Abstract
Erwinia chrysanthemi causes soft-rot diseases of various plants by enzymatic degradation of the pectin in plant cell walls. Pectin is a complex polysaccharide. The main chain is constituted of galacturonate residues, and some of them are modified by methyl and/or acetyl esterification. Esterases are necessary to remove these modifications and, thus, to facilitate the further degradation of the polysaccharidic chain. In addition to PaeY, the first pectin acetylesterase identified in the E. chrysanthemi strain 3937, we showed that this bacterium produces a second pectin acetylesterase encoded by the gene paeX. The paeX open reading frame encodes a 322-residue precursor protein of 34,940 Da, including a 21-amino-acid signal peptide. Analysis of paeX transcription, by using gene fusions, revealed that it is induced by pectic catabolic products and affected by catabolite repression. The expression of paeX is regulated by the repressor KdgR, which controls all the steps of pectin catabolism; by the repressor PecS, which controls most of the pectinase genes; and by catabolite regulatory protein, the global activator of sugar catabolism. The paeX gene is situated in a cluster of genes involved in the catabolism and transport of pectic oligomers. In induced conditions, the two contiguous genes kdgM, encoding an oligogalacturonate-specific porin, and paeX are both transcribed as an operon from a promoter proximal to kdgM, but transcription of paeX can also be uncoupled from that of kdgM in noninduced conditions. PaeX is homologous to the C-terminal domain of the Butyrivibrio fibriosolvens xylanase XynB and to a few bacterial esterases. PaeX contains the typical box (GxSxG) corresponding to the active site of the large family of serine hydrolases. Purified PaeX releases acetate from various synthetic substrates and from sugar beet pectin. The PaeX activity increased after previous depolymerization and demethylation of pectin, indicating that its preferred substrates are nonmethylated oligogalacturonides. PaeX is mostly found in the periplasmic space of E. chrysanthemi. These data suggest that PaeX is mainly involved in the deacetylation of esterified oligogalacturonides that enter the periplasm by the KdgM porin.
Plant cell walls contain up to 7% O-bound acetyl groups by dry weight. These acetyl moieties are mostly linked to two polysaccharides, xylan and pectin. Efficient breakdown of plant polysaccharides is achieved by the concerted action of several enzymes. Esterases are required for the removal of acetyl groups esterified to the O-2 and/or O-3 position of sugars. The action of depolymerases is often precluded by the presence of these side groups, whose removal is necessary prior to, or concomitantly with, the action of depolymerases.
Pectins are important structural constituents of plant cell walls, and they play a key role in plant physiology and plant pathology. The general structure of pectic polymers consists of linear polygalacturonate chains interspersed with highly branched rhamnogalacturonan chains. Some of the galacturonate residues are modified by methyl esterification at the carboxyl group or acetyl esterification on the hydroxyl groups. The degrees of methylation and acetylation vary greatly, depending on the source of pectin. Many saprophytic and plant-pathogenic microorganisms secrete enzymes involved in pectin degradation. While several xylan acetylesterases have been characterized for a variety of fungi and bacteria, very little genetic and biochemical information is available on pectin acetylesterases. PaeY of Erwinia chrysanthemi is the only bacterial representative that has been identified (27). A fungal gene encoding a rhamnogalacturonan acetylesterase has been isolated (25), and biochemical data have been obtained from plant pectin acetylesterases (4, 5, 33).
E. chrysanthemi is a phytopathogenic bacterium which causes soft-rot disease of various plants. This bacterium is capable of using pectin as its sole carbon source for growth. E. chrysanthemi effects pectin depolymerization through the production of multiple pectate lyases (1, 14, 19, 32). The depolymerization of pectin is favored by the previous action of esterases, since pectate lyases exhibit a reduced activity on acetylated or methylated substrates (27, 32). In E. chrysanthemi 3937, the pectin methylesterase activity is mainly due to the secreted protein PemA, while a second isoenzyme, PemB, is a cell-linked outer membrane lipoprotein (26). The pectin acetylesterase PaeY was shown previously to act in synergy with pectate lyases and pectin methylesterases for an efficient breakdown of pectin (27).
Most of the pectinase genes are organized in clusters, and their transcription is induced in the presence of pectin (10, 11, 27, 28). This induction results from the intracellular formation of catabolic products such as 2-keto-3-deoxygluconate (KDG) and is mainly mediated by the specific repressor KdgR (17). In addition, the regulation of the pectinase genes involves various global transcriptional regulators, such as the repressors PecS and PecT, and the activator catabolite regulatory protein (CRP) (21, 22, 30).
During the study of PaeY, we developed a method for the direct detection of acetylesterases after electrofocusing, using a chromogenic substrate (27). Analysis of E. chrysanthemi culture supernatants revealed the presence of several acetylesterases (27). The induction of one of these esterases in the presence of pectin suggested that it could be involved in pectin degradation. In this paper, we report the identification and characterization of this second pectin acetylesterase, PaeX.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. The paeX::uidA-Km fusion and the paeX-Cm mutation were transduced into various strains by using the φ-EC2 generalized transducing phage (20).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or phenotype | Reference or origin |
|---|---|---|
| E. chrysanthemi strains, 3937 derivatives | ||
| A350 | lmrTclacZ2 | Laboratory collection |
| A576 | lmrTclacZ2 arg10 kdgK | Laboratory collection |
| A1077 | lmrTclacZ2 kdgR::Mu Cm | Laboratory collection |
| A1091 | lmrTclacZ2 arg10 kdgK paeY::lacZ Km | 27 |
| A1524 | lmrTclacZ2 pecS::Mu Cm | 22 |
| A2174 | lmrTclacZ2 pecT::Cm | 30 |
| A2507 | lmrTclacZ2 crp::Cm | 21 |
| A3433 | lmrTclacZ2 arg10 kdgK togM::uidA Km | 9 |
| A3719 | lmrTclacZ2 kdgR::Sm | Laboratory collection |
| A3785 | lmrTclacZ2 paeX::uidA Km | This work |
| A3816 | lmrTclacZ2 arg10 kdgK kdgM::uidA Km | 3 |
| A3910 | lmrTclacZ2 arg10 kdgK paeX::uidA Km | This work |
| A3911 | lmrTclacZ2 kdgR::Mu Cm paeX::uidA Km | This work |
| A3912 | lmrTclacZ2 pecS::Mu Cm paeX::uidA Km | This work |
| A3913 | lmrTclacZ2 pecT::Cm paeX::uidA Km | This work |
| A3914 | lmrTclacZ2 crp::Cm paeX::uidA Km | This work |
| A3922 | lmrTclacZ2 arg10 kdgK paeX::uidA Km outD::Cm | This work |
| A4026 | lmrTclacZ2 paeX-Cm | This work |
| A4027 | paeY::lacZ Km | This work |
| A4029 | paeX-Cm | This work |
| A4030 | paeX-Cm paeY::lacZ Km | This work |
| A4039 | kdgK::Cm | T. Franza |
| A4043 | lmrTclacZ2 kdgR::Sm pecS::Mu Cm | Laboratory collection |
| A4122 | lmrTclacZ2 paeY::lacZ Km | This work |
| A4123 | lmrTclacZ2 paeX-Cm paeY::lacZ Km | This work |
| Other E. chrysanthemi strains | ||
| B374 | Wild type | A. Toussaint |
| EC16 | Wild type | N. T. Keen |
| ENA49 | Wild type | Laboratory collection |
| EP2 | Wild type | M. Boccara |
| E. carotovora subsp. carotovora strains | ||
| SCRI193 | Wild type | M. Perombelon |
| CC3-2 | Wild type | Laboratory collection |
| E. carotovora subsp. atroseptica strains | ||
| SCRI31 | Wild type | M. Perombelon |
| CA36A | Wild type | Laboratory collection |
| Plasmids | ||
| pBSCm | Bluescript KS(+) Cm | Stratagene |
| pN2101 | pBSCm derivative with a 1.4-kb NarI-EagI fragment; paeX+ | This work |
| pL306 | pN2101 derivative with a uidA-Km cassette in the MluI site; paeX::uidA Km | This work |
| pT7-5 | φ10 promoter; Ap | 31 |
| pN2170 | pT7-5 derivative with a 1.4-kb NarI-EagI fragment; paeX+ | This work |
Media and growth conditions.
