Abstract
Single-walled carbon nanotubes (SWNTs) have been considered as the leading candidate for nanodevice applications ranging from gene therapy and novel drug delivery to membrane separations. The miniaturization of DNA-nanotube devices for biological applications requires fully understanding DNA-nanotube interaction mechanism. We report here, for the first time, that DNA destabilization and conformational transition induced by SWNTs are sequence-dependent. Contrasting changes for SWNTs binding to poly[dGdC]:poly[dGdC] and poly[dAdT]:poly[dAdT] were observed. For GC homopolymer, DNA melting temperature was decreased 40°C by SWNTs but no change for AT-DNA. SWNTs can induce B–A transition for GC-DNA but AT-DNA resisted the transition. Our circular dichroism, competitive binding assay and triplex destabilization studies provide direct evidence that SWNTs induce DNA B–A transition in solution and they bind to the DNA major groove with GC preference.
INTRODUCTION
Single-walled carbon nanotubes (SWNTs) have been considered as the leading candidate for nanodevice applications because of their one-dimensional electronic band structure, molecular size, biocompatibility, controllable property of conducting electrical current and reversible response to biochemical reagents (1–6). These potential applications range from gene therapy and drug delivery to membrane separations (4–16). Among the molecules that can non-covalently bind to the surface of SWNTs, DNA has been the research focus (7–16), which adsorbs as a single-strand or double-strand complexes. By screening a library of oligonucleotides, previous reports have shown that a particular sequence of single stranded DNA self-assembles into a helical structure around individual carbon nanotubes. Since carbon nanotube–DNA hybrids have different electrostatic properties that depend on the diameter of the nanotubes and electronic properties, they can be separated and sorted using anion exchange chromatography (11,12). Carbon nanotubes are able to condense double stranded plasmid DNA to varying degree and exhibit upregulation of marker gene expression over naked DNA using a mammalian (human) cell line, a nanotube-based gene-delivery vector has been reported (16). In a recent report, a piece of double-stranded DNA wrapped on the surface of a single-walled carbon nanotube can serve as sensors in living cells (17) and the heart of the new optical detection system is based on the transition of DNA secondary structure from the native, right-handed ‘B’ form to the alternate, left-handed ‘Z’ form which was modulated by metal ions. Therefore, it is important and fundamental to understand the interaction mechanism of SWNT with double-stranded DNA for nanodevice application.
In this report, SWNTs DNA binding mode, binding preference and the impact on DNA stability and conformation were studied. Contrasting changes for SWNTs binding to poly[dGdC]:poly[dGdC] and poly[dAdT]:poly[dAdT] were observed. We report here, for the first time, that DNA condensation, destabilization and conformational transition induced by SWNTs are sequence-dependent. Our circular dichroism (CD), competitive binding assay and triplex destabilization studies provide direct evidence that SWNTs induce DNA B–A transition in solution and they bind to the DNA major groove with GC preference. The sequence-dependent condensation and B–A transition by SWNTs shed light on the design of miniature of optical devices and label-free detection of specific genes.
MATERIALS AND METHODS
SWNTs (φ = 1.1nm, purity >90%) were purchased from Aldrich and purified as described previously by sonicating SWNTs in a 3:1 v/v solution of concentrated sulfuric acid (98%) and concentrated nitric acid (70%) for 24h at 35–40°C and washed with water, leaving an open hole in the tube side and functionalized the open end of SWNTs with carboxyl group to increase their solubility in aqueous solution (2). The stock solution of SWNTs (0.15 mg mL−1) was obtained by sonicating the SWNTs for 8 h in pH 7.0 aqueous solution. Calf thymus DNA (ct-DNA) was obtained from Sigma and purified as described earlier (18). Poly[dGdC]:poly[dGdC] (GC-DNA), poly[dAdT]:poly[dAdT] (AT-DNA), polydA, and polydT were purchased from Pharmacia. The concentration of ct-DNA, GC-DNA and AT-DNA were determined by ultraviolet absorbance measurements using the extinction coefficient: ɛ260 = 12 824 M−1 cm−1, ɛ262 = 16 800 M−1 cm−1, ɛ254 = 13200 M−1 cm−1, respectively (18). Triplex DNA (polydA:[polydT]2) was prepared as described previously (19–21). Ethidium bromide (EB), hoechst 33258 and daunomycin (DM) were purchased from Sigma, methylene green was purchased from Aldrich and were used without further purification. Their concentrations were determined by absorbance measurements using the extinction coefficient: ɛ480 = 5600 M−1 cm−1, ɛ338 = 42 000 M−1 cm−1, ɛ480 = 11 500 M−1 cm−1 for EB, Hoechst 33 258 and DM, respectively (22). All the experiments were carried out in Tris buffer (10 mM Tris, pH = 7.1) unless stated otherwise. In sodium iodide fluorescence quenching experiments, the ionic strength was kept constant.
