosses indicate presence and absence of the triplex formation, respectively. (c) Schematic presentation of the first exons of Txndc5 clones. To clarify the figure, 15 clones were randomly selected. The probe 2 designed on exon 1 is indicated with the line in gray. The clones 7, 9 and 10 with underlines were proved to use an alternative exon downstream as a first exon.
In this study, the specificity was defined as ‘the number of assays in which the presence/absence of probe sequences was correctly detected by our method’ over ‘the number of total assays’. Concerning the sensitivity, it was defined as ‘the number of assays in which triple-stranded complex formations were detected by RecA-mediated method’ over ‘the number of assays in which triplex formations should be detected due to full-length sequencing’. With these definitions, the specificity and the sensitivity of our RecA-mediated method were calculated as 93.3% [(1520) − (15 + 86)]/(1520) and 91.9% (976/1062), respectively. Result is summarized in Table 2.
Table 2.
Summary of exon profiling on Thioredoxin domain containing 5 locus
| Thioredoxin domain containing 5 | ||
|---|---|---|
| Specificity | 1419/1520 | 93.3% |
| Sensitivity | 976/1062 | 91.9% |
| Number of newly identified splice patterns | 13 | |
| Number of newly identified splice variants | 25 |
Identification of alternative splice variants on three selected gene loci
Since the reliability of this method could be confirmed as above, we further applied this method to three other selected gene loci, IL-1β, IL-1F6 and glutamine-rich hypothetical protein. As for these three gene loci, only splice variant candidates which were selected by the RecA-mediated method were fully sequenced. The result of the exon profiling on these three loci was summarized in Table 3.
Table 3.
Summary of exon profiling on IL-1β, IL-1F6 and glutamine-rich hypothetical protein
| Locus name | IL-1β | IL-1F6 | GR |
|---|---|---|---|
| Number of splice variant candidates | 3 | 2 | 7 |
| Number of newly identified splice patterns | 2 | 1 | 3 |
| Number of newly identified splice variants | 2 | 2 | 4 |
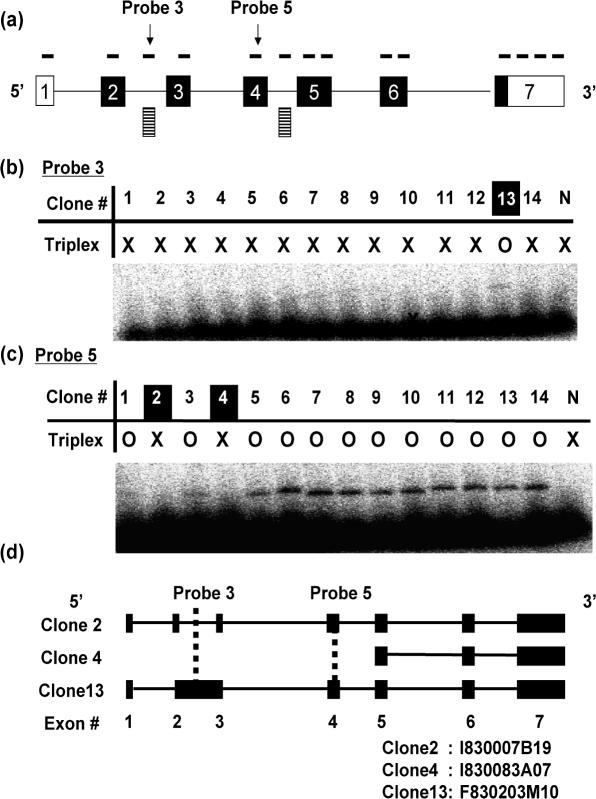
In IL-1β locus, 12 probes for known exons and 2 probes for novel exon candidates were designed (Figure 5a), and 14 cDNA clones were subjected to exon profiling. The probe sequences and the list of clone IDs are provided in Supplementary Tables S4 and S8, respectively. The exon profiling experiment suggested that three clones were alternative splice variants. In one of these splice variant candidates, triplex formation was detected for the probe 3, which was designed on a predicted novel exon (Figure 5b, lane 13). Following full-length sequencing demonstrated that an intron retention phenomenon was successfully detected (clone 13 in Figure 5b–d), and that this cDNA clone (F830203M10) was confirmed to be a novel alternative splice variant. It should be a significant example for the combination of RNA mapping with RecA-mediated exon profiling method. Concerning the remaining two candidates, the probe (Probe 5) designed on the known exon (Exon 4) did show no triplex formation, implying the lack of the corresponding exon (Figure 5a and c, lanes 2 and 4). However, sequencing data showed that one of the clones (I830007B19) was wrongly selected by the RecA-mediated method and was identical to the known cDNA, and that other one (I830083A07) lacked first four exons out of seven exons and was proved to be a genuine splice variant (Figures 5d and 6a).