Bacteria were grown in complete Luria-Bertani (LB) medium or in synthetic M63 medium (15). When required, the media were solidified with Difco agar (15 g · liter−1). E. chrysanthemi cells were usually incubated at 30°C, and Escherichia coli cells were incubated at 37°C. Carbon sources were added at 2 g · liter−1, except for polygalacturonate and pectin, added at 4 g · liter−1. Polygalacturonate (grade II) was obtained from Sigma Chemical Co. Sugar beet pectin was a gift from Copenhagen Pectin (degree of acetylation, 23%; degree of methylation, 56%; mean molecular weight, 62,000; content in galacturonic acid residues, 66%). When required, antibiotics were added at the concentrations indicated: kanamycin, 20 μg · ml−1; ampicillin, 50 μg · ml−1; chloramphenicol, 20 μg · ml−1; streptomycin, 100 μg · ml−1.
Recombinant DNA techniques.
Preparation of plasmid DNA, restriction digestions, ligations, DNA electrophoresis, and bacterial transformations were carried out according to classical methods (24). Deletions for nucleotide sequencing were generated with restriction endonucleases, and the sequences were determined by Genome Express SA (Grenoble, France).
The uidA-Km cassette was liberated by SmaI digestion of pUIDK1 (2) and inserted into the MluI site of plasmid pN2101, previously filled with Klenow enzyme. In one of the recombinant plasmids, pL306, uidA is oriented in the same direction of transcription as paeX, giving rise to a paeX::uidA fusion. The pL306 plasmid was introduced into E. chrysanthemi cells by electroporation. The paeX mutation was then introduced into the E. chrysanthemi chromosome by marker-exchange recombination after successive cultures in low-phosphate medium in the presence of kanamycin (23).
Overproduction and purification of PaeX.
Overexpression of the paeX gene was obtained by using the T7 promoter-T7 RNA polymerase system (31). The paeX gene was subcloned into the pT7-5 expression vector under the T7 promoter (pN2170). The plasmid pN2170 was introduced in E. coli BL21(DE3), which contains a single chromosomal copy of the gene encoding T7 RNA polymerase under the control of the lacUV5 promoter (29). The BL21(DE3)/pN2170 cells were grown at 30°C in LB medium supplemented with ampicillin (150 μg · ml−1). At an optical density at 600 nm of 0.8 to 1, the synthesis of T7 RNA polymerase was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside and cells were grown for an additional 2 to 3 h.
Cells were harvested by centrifugation for 10 min at 5,000 × g at 4°C and then frozen at −80°C. The periplasmic fraction was extracted from cells by three cycles of freezing-thawing (12). Proteins were concentrated by 85% ammonium sulfate precipitation. The pellet was solubilized in 50 mM sodium phosphate buffer (pH 7) containing 5 mM EDTA and 1.5 M ammonium sulfate and loaded onto a Phenyl-TSK-Gel column equilibrated with the same buffer. The column was extensively washed with buffer containing 1 M ammonium sulfate. Upon application of a 1 to 0 M ammonium sulfate linear gradient, the PaeX protein was eluted at about an 0.4 M ammonium sulfate concentration. The fractions containing PaeX were pooled and concentrated with Centricon 10 (Amicon). During the purification steps, the presence of acetylesterase activity was monitored by the X-acetate (5-bromo-6-chloro-3-indolylacetate) assay.
Cellular fractionation and protein analysis.
The release of periplasmic proteins from E. coli cells was realized by osmotic shock (7).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on slab gels (4% stacking gel and 12% separating gel) with the Mini-Protean II system (Bio-Rad Laboratories). Proteins were stained with Coomassie blue G-250. Electrofocusing was performed in a 3 to 10 pH gradient with Pharmalytes. To detect acetylesterase activity, the gel was incubated for 5 to 30 min in 0.1 M Tris-HCl (pH 7.5) buffer containing 1 mM X-acetate. Proteins with acetylesterase activity form blue bands.
Purified PaeX protein was used to immunize a rabbit to obtain PaeX antiserum. Immunoblotting was performed as described previously (26). PaeX antiserum was diluted 1:5,000. Detection was performed with the ECL detection kit (Amersham).
The PaeX protein was subjected to N-terminal sequencing by Edman degradation at the Institut de Biologie et Chimie des Proteines (Lyon, France).
Enzyme assays.
Acetylesterase activity was measured with the synthetic substrates p-nitrophenyl acetate (pNPA), triacetin, and X-acetate (Sigma Chemical Co.). Hydrolysis of pNPA was monitored spectrophotometrically at 400 nm by the formation of p-nitrophenol. The assay mixture (1 ml) containing 5 mM pNPA in 0.1 M sodium acetate buffer (pH 5.8) was incubated at 30°C with 1 to 10 μg of the pure enzyme. Cleavage of X-acetate was estimated spectrophotometrically at 650 nm by the formation of the blue product. X-acetate (1 mM) was incubated with enzyme at 30°C in 0.1 M Tris-HCl buffer, pH 7.5. During the 5- to 60-min incubation period, samples of the reaction mixture were removed and added to 3 volumes of methanol and the absorbance at 650 nm was measured immediately. Acetylesterase activity was also determined by measuring the release of acetate from triacetin with the acetic acid kit (Boehringer Mannheim Roche). Triacetin (50 mM) was incubated with enzyme at 30°C in 0.1 M Tris-HCl buffer, pH 7.5. Specific activity is expressed in micromoles of product liberated per minute per microgram of protein.
Further measurements of pectin acetylesterase activity were performed on sugar beet pectin (degree of acetylation, 23%; degree of methylation, 56%). The pectin (10 mg · ml−1) was incubated with enzyme at 30°C in 0.1 M Tris-HCl buffer, pH 7.5. Every 30 min, samples were removed, and after heat inactivation, the concentration of released acetate was determined with the acetic acid kit (Boehringer Mannheim Roche). Specific activity is expressed in micromoles of acetate liberated per minute per microgram of protein.