An AFM (Nanoscope IIIa, Digital Instruments, Santa Barbara, CA) was used to image all DNAs in the presence or absence of SWNTs. The sample solution was deposited onto a piece of freshly cleaved mica and rinsed with water and dried before measurements (23). Tapping mode was used to acquire the images under ambient condition. CD spectra were measured at 20°C on a JASCO J-810 spectropolarimeter with a computer-controlled water bath (24). The optical chamber of CD spectrometer was deoxygenated with dry purified nitrogen (99.99%) for 45 min before use and kept the nitrogen atmosphere during experiments. Three scans were accumulated and automatically averaged. Absorbance measurements and melting experiments were made on a Cary 300 UV/Vis spectrophotometer, equipped with a Peltier temperature control accessory (25). All UV/Vis spectra were measured in 1.0 cm path length quartz cuvettes with the same concentration of SWNTs aqueous solution as the reference. Absorbance changes at 260 nm versus temperature were collected at a heating rate of 0.5°C min−1 for DNA melting experiments. Primary data were transferred to the graphics program Origin for plotting and analysis. Fluorescence experiments were carried out on a JASCO FP-6500 spectrofluorometer at 20°C (24).
RESULTS AND DISCUSSION
SWNT inducing sequence-dependent DNA condensation
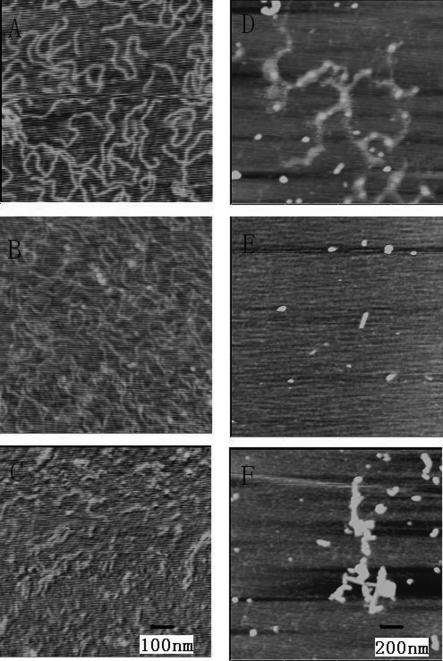
AFM studies showed that DNA condensed when they bound to carboxyl-modified SWNTs (2) and the condensation was dependent on DNA composition. In the absence of SWNTs, linear GC-DNA, AT-DNA and natural DNA, purified calf thymus DNA (ct-DNA) were observed (Figure 1A–C). When they bound with carboxyl-modified SWNTs, however, striking differences were observed among these DNA molecules (Figure 1D–F). SWNTs condense DNA depending on DNA GC content: GC homopolymer, GC-DNA was condensed (23) as a network (Figure 1D). For calf thymus DNA (ct-DNA) and AT-DNA, they looked like forming DNA-wrapped complexes and condensed slightly (Figure 1E and F) showing that GC-DNA was more easily condensed than AT-DNA and ct-DNA. Premilat et al. have measured the major groove width of GC-DNA (∼1.35 nm) and AT-DNA (∼1.75 nm) through fiber X-ray diffraction (26,27). SWNTs (φ = 1.1nm) we used were modified with carboxyl group. Carboxyl groups at the open end of SWNTs greatly increased their water solubility and may impact DNA binding to the modified nanotube surface. Based on SWNTs size, hydrophobic property and their improved solubility, SWNTs should not bind to DNA minor grooves due to the narrower groove width. Alternatively, SWNTs may bind to the major groove and would fit better to GC-DNA major groove because AT-DNA major groove is too wide for SWNTs binding. Hansma and Kankia have reported that condensation of DNA by metal ions is sequence dependent owing to the difference of GC and AT sequence in their extent of dehydration (23,28,29). GC-rich regions are being more heavily dehydrated than AT-rich regions, this can be the reason why GC-DNA was easily condensed by SWNTs and this will be further discussed in our CD studies. SWNTs, as gene-delivery vector (16), can take advantage to select highly GC-content gene for delivery and this needs to be verified in various gene expression systems.
Figure 1.
DNA AFM images in the absence or presence of SWNTs: (A) GC-DNA alone; (B) ct-DNA alone; (C) AT-DNA alone; (D) GC-DNA + 40 µg ml−1 SWNTs; (E) ct-DNA + 40 µg ml−1 SWNTs; (F) AT-DNA + 40 µg ml−1 SWNTs. The DNA concentration used in all experiments was 19.5 µg ml−1. All the AFM images are captured on freshly cleaved mica. The image for (A–C) is 500 nm × 500 nm and the same scale bar. The image for (D–F) is 1 µm × 1 µm and with the same scale bar.