Figure 5.
Exon profiling of IL-1β. (a) Exon structure of a representative clone (GeneID: 16176) in IL1-β locus. Boxes and lines represent exons and introns, respectively. Closed boxes indicate the region of protein-coding sequence. Solid lines above exonic regions indicate probes. Striped boxes represent novel exon candidates. (b) Typical result of RecA-mediated exon profiling method. The probe 3 was designed on a novel exon candidate. Circles and crosses indicate the presence and the absence of the triplex formation, respectively. (c) Typical result of RecA-mediated exon profiling method. The probe 5 was designed on a known exon (exon 4). (d) Exon structures of three alternative splice candidate transcripts (clones 2, 4 and 13). Closed boxes and lines indicate exons and introns, respectively. The clone IDs for the clones 2, 4 and 13 are indicated at the bottom.
Figure 6.
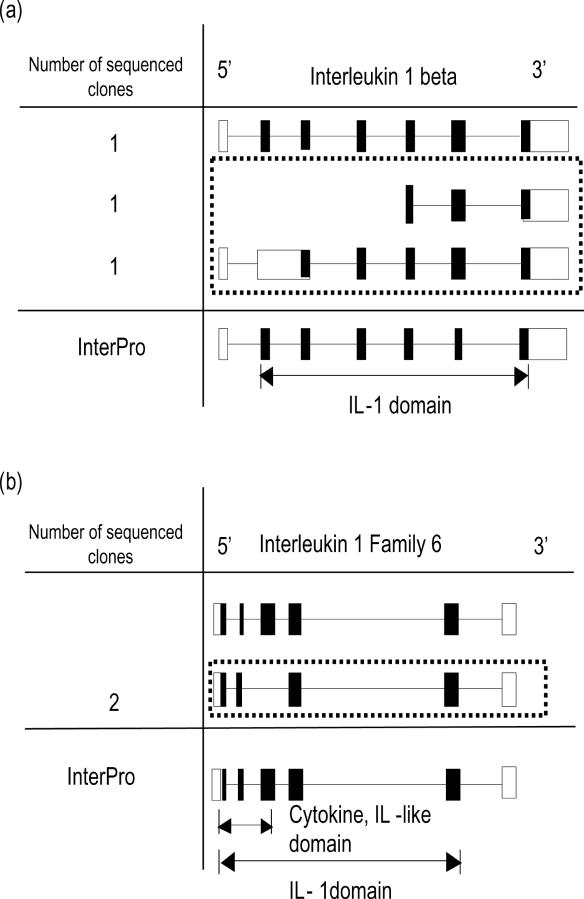
Exon structures of known transcript and of sequenced alternative splice variant candidates. Known transcript is represented at top. Boxes and lines represent exons and introns, respectively. Closed boxes indicate the region of protein-coding sequence. Squares with dotted lines indicate genuine alternative splice variants. Positions of domains predicted by InterPro are indicated with double-headed arrows. (a) In IL1-β locus. (b) In IL-1F6 (GeneID: 54448) locus.
In IL-1F6 locus, 11 probes on 6 known exons and 3 probes on 3 transfrags, and 17 cDNA clones were employed to exon profiling (Supplementary Tables S5 and S9). Out of 17 clones, 2 clones were detected as alternatively splice candidates with lacking the sequence of probe 6 designed on exon 4. Full-length sequence demonstrated that the two clones (Supplementary Figure S2, lanes 1 and 11) lacking the exon 4 were identical to each other even at the nucleotide level. Compared with the sequence of the known cDNAs, these two were proved to be novel splice variants (Figure 6b).
Concerning the glutamine-rich hypothetical protein locus, 13 cDNA clones were clustered into this locus, and 22 probes on 22 known exons and 28 probes on 28 novel exon candidates were designed for exon profiling (Supplementary Tables S6 and S10). In this locus, eight different splice patterns were already reported. The RecA-mediated exon profiling suggested that seven clones had different splice patterns from the reported ones. Out of these seven clones, four clones were bona fide novel splice variants. Two of the four clones showed the same splice pattern each other, resulting in identifying three novel splice patterns (Supplementary Figure S3).