Pectate lyase activity was determined by monitoring spectrophotometrically the formation of unsaturated products from polygalacturonate at 230 nm (16). When specified, sugar beet pectin was used instead of polygalacturonate. Pectate lyase specific activity is expressed in micromoles of unsaturated products per minute per milligram (dry weight) of bacteria.
β-Glucuronidase was measured by monitoring the formation of p-nitrophenol from p-nitrophenyl-β-d-glucuronide at 405 nm (2). β-Glucuronidase specific activity is expressed as nanomoles of product liberated per minute per milligram (dry weight) of bacteria.
Pathogenicity tests.
Chicory leaves were slightly wounded prior to inoculation. For each strain, 16 leaves were infected with 106 bacteria per inoculation site. After incubation in a dew chamber for 24 h, the length of rotted tissue was measured to estimate the disease severity. The experiment was repeated three times.
Nucleotide sequence accession number.
Sequence data reported in this paper will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under the accession number AJ507215.
RESULTS AND DISCUSSION
Determination of the paeX nucleotide sequence.
We recently identified a cluster of genes involved in the catabolism and uptake of oligogalacturonides resulting from extracellular pectin degradation (9). To determine whether this region contains other genes involved in pectin degradation, we determined the nucleotide sequence of the DraI-EagI fragment of 1,280 nucleotides (nt) situated downstream of the kdgM gene (accession number AJ507215). Sequence analysis revealed the presence of a unique complete open reading frame (ORF) that begins with an ATG codon at position 150 and ends with TAA at position 1116. The ATG start is preceded (8 nt) by the potential ribosome-binding site GGGTAAA. This ORF, named paeX, encodes a predicted 322-amino-acid protein. The N-terminal sequence presents the essential features of signal peptides: a short positively charged region (residues 1 to 5) is followed by a long hydrophobic region (residues 6 to 21). The existence of this signal sequence was confirmed by determination of the N-terminal sequence of the purified PaeX protein (see below), indicating that the mature form begins at residue 22. The precursor and mature forms of PaeX have a predicted molecular mass of 34,940 and 32,716 Da, respectively.
Comparison of the deduced amino acid sequence of PaeX with proteins present in databases revealed homology with a hypothetical protein of Caulobacter crescentus (GenBank accession number AAK24284.1) (46% over the entire length of the proteins) and with the C-terminal domain of the endo-1,4-β-xylanase XynB of Butyrivibrio fibriosolvens (37% identity over 208 residues) (GenBank accession number X61495). Xylanases often have a modular organization, and XynB is a large protein of 635 residues, supposed to contain two domains (13). The xylanase activity is due to the N-terminal domain of XynB (residues 1 to 337), which is homologous to family 10 of glycosyl hydrolases (P. M. Coutinho and B. Henrissat, http://afmb.cnrs.mrs.fr/∼cazy/CAZY/index.html, 1999). Homology with PaeX is found in the XynB C-terminal domain of unknown function, from residues 405 to 609 (Fig. 1). Since enzymatic analysis revealed that PaeX has a pectin acetylesterase activity (see below), the homology data indicate that the xylanase XynB is likely to be a bifunctional enzyme, with associated hydrolase and esterase activities.
FIG. 1.
Comparison of PaeX with its homologues. The PaeX sequence, from residues 80 to 322 (“PaeX E. ch”), is aligned with the C-terminal domain of a B. fibriosolvens xylanase (X61495) (“XynB B. fi”) and with a hypothetical protein of C. crescentus (AAK24284.1) (“Hyp C. cr”). The residues conserved in all the sequences are given below the alignment. The residues corresponding to the G-S-G motif of serine hydrolases are underlined. The arrowheads point to the conserved residues, S, D, and H, predicted to constitute the catalytic triad of a serine hydrolase.
Other proteins showing homology with PaeX were found, including the heroin esterase Her of Rhodococcus sp. (24% identity), the esterase EstB of Thermotoga maritima (23% identity), and several hypothetical proteins from Agrobacterium tumefaciens (41% of identity), C. crescentus (34% identity), Deinococcus radiodurans (32 and 27% identity), Clostridium acetobutylicum (32% identity), Lactococcus lactis (31% identity), and Bacillus subtilis (YtmA, 28% identity). The most highly conserved region among PaeX, XynB, and their homologues overlaps a sequence (GxSxG) typical of the active site of a large and diverse family of serine hydrolases, including carboxylesterases, lipases, and proteases. The reaction mechanism of these enzymes involves a catalytic triad with Ser, Asp, and His residues. Visual inspection of the sequence alignments revealed the presence of conserved Ser, Asp, and His in PaeX and homologous proteins. From analogy with the other serine hydrolases, Ser161 could be the active site residue of PaeX, Asp251 and His283 being the two other residues of the catalytic triad (Fig. 1).
A recent classification based on sequence similarity distinguished 13 families of carbohydrate esterases (P. M. Coutinho and B. Henrissat, http://afmb.cnrs.mrs.fr/∼cazy/CAZY/index.html,1999). PaeY of E. chrysanthemi and the rhamnogalacturonan acetylesterase of Aspergillus aculeatus belong to family 12. PaeX shows no significant homology with any members of these 13 families. Thus, PaeX and XynB appear to form a new family of carbohydrate esterases.
Transcription of the paeX gene.
To analyze paeX expression, we constructed a paeX::uidA transcriptional fusion by inserting an uidA-Km cassette into the unique MluI site located in paeX. In the absence of inducer, paeX showed a low basal level of expression, and its transcription was stimulated about fourfold in the presence of polygalacturonate (Table 2). The presence of a readily utilizable carbon source, such as glucose, provoked a twofold decrease in paeX transcription (Table 2).
TABLE 2.
Expression of the paeX::uidA fusion
| Strain (main genotype) | Carbon source(s)a | Sp actb
|
|
|---|---|---|---|
| Pectate lyase | β-Glucuron- idase | ||
| A3785 (paeX::uidA) | Glycerol | 0.06 | 56 |
| Glycerol + PGAc | 3.26 | 207 | |
| Glucose | 0.04 | 29 | |
| Glucose + PGA | 1.25 | 109 | |
| A3910 (kdgK paeX::uidA) | Glycerol | 0.04 | 55 |
| Glycerol + PGA | 51.95 | 1,247 | |
| A3911 (kdgR paeX::uidA) | Glycerol | 0.63 | 327 |
| Glycerol + PGA | 2.81 | 646 | |
| A3912 (pecS paeX::uidA) | Glycerol | 0.66 | 110 |
| Glycerol + PGA | 8.70 | 389 | |
| A3913 (pecT paeX::uidA) | Glycerol | 0.97 | 52 |
| Glycerol + PGA | 8.01 | 184 | |
| A3914 (crp paeX::uidA) | Glucose | 0.01 | 25 |
| Glucose + PGA | 0.02 | 28 | |
Cultures were grown in M63 minimal medium to late log phase, in the absence or presence of polygalacturonate.
β-Glucuronidase specific activity reflects the expression of the paeX::uidA fusion and is expressed as nanomoles of product liberated per minute per milligram (dry weight) of bacteria. Pectate lyase specific activity is expressed as micromoles of product liberated per minute per milligram (dry weight) of bacteria. The results reported are the averages of at least three independent experiments, and the standard deviation was, in each case, less than 10% for β-glucuronidase and less than 20% for pectate lyase assays.