SWNTs selective destabilization of DNA
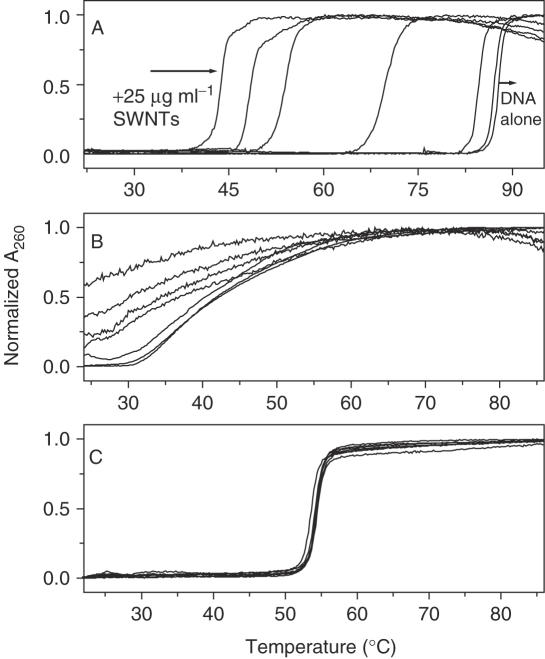
Figure 2 shows DNA UV melting profiles in the absence or presence of SWNTs. Contrasting changes for SWNTs binding to GC-DNA and AT-DNA were observed. It is obvious that GC-DNA and ct-DNA became unstable in the presence of SWNTs. Melting temperature Tm decreased 40°C for GC-DNA when SWNTs at 25 µg/ml (Figure 2A). The absorption after 80°C decreased showing the strong interaction of single-strand DNA with SWNTs (11,12). Since the melting temperature of GC-DNA in the presence of 15 µg ml−1 or higher SWNTs concnentration was much lower than 60°C, it seems unlikely that the destabilization was due to pH change with temperature. On the contrary to GC-DNA and ct-DNA, SWNTs did not influence AT-DNA stability even at high concentration of SWNTs (Figure 2C). At 20 mM NaCl, similar trend for SWNTs bound to DNA was observed (data not shown). We also studied SWNTs bound to triplex DNA at high salt conditions which will be addressed in triplex destabilization section. These results showed that the binding preference of SWNTs was: GC-DNA>ct-DNA>AT-DNA, consistent with the AFM and CD results which will be addressed next.
Figure 2.
UV melting profiles of DNA: (A) poly[dGdC]:poly[dGdC], (B) ct-DNA, (C) poly[dAdT]:poly[dAdT] in the absence or presence of SWNTs. From right to left: 0, 1, 5, 10, 15, 20, 25 µg ml−1 SWNTs in pH = 7 solution. Normalized absorption changes at 260 nm were plotted against temperature. The data were collected at a heating rate of 0.5°C min−1. Details see Materials and Methods.
SWNTs making DNA B–A transition in solution
As DNA bound to carboxyl-modified SWNTs, various interactions of DNA bases and backbone with SWNTs, such as hydrophobic interactions, van der Waals, electrostatic interactions, can take place (15). The strong interactions between SWNTs and DNA can disturb DNA hydration layer, even DNA structure (22,29).
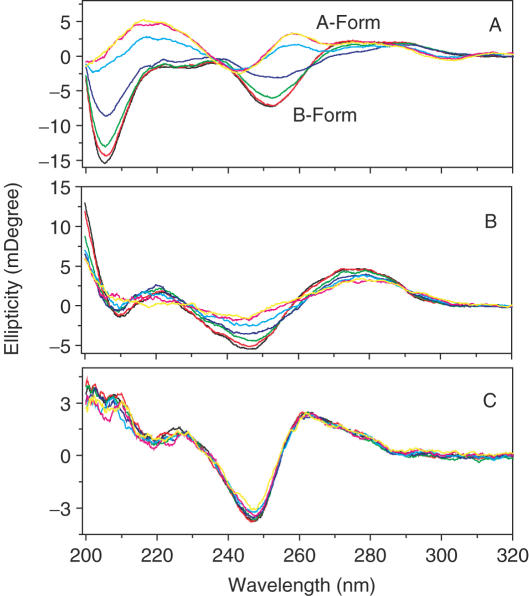
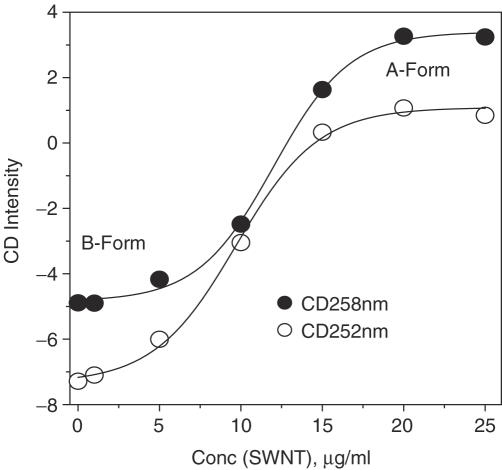
CD spectra showed these DNA molecules were in B-form (Figure 3) with a positive band near 270 nm and a negative band near 250 nm in the absence of SWNTs (18,24). Upon addition of SWNTs (Figure 3A), the canonical B form of GC-DNA altered with a positive band near 258 nm and a negative band near 242 nm indicating that B–A transition (28–33) occurred. The transition was cooperative and the transition midpoint was at 10 µg/ml SWNTs (Figure 4). CD spectroscopy provided the direct evidence that SWNTs could make DNA B–A transition in solution, consistent with previous molecular dynamics simulation results which show B–A transition when DNA encapsulated in carbon nanotube or on gold surface (7,9). The induced B to A transition was due to SWNTs bound to the major groove resulting in deepening and narrowing the major groove while widening the minor groove, which was coincident with the previous simulation results (33).