Altogether, RecA-mediated exon profiling analysis could identify eight alternative splice variants from two, two and four clones of IL1β, IL-1F6 and glutamine-rich hypothetical protein, respectively.
Characterization of functionally altered variants
Following the confirmation of newly identified variants by sequencing, we characterized them with several analyses.
The InterPro analysis indicated that the known Txndc5 cDNA has a Thioredoxin domain 2 (IPR: 006663), expanding between exon 1 and exon 9. Out of the 25 clones clustered in this locus, on the other hand, 21 clones lacked the exon 1 or used an alternative exonic region as exon 1 (5′-most exon), implying the alteration of function for novel splice variants. Consistent with this observation, the SOSUI program predicted that the cDNA containing the known exon 1 carried a signal peptide and was an insoluble protein, but the splice variant lacking the exon 1 became soluble without the signal peptide.
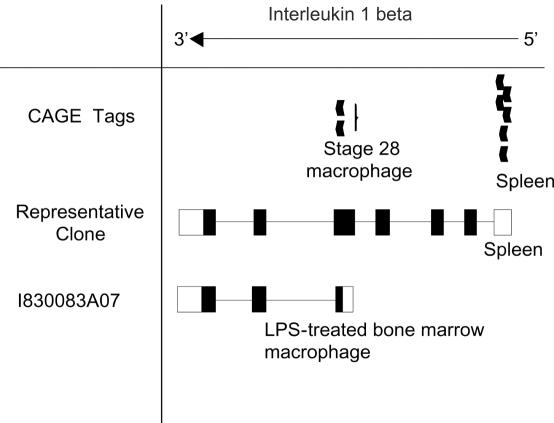
IL-1β is a well-known cytokine with various important biological functions. It has been reported to be a central mediator of inflammation and connective tissue destruction in rheumatoid arthritis. The IL-1β activates articular chondrocytes to produce matrix metalloproteinase-1 (MMP-1), which is capable of dismantling the collagen scaffold of articular cartilage (39). Within the IL-1β locus, two novel splice patterns were identified as mentioned above. One of these lacked first 4 exons, implying a truncated clone. To make sure whether it is a truncated cDNA or not, the sequence of this clone was compared with the information on transcription start sites derived from CAGE and GIS tags (40,41). As a result, two CAGE tags corresponding to the position of 5′ end of splice variant, suggested that this clone should be a full-length cDNA. In addition, the InterPro analysis indicated that this short clone lacked the IL-1β domain and had an ‘unintegrated transmembrane region’ instead (Figures 6a and 7).
Figure 7.
Alignment of known transcript, newly identified splice variant and CAGE tags. CAGE tags are represented with closed arrowheads. Boxes and lines represent exons and introns, respectively. Closed boxes indicate the region of protein-coding sequence. Tissue names of origin are indicated.
IL-1F6 is involved in a wide array of biological activities that initiate and promote the host response to injury or infection by activating a set of transcription factors (42). From our analysis, we found that two clones, which were identical each other, lack a cytokine IL-1-like domain (IPR: 008996). The cytokine IL-1 domain is known to have a function in stimulating mRNA and protein synthesis level of insulin-like growth factor 1 (IGF-1), possibly promoting the intimal hyperplasia (43). The novel splice variant, lacking the Cytokine IL-1-like domain may lead to alteration of transcriptional level or even translational level of IGF-1.
DISCUSSION
In this study, we have developed an effective exon profiling method by utilizing RecA protein. This method allows us to screen alternatively spliced transcripts/cDNAs from a cDNA clone set with high accuracy. With this method, we successfully identified splice variants from four independent gene loci.
Recent cDNA projects largely contributed to understand the complexity of transcriptome to describe its detailed picture. However, limited number of cDNAs was fully sequenced, and most of collected cDNAs were clustered into TUs with their EST information and stored without being fully sequenced, implying that many of novel splice variants remain to be sequenced. In our RIKEN cDNA clone bank, a few million of cDNAs were clustered into 28 163 TUs, and 53% (14 994 TUs) of all TUs include more than two clones within each TU. In addition, 18% (5019 TUs) of all TUs contain >10 clones/TU, and the number of clones for 163 TUs is >100 entries. In this study, we used 96-well plates for exon profiling, and were able to profile all exon structures of 95 clones easily. This fact strongly suggests that our RecA-mediated exon profiling method is applicable to most TUs of RIKEN clone bank. In other words, our method should be a considerable option for the first screening to identify novel splice variants, especially in the case that one looks for splice variants of certain genes.