PGA, polygalacturonate.
The paeX::uidA fusion was transduced into strains containing regulatory mutations affecting pectinase production, namely, crp, pecS, pecT, and kdgR (Table 2) (10). The expression of the paeX::uidA fusion appeared to be unaffected by the pecT mutation. In the crp mutant, expression of the paeX::uidA fusion decreased, demonstrating that CRP activates paeX transcription. The pecS mutation provoked an increase in paeX::uidA transcription, indicating that the protein PecS, which represses the transcription of most pectinase genes, acts also as a repressor of paeX expression. The paeX::uidA expression was clearly affected by kdgR and kdgK mutations (Table 2). The kdgR gene has a major role in the regulation of pectin catabolism. The presence of pectin catabolic products, mainly KDG, provokes the dissociation of the repressor from its operators (18). Both paeX expression and pectate lyase activity are greatly increased in the kdgR mutant in the absence of polygalacturonate, due to inactivation of the repressor KdgR, and in the kdgK mutant in the presence of polygalacturonate, due to the accumulation of the intracellular inducer KDG (Table 2). These results indicate that paeX induction is dependent on the KdgR-KDG couple. Thus, the regulation of paeX expression resembles that of paeY and of other E. chrysanthemi pectinase genes (10). The paeX transcription is (i) subject to pectin induction mediated by the repressor KdgR, (ii) regulated by the repressor PecS, and (iii) subject to catabolite repression mediated by the activator CRP.
The paeX ORF begins 272 nt after the kdgM stop codon. A computer search performed to identify potential promoter sequences in the 5′ noncoding region of paeX revealed no sequence presenting a significant level of homology with the classical consensus. Moreover, no sequence presenting a significant homology either with the CRP binding site or with the KdgR binding site was observed in the 5′ untranslated region of paeX, while analysis of gene fusions indicated transcriptional control by these two regulators. For some pectinase genes, such as paeY (27), the apparently indirect control by KdgR was shown to be due to the transcription of a polycistronic mRNA from a distant KdgR-controlled promoter.
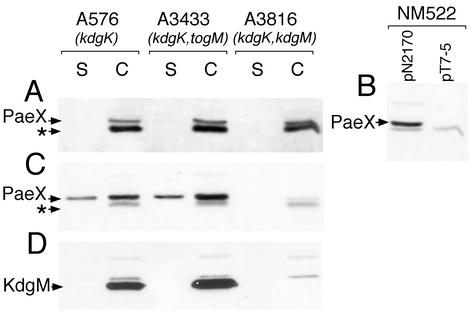
To determine whether the paeX gene could be part of an operon, we analyzed the effect of polar insertion mutations in the preceding genes, togM and kdgM, on PaeX production. PaeX and KdgM production was detected by immunoblotting on extracts of strains derived from the E. chrysanthemi kdgK mutant A576. In the presence of a kdgK mutation, the expression of pectin-induced genes is strongly increased in the presence of polygalacturonate or galacturonate. In the absence of inducer in the medium, the same basal level of PaeX was detected in the kdgM and togM mutants and in the parental strain (Fig. 2A). Thus, paeX is preceded by a functional promoter region. We verified the promoter activity of the kdgM-paeX intergenic region by introduction into E. coli NM522, which does not express the T7 RNA polymerase, of the plasmid pN2170 carrying the paeX gene with a 246-nt upstream region cloned into the pT7-5 vector. The production of PaeX in this strain confirmed that a functional promoter is situated upstream of paeX (Fig. 2B).
FIG. 2.
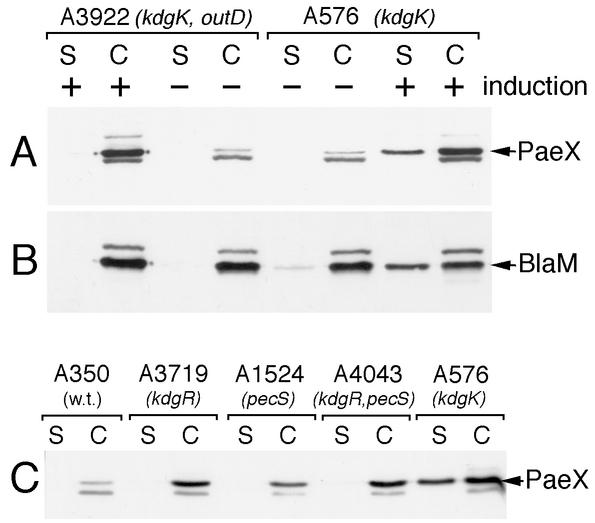
Effect of the kdgM and togM polar mutations on PaeX production. E. chrysanthemi A576 (kdgK), A3433 (kdgK togM), and A3816 (kdgK kdgM) were grown in LB medium (A) or in LB medium supplemented with 0.2% galacturonate (C and D) until the early stationary phase. The culture supernatant (S) and whole-cell (C) fractions were separated by SDS-PAGE and analyzed by immunoblotting with either PaeX (A, B, and C) or KdgM (D) antibodies. Immunoblotting analysis with PaeX antibodies was also performed on the whole-cell extracts of E. coli NM522 carrying either pT7-5 or pN2170 (pT7-5 with paeX) (B). The arrows indicate the positions of PaeX, KdgM, and an unknown protein (*) cross-reacting with the PaeX antiserum.
When the E. chrysanthemi kdgK mutants were grown in the presence of galacturonate in the medium, PaeX production increased strongly, except in the kdgM mutant (Fig. 2C). Thus, in induced conditions, paeX is transcribed in the polycistronic mRNA originating from the kdgM promoter. In contrast, neither PaeX nor KdgM production is affected by a mutation in togM, which is a part of the operon situated upstream of kdgM (3) (Fig. 2D). The synthesis of a polycistronic mRNA explains the regulation of paeX by KdgR, PecS, and CRP, which occurs as a consequence of the interaction of these proteins with their binding sites situated upstream of kdgM (3).
Therefore, paeX can be transcribed both, at a low level, in a monocistronic mRNA and, at a high level in induced conditions, in a polycistronic mRNA originating from the kdgM promoter. This type of regulation has been previously observed for pectinase genes. For instance, in induced conditions the contiguous genes pelD, paeY, and pemA are transcribed as an operon, with a polycistronic mRNA originating from a pelD proximal promoter. In noninduced conditions, a monocistronic mRNA is formed from an internal promoter. The simultaneous synthesis of the three pectinases PelD, PaeY, and PemA reflects their synergistic action on pectin (27). A similar organization for paeX and kdgM suggests that the action of the pectin acetylesterase PaeX could be coupled to that of the oligogalacturonate-specific porin, KdgM, to ensure a deacetylation of oligogalacturonides concomitant with their transport into the periplasm.
Role of paeX in the pathogenicity of E. chrysanthemi and in pectin catabolism.