Figure 3.
CD spectra of DNA. (A) poly[dGdC]:poly[dGdC] (B) ct-DNA (C) poly[dAdT]:poly[dAdT] in the absence (black) or presence of SWNTs: SWNTs 1 (red), 5 (green), 10 (blue), 15 (cyan), 20 (magenta) and 25 µg ml−1 (yellow) in Tris buffer (pH = 7.1) at 20°C. CD spectra were measured on a JASCO J-810 spectropolarimeter with a computer-controlled water bath as described in Materials and Methods. Three scans were accumulated and automatically averaged.
Figure 4.
Plot of CD intensity versus concentration of SWNTs. CD intensity at 258 nm (solid circles) and at 252 nm (open circles). The data were adopted from Figure 3A.
Calf thymus DNA (ct-DNA, 42% GC and 58%AT) was induced to A-form by SWNTs but not as easily as GC-DNA while AT-DNA persisted in B-form (Figure 3B and C), showing that B–A transition was dependent on the G-C content of the DNA helix (28–33). Ivanov and Krylov (34) have reviewed the cooperative character of B–A transition and determined the cooperative width of B–A transition with three different methods, and confirm that the cooperative width of B–A transition for DNA with mixed sequence is in the range of 10–30 bp. Since GC-DNA was more easily condensed by SWNTs, the width of the transition for GC homopolymer should be lower than that for the DNA with mixed sequence (31), such as ct-DNA. Since the water activity is an apparent driving force for B–A transition and the water activity (31) of GC-rich region (81.2) is lower than AT-rich region (81.5), GC-DNA undergoes the B to A transition most easily, whereas AT-DNA resists the B to A transition. Previous studies show that GC homopolymer have a stronger tendency for aggregation than AT homopolymer, and GC homopolymer can undergo B–Z–A transition by reducing water activity (35). Under the usual experimental conditions for B–A transition, GC homopolymer will aggregate (36) while AT homopolymer would remain in the B-form even at conditions which normally favors the A-form (36,37). These results further support that GC homopolymer was more easily condensed by SWNTs than AT homopolymer as shown in our AFM studies.
A-DNA is biologically relevant and has 11 bp per helical turn, base pairs are tilted to ∼20°C with respect to the helical axis (38), the grooves are not as deep as those in B-DNA, the sugar pucker is C3′ endo compared with C2′ endo for B-DNA. Like B–Z DNA transition (17,18,24,39,40), the transition from the B-DNA double helix to the A-form is essential for biological function (28–33), as shown by the existence of the A-form in many protein–DNA complexes, increasing the fidelity of DNA and RNA synthesis and protection from DNA damage.
Fluorescence competitive binding assay and triplex DNA destabilization by SWNTs
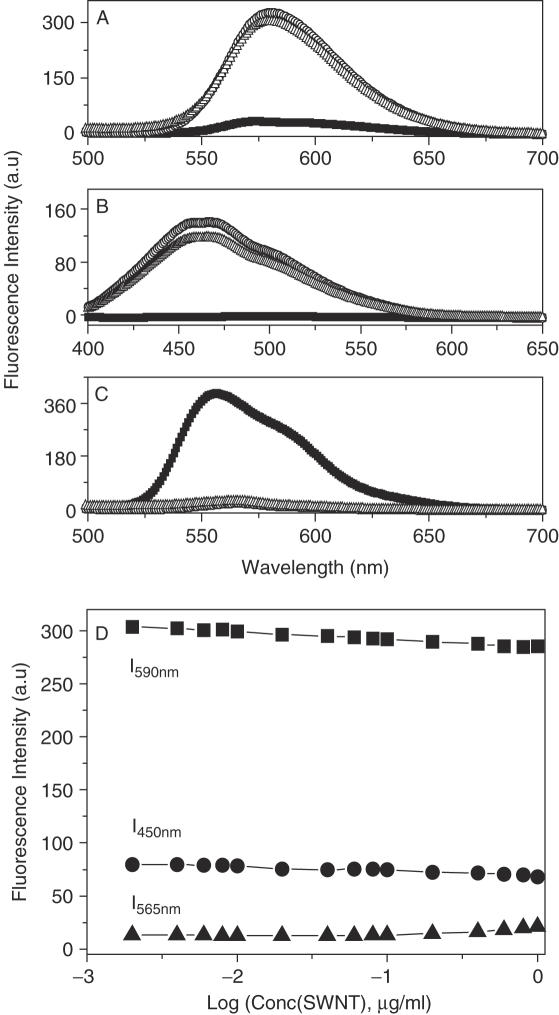
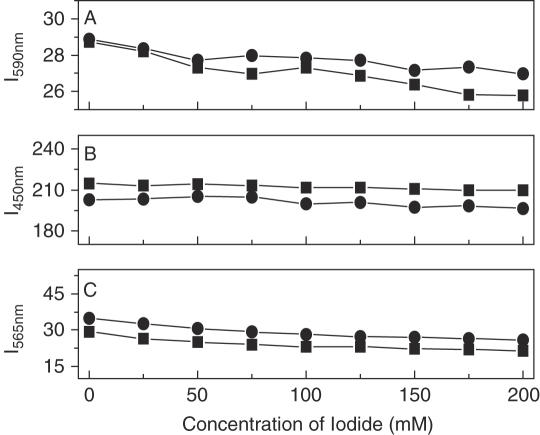
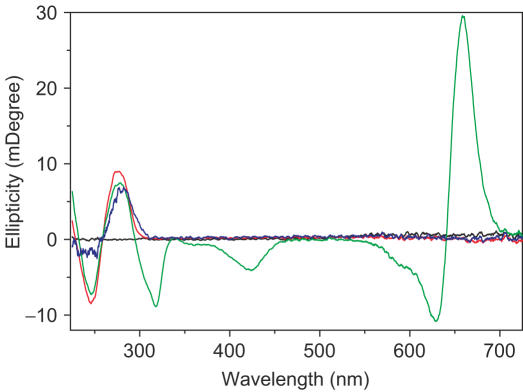