To consider exon profiling, PCR technique can be another possible option. When sequence information about known exons and candidate exons are available, exon profiling can be carried out by PCR with primers corresponding to exonic regions. Designing primers for PCR, however, requires considering the melting temperature of primers and/or careful selection to avoid mis-priming of primers. In addition, ends of exonic regions tend to be conserved (44), and this fact might cause difficulties on designing primers for PCR. In contrast, our detection method using RecA protein allows us to design probe DNAs more flexibly because of its little sequence restriction. Moreover, regardless of probe sequences, RecA reaction can be simply performed with incubating reaction mixture at 37°C. This flexibility and easiness of the RecA-mediated method should be advantageous to profile exon structures.
It should be noted that the RecA-mediated method has some limitations for exon profiling. One possible limitation is the difficulty on designing probes for unknown/novel exons. In our analysis, novel exon candidates were experimentally extracted from the microarray-based RNA mapping since the RNAs utilized for cDNA library construction were available. This successfully led us to identify a novel splice variant of IL-1β. In contrast, full-length sequencing for Txndc5 gene showed that two novel splice variants harboring unknown exons were not detected by the RecA-mediated method because the corresponding exons were not predicted as novel exons through RNA mapping. In addition, out of total 36 novel exon candidates, the sequences of 35 candidates could not be found within any fully sequenced cDNAs from four selected genes. It implies the possibility that some rare transcripts were diluted by mixing RNAs and signals for these transcripts were below the detection limit of microarray. Better prediction for novel exon candidates is required. In the case that target tissues and genes are determined, the number of RNA pools to mix can be kept as less as possible, it would help to improve the prediction for novel exon candidates. However, this might increase the risk of cross hybridization instead. To compensate, exon prediction programs such as GENSCAN (45) and Exoniphy (46) should be utilized for predicting novel exon candidates.
Another possible limitation is the difficulty on detecting exons, which are different in length. It has been known that many splice variants carry exons, especially 5′- and 3′-most exons, which are different in length. In our analysis, one probe per 100 bp was designed, and our method failed to detect the difference of exon length for the 5′-most and 3′-most exons in Txndc5 at 6% (12/190). To detect this kind of exons more precisely, probes should be designed more frequently with the help of statistical model (47).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors express their gratitude for Ann Karlsson for helpful comments and manuscript editing. We would also like to thank C. Kai, Y. Takeda, E. Shibazaki, H. Nishida, S. Komatsu, T. Suzuki, M. Nishikawa and all the other members of the RIKEN Genome Exploration Research Group for their direct and indirect contributions to this experiment. This work was supported in part by a FY2005 DRI Research Grant from RIKEN institute to N.M., a Research Grant for the Genome Network Project, the Functional RNA project; The Strategic Programs for R&D of RIKEN and Grant from the RIKEN Frontier Research System and a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science, and Technology of Japanese Government to Y.H. Funding to pay the Open Access publication charges for this article was provided by Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Edu-cation, Culture, Sports, Science, and Technology of Japanese Government to Y.H.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ewing B., Green P. Analysis of expressed sequence tags indicates 35 000 human genes. Nature Genet. 2000;25:232–234. doi: 10.1038/76115. [DOI] [PubMed] [Google Scholar]
- 2.Mironov A.A., Fickett J.W., Gelfand M.S. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 5.Lee L.N., Kuo S.H., Lee Y.C., Chang Y.L., Chang H.C., Jan I.S., Yang P.C. CD44 splicing pattern is associated with disease progression in pulmonary adenocarcinoma. J. Formos. Med. Assoc. 2005;104:541–548. [PubMed] [Google Scholar]
- 6.McEachern K.A., Archey W.B., Douville K., Arrick B.A. BRCA1 splice variants exhibit overlapping and distinct transcriptional transactivation activities. J. Cell Biochem. 2003;89:120–132. doi: 10.1002/jcb.10487. [DOI] [PubMed] [Google Scholar]