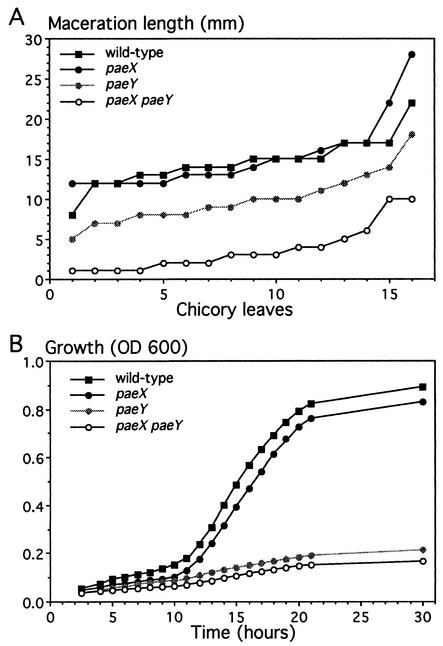
Pectate lyases are directly responsible for the symptom of maceration observed in the plant tissues infected with E. chrysanthemi. The pectin acetylesterase action probably favors pectin degradation by making the substrate more readily available for cleavage by pectate lyases. We first compared the pathogenic behavior of the E. chrysanthemi paeX mutant with that of the wild-type strain (Fig. 3A). We observed no significant difference between the two strains. Since there are two pectin acetylesterases in E. chrysanthemi 3937, we also infected chicory leaves with a paeY mutant and a paeY paeX double mutant. As shown previously, we observed that the virulence of the paeY mutant is attenuated (27). Moreover, the paeY paeX double mutant showed only a very reduced maceration of plant tissue compared to any other strain. The clear difference between the paeY single mutant and the paeY paeX double mutant indicates that both E. chrysanthemi pectin acetylesterases are important for the development of soft rot.
FIG. 3.
Analysis of the pae mutants. (A) Effect of the pae mutations on virulence. Sixteen chicory leaves were infected for each strain. After incubation at 30°C for 24 h, the length of rotted tissue was measured to estimate the disease severity. Distribution of the extent of maceration is represented, each point corresponding to one leaf. Comparison of the distribution obtained for each strain by the statistical chi-square test indicates a significant difference between pairs of strains, except between the paeX mutant and the wild-type strain. This experiment was repeated three times, and the different experiments gave similar results. (B) Growth of E. chrysanthemi pae mutants on pectin. The E. chrysanthemi strains 3937, A4027 (paeY), A4029 (paeX), and A4030 (paeX paeY) were grown in M63 minimal medium supplemented with 0.4% sugar beet pectin (degree of acetylation, 23%). Cell density was estimated by measurement of the optical density at 600 nm.
To estimate the contribution of both pectin acetylesterases to pectin degradation by E. chrysanthemi, we analyzed whether its ability to utilize pectin as a carbon source for growth is affected by the corresponding mutations. We monitored the growth of single paeX and paeY mutants, the double paeX paeY mutant, and the wild-type strain, using a 23% acetylated sugar beet pectin as the sole carbon source (Fig. 3B). Compared to the parental strain, the mutants showed a longer lag time, a lower growth rate, and a lower final total growth. The paeY mutant showed pronounced growth limitation. The growth of the paeX mutant is weakly, but consistently, affected. Moreover, the growth of the double paeX paeY mutant appeared to be affected more than that of the paeY mutant. Therefore, pectin utilization by E. chrysanthemi involves the complementary actions of PaeY and PaeX.
Characterization of the PaeX protein.
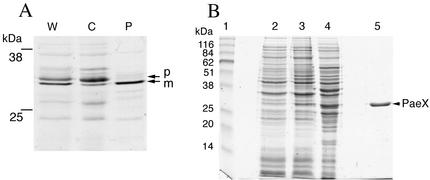
The paeX gene was inserted in a pT7 vector, allowing specific expression of the cloned gene by T7 RNA polymerase. Specific labeling of the plasmid-encoded proteins with [35S]cysteine-methionine, followed by subcellular fractionation, showed that the precursor form of PaeX (32 kDa) is not released by osmotic shock while the mature form (30 kDa) is found in the periplasmic fraction (Fig. 4A). After optimization of the overproduction conditions, the mature PaeX was purified from the recombinant E. coli cells after ammonium sulfate fractionation and chromatography on a hydrophobic interaction column (Fig. 4B). The N-terminal sequence of purified mature PaeX was determined (DTIFPIWP), confirming the presence of a N-terminal signal sequence.
FIG. 4.
Fractionation and purification of the PaeX protein. (A) Cellular localization of PaeX in E. coli. Cells of E. coli K38/pGP1.2/pN2170 were labeled with [35S]cysteine-methionine. The periplasmic proteins were extracted from the labeled cells by osmotic shock. W, whole cells; P, periplasmic fraction; C, osmotically shocked cell fraction. The proteins were separated by SDS-PAGE, and the gels were autoradiographed. Precursor (p) and mature (m) forms of PaeX are indicated. (B) Purification of PaeX. Shown are whole-cell lysates of E. coli BL21(DE3)/pN2170 before (lane 2) and after (lane 3) induction, extract from induced cells (lane 4), and purified PaeX (lane 5). The proteins were separated by SDS-PAGE and stained with Coomassie blue G-250. Apparent molecular masses of the standards (lane 1) are indicated. The position of PaeX is indicated.
The purified PaeX protein was tested for acetylesterase activity. Previously characterized pectin acetylesterases release acetyl groups from pectin and also from a range of synthetic substrates, such as pNPA, triacetin, or X-acetate. Incubation of PaeX with the chromogenic substrate pNPA or X-acetate showed that PaeX is able to cleave these compounds, liberating acetate and the colored products (Table 3). Incubation of PaeX with triacetin, followed by the detection of liberated acetate, demonstrated that PaeX also deacetylates this substrate. PaeX removed up to 20% of the acetyl groups from triacetin. With sugar beet pectin (23% acetylated) as the substrate, PaeX removed about 2 to 3% of the total acetyl groups after prolonged incubation (Table 3). This activity increased when sugar beet pectin was demethylated by PemA and cleaved by PelB (see below), indicating that nonmethylated oligomers are better PaeX substrates than is the polymer.
TABLE 3.
Acetylesterase activity of PaeX with various substrates
| Substrate | Sp acta (U · μg of protein−1) | Final acetate releaseb (% of total content) |
|---|---|---|
| pNPA | 15 ± 3 | NDc |
| X-acetate | 5 ± 1 | ND |
| Triacetin | 0.50 ± 0.20 | 30 ± 5 |
| Sugar beet pectin | ND | 2 ± 1 |
| Sugar beet pectin treated with PemA and PelBd | 0.40 ± 0.15 | 25 ± 4 |
The initial rate of the reaction was measured at 30°C by monitoring the release of the colored compound (pNPA and X-acetate) or by assay of the liberated acetate (triacetin and pectin). Each experiment was performed at least three times, and the data are averages with standard errors.
The final acetate release was determined after incubation of the substrate with PaeX (5 μg/ml) at 30°C for 12 h. The total content of acetate from triacetin and pectin was determined after their chemical saponification. Each experiment was performed at least three times, and the data are averages with standard errors.
ND, not determined (very low activity).