It is well known that EB and DM can intercalate into DNA through the minor groove and Hoechst 33 258 is a classical DNA minor groove binder. When bound to DNA, the fluorescence of EB or Hoechst is greatly enhanced, and DM fluorescence is strongly quenched. With this in mind, if SWNTs competitively bind to the same sites of DNA as EB, Hoechst and DM, the fluorescence of EB and Hoechst would decrease and the fluorescence of DM would increase because the strong binding of SWNTs with DNA should exclude these DNA binders out of their binding sites. The fluorescence competitive binding assay has been widely used to establish DNA binding mode (41–43). As shown in Figure 5A–C, their fluorescence hardly changed, even titrated by SWNTs (Figure 5D), showing that SWNTs do not competitively bind to the same sites. To further identify their different binding sites, we carried out NaI quenching experiments. Iodide ions cannot quench the fluorescence of these dye molecules when they are bound to DNA (43). If SWNTs could replace these molecules, their fluorescence should be quenched. Figure 6 showed that the fluorescence was not quenched by iodide at all. However, we found that SWNTs could exclude methylene green, a proven DNA major groove binder (44,45), out of DNA. Figure 7 showed CD spectral changes in the presence of SWNTs. For methylene green, there was no CD signal in our experimental conditions. When methylene green bound to DNA, three induced CD signals characteristic of bound methylene green around 310 nm, 430 nm, 650 nm were observed (44,45). With addition of SWNTs, the induced CD intensity was decreased and even disappeared, typical data was shown in Figure 7 which was consistent with previous reports that methylene green can be excluded out of DNA major groove (44,45). DNA CD signals were also changed in the presence of SWNTs (Figure 7). When methylene green was out of DNA major groove, DNA CD spectrum was like the one for DNA–SWNTs in the absence of methylene green (Figure 3B), further supporting that SWNTs bound to DNA in the major groove by replacement of methylene green molecules. In combination with CD data, thermal denaturation, competitive binding assay and condensation results, SWNTs bound to the DNA major groove but not the minor groove, in agreement with SWNT–DNA simulation results which show that SWNTs bind to DNA in the major groove in the vacuum (14). Another evidence of SWNTs bound to the major groove was from the competitive binding between SWNTs and the third strand to duplex DNA in the major groove.
Figure 5.
Fluorescence emission spectra of (A) 1 µM EB (Excitation wavelength: 480 nm, Slit: 10 nm); (B) 1 µM Hoechst 33 258 (Excitation wavelength: 355 nm, Slit: 3 nm); (C) 1 µM DM (Excitation wavelength: 480 nm, Slit: 10 nm): 1µM fluorophore (EB, Hoechst 33 258, DM) alone (black curves), 1 µM fluorophore + 30 µg ml−1 CT-DNA (open circles), 1 µM fluorophore + 30 µg ml−1 CT-DNA + 1 µg ml−1 SWNTs (upper triangles). (D) Fluorescence intensity as a function of concentration of SWNTs: 1 µM EB + 30 µg ml−1 CT-DNA (squares), 1 µM Hoechst 33 258 + 30 µg ml−1 CT-DNA (circles), 1 µM DM + 30 µg ml−1 CT-DNA (triangles). The experiments were carried out in Tris buffer (pH = 7.1) at 20°C.
Figure 6.
Plot of fluorescence intensity of fluorephore-DNA (squares) or fluorephore-DNA–SWNTs (circles) versus NaI concentration. The ionic strength was kept constant: (A) 10 µM EB (Excitation wavelength: 480 nm, Emission wavelength: 590 nm Slit: 5 nm); (B) 10 µM Hoechst 33 258 (Excitation wavelength: 355 nm, Emission wavelength: 450 nm Slit: 3 nm); (C) 10 µM DM (Excitation wavelength: 480 nm, Emission wavelength: 565 nm Slit: 10 nm). Experimental details described in the experimental section.
Figure 7.