- 7.Baross A., Butterfield Y.S., Coughlin S.M., Zeng T., Griffith M., Griffith O.L., Petrescu A.S., Smailus D.E., Khattra J., McDonald H.L., et al. Systematic recovery and analysis of full-ORF human cDNA clones. Genome Res. 2004;14:2083–2092. doi: 10.1101/gr.2473704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashizaki Y., Kanamori M. Dynamic transcriptome of mice. Trends Biotechnol. 2004;22:161–167. doi: 10.1016/j.tibtech.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Imanishi T., Itoh T., Suzuki Y., O'Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., et al. Integrative annotation of 21 037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kan Z., Rouchka E.C., Gish W.R., States D.J. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modrek B., Resch A., Grasso C., Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schena M., Heller R.A., Theriault T.P., Konrad K., Lachenmeier E., Davis R.W. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 1998;16:301–306. doi: 10.1016/s0167-7799(98)01219-0. [DOI] [PubMed] [Google Scholar]
- 14.Young R.A. Biomedical discovery with DNA arrays. Cell. 2000;102:9–15. doi: 10.1016/s0092-8674(00)00005-2. [DOI] [PubMed] [Google Scholar]
- 15.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 16.Castle J., Garrett-Engele P., Armour C.D., Duenwald S.J., Loerch P.M., Meyer M.R., Schadt E.E., Stoughton R., Parrish M.L., Shoemaker D.D., et al. Optimization of oligonucleotide arrays and RNA amplification protocols for analysis of transcript structure and alternative splicing. Genome Biol. 2003;4:R66. doi: 10.1186/gb-2003-4-10-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 18.Kapranov P., Sementchenko V.I., Gingeras T.R. Beyond expression profiling: next generation uses of high density oligonucleotide arrays. Brief Funct. Genomic Proteom. 2003;2:47–56. doi: 10.1093/bfgp/2.1.47. [DOI] [PubMed] [Google Scholar]
- 19.Mockler T.C., Chan S., Sundaresan A., Chen H., Jacobsen S.E., Ecker J.R. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85:1–15. doi: 10.1016/j.ygeno.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 21.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 22.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 23.Zavolan M., Kondo S., Schonbach C., Adachi J., Hume D.A., Hayashizaki Y., Gaasterland T. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogawa T., Yu X., Shinohara A., Egelman E.H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 26.Rao B.J., Radding C.M. Formation of base triplets by non-Watson–Crick bonds mediates homologous recognition in RecA recombination filaments. Proc. Natl Acad. Sci. USA. 1994;91:6161–6165. doi: 10.1073/pnas.91.13.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata T., Cunningham R.P., DasGupta C., Radding C.M. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc. Natl Acad. Sci. USA. 1979;76:5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X., Adzuma K. DNA strand exchange mediated by the Escherichia coli RecA protein initiates in the minor groove of double-stranded DNA. Biochemistry. 1997;36:4650–4661. doi: 10.1021/bi9630063. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Lipman D.J. Gapped BLAST and PHI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 31.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 32.Andreeva A., Howorth D., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Holich V., Lassmann T., Moxon S., Marchall M., Khanna A., Durbin R., et al. Pfam: clons, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 37.Ferrin L.J., Camerini-Otero R.D. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- 38.Kiianitsa K., Stasiak A. Helical repeat of DNA in the region of homologous pairing. Proc. Natl Acad. Sci. USA. 1997;94:7837–7840. doi: 10.1073/pnas.94.15.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raymond L., Eck S., Mollmark J., Hays E., Tomek I., Kantor S., Elliott S., Vincenti M. Interleukin-1 beta induction of matrix metalloproteinase-1 transcription in chondrocytes requires ERK-dependent activation of CCAAT enhancer-binding protein-beta. J. Cell Physiol. 2006;207:683–688. doi: 10.1002/jcp.20608. [DOI] [PubMed] [Google Scholar]
- 40.Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nature Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- 41.Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Towne J.E., Garka K.E., Renshaw B.R., Virca G.D., Sims J.E. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J. Biol. Chem. 2004;279:13677–13688. doi: 10.1074/jbc.M400117200. [DOI] [PubMed] [Google Scholar]
- 43.Glazebrook H., Hatch T., Brindle N.P. Regulation of insulin-like growth factor-1 expression in vascular endothelial cells by the inflammatory cytokine interleukin-1. J. Vasc. Res. 1998;35:143–149. doi: 10.1159/000025577. [DOI] [PubMed] [Google Scholar]
- 44.Patel A.A., Steitz J.A. Splicing double: insights from the second spliceosome. Nature Rev. Mol. Cell. Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 45.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 46.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chern T.M., van Nimwegen E., Kai C., Kawai J., Carninci P., Hayashizaki Y., Zavolan M. A simple physical model predicts small exon length variations. PLoS Genet. 2006;2:e45. doi: 10.1371/journal.pgen.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.