Sugar beet pectin (23% acetylated, 56% methylated), 10 g·liter−1 in 0.1 M Tris-HCl buffer (pH 8.0), was treated with the combination of PemA (2 μg· ml−1) and PelB (1 μg·ml−1). After incubation at 30°C for 12 h, this mixture was used as substrate for PaeX.
X-acetate was chosen as the substrate to determine the PaeX biochemical characteristics. The optimal pH for PaeX activity was 8.7. Addition of the chelating agent EDTA or of cations (Ca2+, Co2+, Mg2+, Mn2+, Ba2+, Sn2+, and Fe2+) at a 1 mM concentration did not affect PaeX activity. PaeX was completely inactive in the presence of 0.01% SDS. The addition of N-lauroylsarcosine or Nonidet P-40, at the same concentration, increased the PaeX activity by fivefold and twofold, respectively. Other detergents, such as Triton X-100, Triton X-114, or Tween 20, had no significant influence on PaeX activity. The positive influence of detergents was not observed when triacetin was used as the substrate; thus, this probably resulted from the effect of detergents on the X-acetate solubility rather than on PaeX activity.
Cellular localization of PaeX.
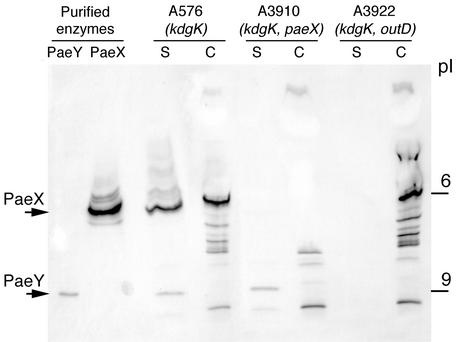
We developed a method for the direct detection of acetylesterases after electrofocusing, using the synthetic substrate X-acetate (27). The apparent isoelectric point of the purified PaeX protein was about 6 (Fig. 5). To analyze the different acetylesterase(s) specifically produced by E. chrysanthemi in the presence of pectin, we used the kdgK mutant A576, in which the expression of pectin-induced genes is strongly increased in the presence of galacturonate. In the supernatant of the kdgK mutant, bands corresponding to PaeX and PaeY were observed at pI 6 and 9, respectively. The band corresponding to PaeX was also detected in the periplasm of the kdgK mutant. Analysis of the paeX mutant confirmed that the bands at pI 6 disappear from both the supernatant and the periplasm (Fig. 5).
FIG. 5.
Electrofocusing followed by specific detection of acetylesterase activity. E. chrysanthemi A576 (kdgK), A3910 (kdgK paeX), and A3922 (kdgK outD) were grown in LB medium supplemented with 0.2% galacturonate. The culture supernatant (S) and whole-cell (C) fractions were separated by electrofocusing and stained with X-acetate. Purified PaeY and PaeX proteins were used as controls. The positions of the pI standards and of PaeX and PaeY are indicated.
Immunoblotting detection also confirmed that about 20 to 40% of PaeX is detected in the culture supernatant of the kdgK mutant A576 (Fig. 6A). Most of the E. chrysanthemi pectinases are secreted via the type II Out system. Thus, we analyzed whether the Out system could be involved in the partial secretion of PaeX. In the double kdgK outD mutant A3922, PaeX was completely retained in the cells (Fig. 5 and 6A), indicating that secretion of PaeX is dependent on the presence of the Out system. However, the partial secretion of PaeX is observed only in the kdgK mutants in induced conditions. Other regulatory mutations increasing pectinase synthesis (kdgR, pecS, and the double kdgR pecS) led to an increased PaeX production but did not provoke secretion of this protein in the medium (Fig. 6C). In contrast to the secreted pectinases that are completely, or almost completely, secreted in the medium by the Out system, only 20 to 40% of PaeX was detected in the culture supernatant. Therefore, we controlled the cellular localization of a small periplasmic protein, β-lactamase, in the strains A576 (kdgK) and A3922 (kdgK outD) carrying the pT7-6 plasmid. In noninduced conditions, both β-lactamase and PaeX were detected only within the cells of the two strains (Fig. 6A and B). Addition of galacturonate drastically increased PaeX production and led to a partial liberation of both PaeX and β-lactamase in the medium, but only in the A576 Out+ strain. Similar proportions of PaeX and β-lactamase were detected in the culture supernatant of A576. Thus, despite the fact that PaeX liberation in the medium depends on the presence of the Out secretion machinery, this process cannot be considered true Out-dependent secretion. The high intracellular concentration of inducer reached when kdgK mutants are grown in the presence of galacturonate provokes an increased synthesis of most of the Out-secreted proteins. This probably leads to a partial liberation of the proteins retained in the periplasm by an overloaded Out secretion system.
FIG. 6.
Cellular localization of PaeX. (A and B) E. chrysanthemi A576 (kdgK) and A3922 (kdgK outD) carrying the pT7-6 plasmid were grown in LB medium (−) or in LB medium supplemented with 0.2% galacturonate (+) until the early stationary phase. The culture supernatant (S) and whole-cell (C) fractions were separated by SDS-PAGE and analyzed by immunoblotting with either PaeX (A) or BlaM (B) antibodies. (C) The parental E. chrysanthemi strain A350 and different regulatory mutants were grown in LB medium supplemented with 0.2% galacturonate. The culture supernatant (S) and whole-cell (C) fractions were analyzed by immunoblotting. Positions of PaeX and BlaM are indicated by arrows.
Occurrence of PaeX homologues in Erwinia spp.
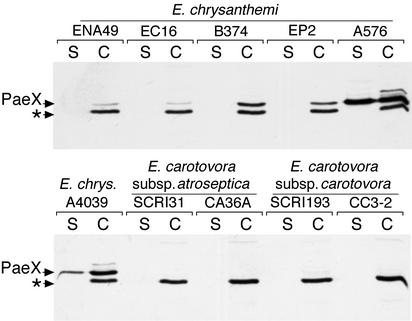
To test the presence of PaeX homologues in other Erwinia species, immunoblotting experiments were performed with culture supernatants and cell extracts of different strains of E. chrysanthemi, Erwinia carotovora subsp. carotovora, and Erwinia carotovora subsp. atroseptica. A band corresponding to a 32-kDa protein was detected in the cell extracts, but not in the culture supernatants, of all the E. chrysanthemi wild-type strains (Fig. 7). As shown previously, the 32-kDa protein was also detected in the supernatant of E. chrysanthemi kdgK mutants (A576 and A4039) (Fig. 7). No specific cross-reaction was detected with the E. carotovora strains. Analysis of the genome of the E. carotovora subsp. atroseptica strain SCRI 1043 (Sanger Institute, unfinished genome) indicated the presence of a potential paeX homologue (85% identity at the protein level). Thus, the negative response obtained by immmunodetection with the E. carotovora subsp. atroseptica extracts is probably due to the absence of cross-reaction of antibodies raised against the E. chrysanthemi protein PaeX. However, the periplasmic pectin acetylesterase PaeX appeared to be a conserved element of the Erwinia pectin degradation complex.
FIG. 7.