Loss of CD signal from calf thymus DNA-methylene green complex upon SWNT–DNA association. Methylene green (black); calf thymus DNA (red); calf thymus DNA-methylene green (green); calf thymus DNA-methylene green after association with SWNTs at binding ratio r = 0.2 (blue). Calf thymus DNA was 50 µM in bp. CD spectra were measured at 20°C on a JASCO J-810 spectropolarimeter with a computer-controlled water bath as described in the experimental section. Three scans were accumulated and automatically averaged.
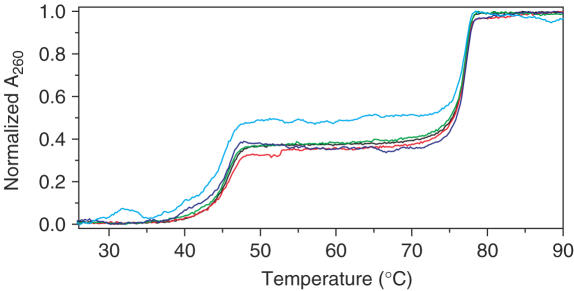
Triplex DNA used was polydT:polydA:polydT (19–21). PolydT can bind to the major groove of polydA:polydT duplex by forming triplex DNA polydA(polydT)2 (19–21). The thermal denaturation profile of free triplex in Tris buffer (10 mM Tris, 200 mM NaCl, pH = 7.1) showed two transitions (Figure 8), the first transition, Tm1 at 46°C and the second transition, Tm2 at 77°C. For TAT triplex DNA, the first transition (Tm1) was attributed to the dissociation of the third strand of polydT with Hoogsteen base pairs from the major groove and the second transition (Tm2) was attributed to the dissociation of Watson–Crick base pairs of duplex polydA:polydT (19–21). Figure 8 showed that Tm1 was decreased in the presence of SWNTs but Tm2 did not change, indicating that SWNTs competed with the third strand binding to the duplex major groove of polydA:polydT, thus decrease the stability of Hoogsteen base pairs but not influence the duplex stability of polydA:polydT, further supporting that SWNTs bound to the duplex major groove with GC preference as shown in duplex melting data of poly[dGdC]:poly[dGdC], calf thymus DNA and poly[dAdT]: poly[dAdT] (Figure 2). Triplex DNA has been the focus of considerable interest because of possible applications in developing new molecular biology tools as well as therapeutic agents, and because of the possible relevance of H-DNA structures in biology system (19–21). However, SWNTs selective destabilization of triplex DNA has not been reported. Based on previous computer simulation results of SWNTs binding to duplex DNA in the major groove (14) and our melting, CD, and competitive binding data, SWNTs probably bound to duplex DNA major groove and had GC preference.
Figure 8.
UV melting profiles of triplex DNA polydA (polydT)2 in the absence (black) or presence of SWNTs: SWNTs 1 µg ml−1 (red); 2 µg ml−1 (green); 5 µg ml−1 (blue); 10 µg ml−1 (cyan) in Tris buffer (10 mM Tris, 200 mM NaCl, pH = 7.1). Normalized absorption changes at 260 nm were plotted against temperature. The data were collected at a heating rate of 0.5°C min−1.
CONCLUSIONS
SWNTs can cause sequence-dependent DNA condensation and strongly destabilize GC-DNA. Contrasting changes for SWNTs binding to GC-DNA and AT-DNA were observed. Our CD, competitive binding and triplex destabilization studies provide direct evidence that SWNTs induce DNA B–A transition in solution and they bind to the DNA major groove with GC preference.
Acknowledgments
The authors are grateful to the referees for their helpful comments on the manuscript. The authors thank Dr L. Wang for his technical assistance on AFM experiments. This project was supported by NSFC (20225102, 20331020, 20473084), and hundred people program from CAS. Funding to pay the Open Access publication charges for this article was provided by the National Natural Science Foundation of China and Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Iijima S. Helical microtubules of graphitic carbon. Nature. 1991;354:56–58. [Google Scholar]
- 2.Liu J., Rinzler A.G., Dai H., Hafner J.H., Bradley R.K., Boul P.J., Lu A., Iverson T., Shelimov K., Huffman C.B., et al. Fullerene pipes. Science. 1998;280:1253–1256. doi: 10.1126/science.280.5367.1253. [DOI] [PubMed] [Google Scholar]
- 3.Keren K., Berman R.S., Buchstab E., Sivan U., Braun E. DNA-templated carbon nanotube field-effect transistor. Science. 2003;302:1380–1382. doi: 10.1126/science.1091022. [DOI] [PubMed] [Google Scholar]
- 4.Wong S.S., Joselevich E., Woolley A.T., Cheung C.L., Lieber C.M. Covalently functionalized nanotubes as nanometre-sized probes in chemistry and biology. Nature. 1998;394:52–55. doi: 10.1038/27873. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y., Taylor S., Li H., Fernando K.A.S., Qu L., Wang W., Gu L., Zhou B., Sun Y-P. Advances toward bioapplications of carbon nanotubes. J. Mater. Chem. 2004;14:527–541. [Google Scholar]