Occurrence of PaeX homologues in different strains of Erwinia. Bacteria were grown in LB medium supplemented with 0.2% galacturonate until the stationary phase. The culture supernatant (S) and whole-cell (C) fractions were separated by SDS-PAGE and analyzed by immunoblotting with PaeX antibodies. The arrows indicate the positions of PaeX and of an unknown protein (*) cross-reacting with the PaeX antiserum.
Combined action of the E. chrysanthemi pectinases.
Synergism is often observed between pectinolytic enzymes which present different modes of action. We therefore tested whether the action of other E. chrysanthemi pectinases, i.e., pectin methylesterase and pectate lyases, favors the action of the pectin acetylesterase PaeX. Digestion of sugar beet pectin with one pectate lyase, either PelB, PelD, or PelL, did not significantly modify PaeX activity (Fig. 8A and data not shown). In contrast, PaeX was more efficient after demethylation of pectin (pretreatment with PemA) and even more efficient after both demethylation and depolymerization of pectin (pretreatment with PemA and PelB) (Fig. 8A). As previously observed for PaeY, PaeX prefers demethylated pectic oligomers as substrate.
FIG. 8.
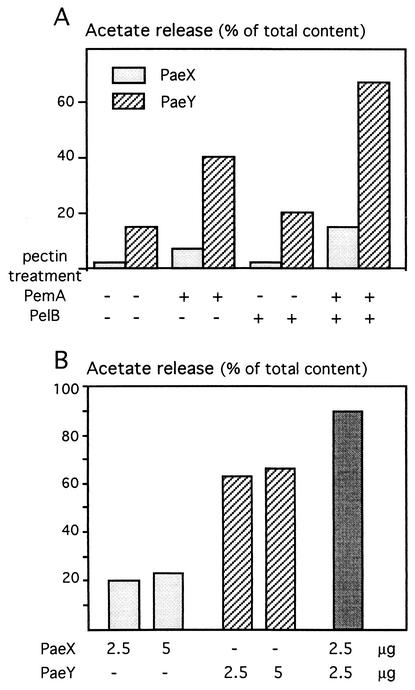
Activity of PaeX on pretreated pectins and in combination with PaeY. (A) Acetylesterase activity on pectin after pretreatment with an endopectate lyase, PelB, and/or with a pectin methylesterase, PemA. Sugar beet pectin (10 g · liter−1) in 0.1 M Tris-HCl buffer (pH 8.0) was pretreated by incubation at 30°C for 12 h with PemA (2 μg · ml−1), with PelB (1 μg · ml−1), or with both PemA and PelB (2 and 1 μg · ml−1, respectively). These mixtures were then used as substrate for PaeX or PaeY. After an additional incubation with 2.5 μg of pectin acetylesterase · ml−1 at 30°C for 12 h, acetate release was quantified. (B) Combined action of PaeX and PaeY. Sugar beet pectin, pretreated as described above with the combination of PemA and PelB, was used as substrate for PaeX and PaeY. Indicated amounts (2.5 or 5 μg · ml−1) of one or both pectin acetylesterases were added, and after an additional incubation at 30°C for 12 h, acetate release was determined.
In addition to their different cellular localizations, the existence of two pectin acetylesterases produced by E. chrysanthemi raises the question of a putative difference in the substrate specificity of each isoenzyme. We analyzed whether combined action of PaeX and PaeY could lead to an improved deacetylation of pectic substances in comparison with the action of each enzyme alone (Fig. 8B). With demethylated oligogalacturonates as a substrate, PaeX was less active than was PaeY (Fig. 8A). However, simultaneous addition of PaeX and PaeY led to a more efficient deacetylation of demethylated oligogalacturonates. The final level of acetate release increased from about 65% for PaeY to about 90% for PaeY and PaeX. This effect did not result from a higher total amount of enzymes, since the addition of twice the amount of PaeY or PaeX alone did not increase the final acetate release (Fig. 8B). Thus, combined action of PaeX and PaeY on demethylated pectic oligomers improved deacetylation in comparison with the action of each enzyme alone. This observation suggests that the two E. chrysanthemi pectin acetylesterases have a difference in their substrate specificity.
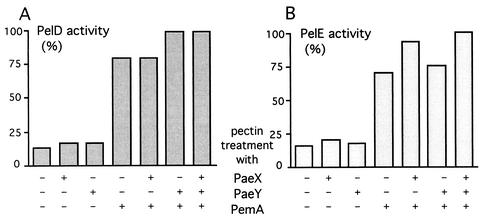
We also analyzed whether the action of the extracellular endopectate lyases could be favored by the deacetylation of pectin. We used the two E. chrysanthemi major pectate lyase isoenzymes PelD and PelE and the two acetylesterases PaeX and PaeY (Fig. 9). Treatment of sugar beet pectin with either of the two acetylesterases led to a slight increase in the pectate lyase activity. Since sugar beet pectin is also methyl esterified (56%), we demethylated the substrate with the pectin methylesterase PemA. This treatment strongly increased the activity of the pectate lyases PelD and PelE. The deacetylation of demethylated pectin led to a further improvement in pectate lyase activity, but this effect depended on the isoenzymes used in the combination (Fig. 9). The PelE activity is further increased by PaeX action while the PelD activity is further increased by PaeY action. These differences in the increase in activity of the two pectate lyases could reflect a difference in the specificity of the two pectin acetylesterases towards the pectic substrates. This is the second observation which suggests that the two pectin acetylesterases of E. chrysanthemi, PaeX and PaeY, possess a difference in their substrate specificity.
FIG. 9.
Pectate lyase activity of PelD and PelE on sugar beet pectin after pretreatment with pectin esterases. Sugar beet pectin (10 g · l−1) in 0.1 M Tris-HCl buffer, pH 8.0, was incubated at 30°C for 12 h with PaeX (2.5 μg · ml−1), PaeY (2.5 μg · ml−1), PemA (2 μg · ml−1), or combinations of these enzymes. After incubation, these mixtures were diluted 1:20 in 0.05 M Tris-HCl buffer (pH 8.0)-0.1 mM CaCl2 and used as substrate for pectate lyase PelD or PelE. The initial rate of reaction was measured at 37°C by monitoring the absorbance at 230 nm during 2 min after the addition of the pectate lyase (0.4 U). The activities are expressed as percentages of the maximal values.
In the case of xylan acetylesterases, two types of isoenzymes were differentiated according to their substrate preference: either short oligomers or long-chain polymers (6). We have shown that both PaeX and PaeY prefer pectic oligomers. Since the acetyl esters in pectin are O-2 or O-3 linked, an evident hypothesis is a specificity in the hydrolysis of only one type of ester. For instance, nuclear magnetic resonance spectrometry analysis suggested that the pectin acetylesterase of Aspergillus niger attacks only one type of acetyl ester of the pectic chain (25). Another type of specificity could be linked to the mechanism of de-esterification. For instance, pectin methylesterases act on pectin either by a processive mechanism, creating blocks of demethylated galacturonate, or by a multichain mechanism, giving a random demethylated polymer (8). Detailed studies of the reaction products resulting from pectin acetylesterase activity will be necessary to clarify the type of specificity, as regards substrates and/or mechanisms, that probably exists among these enzymes.
Acknowledgments
Appreciation is expressed to Valerie James for reading the manuscript. We gratefully acknowledge the members of this laboratory, particularly Sylvie Reverchon, Guy Condemine, and William Nasser, for their helpful discussions.
This work was supported by grants from the Centre National de la Recherche Scientifique and from the Ministère de l'Education Nationale et de la Recherche.
REFERENCES
- 1.Alfano, J. R., J. H. Ham, and A. Collmer. 1995. Use of Tn5tac1 to clone a pel gene encoding a highly alkaline, asparagine-rich pectate lyase isozyme from an Erwinia chrysanthemi EC16 mutant with deletions affecting the major pectate lyase isozymes. J. Bacteriol. 177:4553-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardonnet, N., and C. Blanco. 1992. uidA antibiotic resistance cassettes for insertion mutagenesis, gene fusion and genetic constructions. FEMS Microbiol. Lett. 93:243-248. [DOI] [PubMed] [Google Scholar]
- 3.Blot, N., C. Berrier, N. Hugouvieux-Cotte-Pattat, A. Ghazi, and G. Condemine. 2002. The oligogalacturonate-specific porin KdgM of Erwinia chrysanthemi 3937 belongs to a new porin family. J. Biol. Chem. 277:7936-7944. [DOI] [PubMed] [Google Scholar]
- 4.Breton, C., M. Bordenave, L. Richard, J. C. Pernollet, J. C. Huet, S. Pérez, and R. Goldberg. 1996. PCR cloning and expression analysis of a cDNA encoding a pectinacetylesterase from Vigna radiata L. FEBS Lett. 388:139-142. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, T. M. I. E., J. E. Nielsen, and J. D. Mikkelsen. 1996. Isolation, characterization and immunolocalization of orange fruit acetyl esterase, p. 723-730. In J. Visser and A. G. J. Voragen (ed.), Pectins and pectinases. Elsevier, Amsterdam, The Netherlands.
- 6.Christov, L. P., and B. A. Prior. 1993. Esterases of xylan-degrading microorganisms: production, properties, and significance. Enzyme Microb. Technol. 15:460-475. [DOI] [PubMed] [Google Scholar]
- 7.Copeland, B. R., R. J. Richter, and C. E. Furlong. 1982. Renaturation and identification of periplasmic proteins in two-dimensional gels of Escherichia coli. J. Biol. Chem. 257:15065-15071. [PubMed] [Google Scholar]
- 8.Denes, J., A. Baron, C. Renard, and J. Drilleau. 2000. Different action patterns for pectin methylesterase at pH7.0 and pH4.5. Carbohydr. Res. 327:385-393. [DOI] [PubMed] [Google Scholar]
- 9.Hugouvieux-Cotte-Pattat, N., N. Blot, and S. Reverchon. 2001. Identification of TogMNAB, an ABC transporter which mediates the uptake of pectic oligomers in Erwinia chrysanthemi 3937. Mol. Microbiol. 41:1113-1123. [DOI] [PubMed] [Google Scholar]
- 10.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 11.Hugouvieux-Cotte-Pattat, N., and J. Robert-Baudouy. 1989. Isolation of Erwinia chrysanthemi mutants altered in pectinolytic enzyme production. Mol. Microbiol. 3:1587-1597. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, B. H., and M. H. Hecht. 1994. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Bio/Technology 12:1357-1360. [DOI] [PubMed] [Google Scholar]
- 13.Lin, L.-L., and J. A. Thomson. 1991. Cloning, sequencing and expression of a gene encoding a 73 kDa xylanase enzyme from the rumen anaerobe Butyrivibrio fibriosolvens H17c. Mol. Gen. Genet. 228:55-61. [DOI] [PubMed] [Google Scholar]
- 14.Lojkowska, E., C. Masclaux, M. Boccara, J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1995. Characterization of the pelL gene encoding a novel pectate lyase of Erwinia chrysanthemi 3937. Mol. Microbiol. 16:1183-1195. [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Moran, F., S. Nasuno, and M. P. Starr. 1968. Extracellular and intracellular polygalacturonic acid trans eliminase of Erwinia carotovora. Arch. Biochem. Biophys. 123:298-306. [DOI] [PubMed] [Google Scholar]
- 17.Nasser, W., S. Reverchon, G. Condemine, and J. Robert-Baudouy. 1994. Specific interactions of Erwinia chrysanthemi KdgR repressor with different operators of genes involved in pectinolysis. J. Mol. Biol. 236:427-440. [DOI] [PubMed] [Google Scholar]
- 18.Nasser, W., S. Reverchon, and J. Robert-Baudouy. 1992. Purification and functional characterisation of KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi. Mol. Microbiol. 6:257-265. [DOI] [PubMed] [Google Scholar]
- 19.Pissavin, C., J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1996. Regulation of pelZ, a gene of the pelBC cluster encoding a new pectate lyase in Erwinia chrysanthemi 3937. J. Bacteriol. 178:7187-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resibois, A., M. Colet, M. Faelen, E. Schoonejans, and A. Toussaint. 1984. PhiEC2, a new generalised transducing phage of Erwinia chrysanthemi. Virology 137:102-112. [DOI] [PubMed] [Google Scholar]
- 21.Reverchon, S., D. Expert, J. Robert-Baudouy, and W. Nasser. 1997. The cyclic AMP receptor protein is the main activator of the pectinolysis genes in Erwinia chrysanthemi. J. Bacteriol. 179:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1994. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol. Microbiol. 11:1127-1139. [DOI] [PubMed] [Google Scholar]
- 23.Roeder, D. L., and A. Collmer. 1985. Marker-exchange mutagenesis of pectate lyase isozyme gene in Erwinia chrysanthemi. J. Bacteriol. 164:51-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Searle-van Leeuwen, M. J. F., J.-P. Vincken, D. Schipper, A. G. J. Voragen, and G. Beldman. 1996. Acetyl esterases of Aspergillus niger: purification and mode of action on pectins, p. 793-798. In J. Visser and A. G. J. Voragen (ed.), Pectins and pectinases. Elsevier, Amsterdam, The Netherlands.
- 26.Shevchik, V. E., G. Condemine, N. Hugouvieux-Cotte-Pattat, and J. Robert-Baudouy. 1996. Characterization of pectin methylesterase B, an outer membrane lipoprotein of Erwinia chrysanthemi 3937. Mol. Microbiol. 19:455-466. [DOI] [PubMed] [Google Scholar]
- 27.Shevchik, V. E., and N. Hugouvieux-Cotte-Pattat. 1997. Identification of a bacterial pectin acetyl esterase in Erwinia chrysanthemi. Mol. Microbiol. 24:1285-1301. [DOI] [PubMed] [Google Scholar]
- 28.Shevchik, V. E., J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1997. The pectate lyase PelI of Erwinia chrysanthemi belongs to a new family. J. Bacteriol. 179:7321-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier, W. F., and B. A. Moffat. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 30.Surgey, N., J. Robert-Baudouy, and G. Condemine. 1996. The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J. Bacteriol. 178:1593-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabor, S., and C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tardy, F., W. Nasser, J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1997. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J. Bacteriol. 179:2503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williamson, G. 1991. Purification and characterization of pectin acetylesterase from orange peel. Phytochemistry 30:445-449. [Google Scholar]