- 6.Kam N.W.S., O'Connell M., Wisdom J.A., Dai H.J. Carbon nanotubes as multifunctional biological transporters and near-infrared agents for selective cancer cell destruction. Proc. Natl Acad. Sci. USA. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao H., Kong Y., Cui D. Spontaneous insertion of DNA oligonucleotides into carbon nanotubes. Nano Lett. 2003;3:471–473. [Google Scholar]
- 8.Williams K.A., Veenhuizen P.T.M., de la Torre B.G., Eritja R., Dekker C. Carbon nanotubes with DNA recognition. Nature. 2002;420:761. doi: 10.1038/420761a. [DOI] [PubMed] [Google Scholar]
- 9.Dovbeshko G.I., Repnytaska O.P., Obraztsova E.D., Shtogun Y.V. DNA interaction with single-walled carbon nanotubes: a SEIRA study. Chem. Phys. Lett. 2003;372:432–437. [Google Scholar]
- 10.Hu C., Zhang Y., Bao G., Zhang Y., Liu M., Wang Z. DNA-functionalized single-walled carbon nanotubes for electrochemical detection. J. Phys. Chem. B. 2005;109:20072–20076. doi: 10.1021/jp0550457. [DOI] [PubMed] [Google Scholar]
- 11.Zheng M., Jagota A., Semke E.D., Diner B.A., Mcleen R.S., Lustig S.R., Richardson R.E., Tassi N.G. DNA-assisted dispersion and separation of carbon nanotubes. Nature Mater. 2003;2:338–342. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]
- 12.Zheng M., Jagota A., Strano M.S., Santos A.P., Barone P., Chou S.G., Diner B.A., Dresselhaus M.S., McLean P.S., Onoa G.B., et al. Structure-based carbon nanotube sorting by sequence-dependent DNA assembly. Science. 2003;302:1545–1548. doi: 10.1126/science.1091911. [DOI] [PubMed] [Google Scholar]
- 13.Rajendra J., Baxendale M., Rap L.G.D., Rodger A. Flow linear dichroism to probe binding of aromatic molecules and DNA to single-walled carbon nanotubes. J. Am. Chem. Soc. 2004;126:11182–11188. doi: 10.1021/ja048720j. [DOI] [PubMed] [Google Scholar]
- 14.Lu G., Maragakis P., Kaxiras E. Carbon nanotube interaction with DNA. Nano Letters. 2005;5:897–900. doi: 10.1021/nl050354u. [DOI] [PubMed] [Google Scholar]
- 15.Gao H., Kong Y. Simulation of DNA-nanotube interactions. Annu. Rev. Mater. Res. 2004;34:123–150. [Google Scholar]
- 16.Singh R., Pantarotto D., McCarthy D., Chaloin O., Hoebeke J., Partidos C., Briand J-P., Prato M., Bianco A., Kostarelos K. Binding and condensation of plasmid DNA onto functionalized carbon nanotubes: toward the construction of nanotube-based gene delivery vectors. J. Am. Chem. Soc. 2005;127:4388–4396. doi: 10.1021/ja0441561. [DOI] [PubMed] [Google Scholar]
- 17.Heller D.A., Jeng E.S., Yeung T-K., Martinez B.M., Moll A.E., Gastala J.B., Strano M.S. Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science. 2006;311:508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- 18.Qu X., Trent J.O., Fokt I., Priebe W., Chaires J.B. Allosteric, chiral-selective drug binding to DNA. Proc. Natl Acad. Sci. USA. 2000;97:12032–12037. doi: 10.1073/pnas.200221397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J., Qu X., Dattagupta N., Chaires J.B. Molecular recognition of a RNA:DNA hybrid structure. J. Am. Chem. Soc. 2001;123:6742–6743. doi: 10.1021/ja015649y. [DOI] [PubMed] [Google Scholar]
- 20.Arya D.P., Coffer R.L., Jr, Charles I. Neomycin-induced hybrid triplex formation. J. Am. Chem. Soc. 2001;123:11093–11094. doi: 10.1021/ja016481j. [DOI] [PubMed] [Google Scholar]
- 21.Song G., Xing F., Qu X., Chaires J.B., Ren J. Oxazine 170 induces DNA:RNA:DNA triplex formation. J. Med. Chem. 2005;48:3471–3473. doi: 10.1021/jm050131g. [DOI] [PubMed] [Google Scholar]
- 22.Qu X., Chaires J.B. Hydration changes for DNA intercalation reactions. J. Am. Chem. Soc. 2001;123:1–7. doi: 10.1021/ja002793v. [DOI] [PubMed] [Google Scholar]
- 23.Sitko J.C., Mateescu E.M., Hansma H.G. Sequence-dependent DNA condensation and the electrostatic zipper. Biophysical J. 2003;84:419–431. doi: 10.1016/S0006-3495(03)74862-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H., Yu H., Ren J., Qu X. Reversible B/Z-DNA transition under the low salt condition and Non-B-Form PolydApolydT selectivity by a cubane-like europium-l-aspartic acid complex. Biophysical J. 2006;90:3203–3207. doi: 10.1529/biophysj.105.078402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu X., Ren J., Riccelli P.V., Benight A.S., Chaires J.B. Enthalpy/entropy compensation: influence of DNA flanking sequence on the binding of 7-amino actinomycin D to its primary binding site in short DNA duplexes. Biochemistry. 2003;42:11960–11967. doi: 10.1021/bi0347813. [DOI] [PubMed] [Google Scholar]
- 26.Premilat S., Albiser G. Helix–helix transitions in DNA: fibre X-ray study of the particular cases poly(dG-dC)·poly(dG-dC) and poly(dA)·2poly(dT) Eur. Biophys. J. 1999;28:574–582. doi: 10.1007/s002490050239. [DOI] [PubMed] [Google Scholar]
- 27.Premilat S., Albiser G. A new D-DNA form of poly(dA-dT)·poly(dA-dT): an A-DNA type structure with reversed Hoogsteen pairing. Eur. Biophys. J. 2001;30:404–410. doi: 10.1007/s002490100170. [DOI] [PubMed] [Google Scholar]
- 28.Kankia B.I. Hydration effects of Ni2+ binding to synthetic polynucleotides with regularly alternating purine–pyrimidine sequences. Nucleic Acids Res. 2000;28:911–916. doi: 10.1093/nar/28.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kankia B.I., Buckin V., Bloomfield V.A. Hexamminecobalt (III)-induced condensation of calf thymus DNA: circular dichroism and hydration measurements. Nucleic Acids Res. 2001;29:2795–2801. doi: 10.1093/nar/29.13.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robison H., Wang A.H.J. Neomycin, spermine and hexaamminecobalt (III) share common structural motifs in converting B- to A-DNA. Nucleic Acids Res. 1996;24:676–682. doi: 10.1093/nar/24.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolstorukov M.Y., Ivnov V.I., Malenkov G.G., Jernigan R.L., Zhurkin V.B. Sequence-dependent B↔A transition in DNA evaluated with dimeric and trimeric scales. Biophysical J. 2001;81:3409–3421. doi: 10.1016/S0006-3495(01)75973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jose D., Dietmar Porschke D. Dynamics of the B–A transition of DNA double helices. Nucleic Acids Res. 2004;32:2251–2258. doi: 10.1093/nar/gkh551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng H-L., Dickenson R.E. Mediation of the A/B-DNA helix transition by G-tracts in the crystal structure of duplex CATGGGCCCATG. Nucleic Acids Res. 2002;30:4061–4067. doi: 10.1093/nar/gkf515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanov V.I., Krylov D.Y. A-DNA in solution as studied by diverse approaches. Methods Enzymol. 1992;211:111–127. doi: 10.1016/0076-6879(92)11008-7. [DOI] [PubMed] [Google Scholar]
- 35.Pohl F.M. Polymorphism of a synthetic DNA in solution. Nature. 1976;12:365–366. doi: 10.1038/260365a0. [DOI] [PubMed] [Google Scholar]
- 36.Nara-Inui H., Akutsu H., Kyogoku Y. Alcohol induced BA transition of DNAs with different base compositions studied by circular dichroism. J. Biochem. 1985;98:629–636. doi: 10.1093/oxfordjournals.jbchem.a135319. [DOI] [PubMed] [Google Scholar]
- 37.Arnott S., Selsing E. The conformation of C-DNA. J. Mol. Biol. 1975;98:265–269. doi: 10.1016/s0022-2836(75)80115-x. [DOI] [PubMed] [Google Scholar]
- 38.Neidle S. DNA Structure and Recognition. New York: Oxford University Press Inc.; 1994. [Google Scholar]
- 39.Waring M.J. Facilitating structural transitions in DNA. Proc. Natl Acad. Sci. USA. 2000;97:11685–11687. doi: 10.1073/pnas.97.22.11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rich A., Zhang S. Timeline: Z-DNA: the long road to biological function. Nature Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 41.Boger D.L., Fink B.E., Brunette S.R., Tse W.C., Hedrick M.P. A simple, high resolution method for establishing DNA binding affinity and sequence selectivity. J. Am. Chem. Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 42.Shangguan G., Xing F., Qu X., Mao J., Zhao D, Zhao X., Ren J. DNA binding specificity and cytotoxicity of novel antitumor agent Ge132 derivatives. Bioorg. Med. Chem. Lett. 2005;15:2962–2965. doi: 10.1016/j.bmcl.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 43.Suh D., Chaires J.B. Criteria for the mode of binding of DNA binding agents. Bioorg. Med. Chem. 1995;3:723–728. doi: 10.1016/0968-0896(95)00053-j. [DOI] [PubMed] [Google Scholar]
- 44.Nitta Y., Kuroda R. Quantitative analysis of DNA–porphyrin interactions. Biopolymers. 2006;81:376–391. doi: 10.1002/bip.20430. [DOI] [PubMed] [Google Scholar]
- 45.Tuite E., Sehlstedt U., Hagmar P., Norden B., Takahashi M. Effects of minor and major groove-binding drugs and interactions on the DNA association of minor groove-binding proteins RecA and deoxyribonuclease I detected by flow linear dichroism. Eur. J. Bichem. 1997;243:482–492. doi: 10.1111/j.1432-1033.1997.0482a.x. [DOI] [PubMed] [Google Scholar]