Abstract
A number of groups have developed libraries of siRNAs to identify genes through functional genomics. While these studies have validated the approach of making functional RNAi libraries to understand fundamental cellular mechanisms, they require information and knowledge of existing sequences since the RNAi sequences are generated synthetically. An alternative strategy would be to create an RNAi library from cDNA. Unfortunately, the complexity of such a library of siRNAs would make screening difficult. To reduce the complexity, longer dsRNAs could be used; however, concerns of induction of the interferon response and off-target effects of long dsRNAs have prevented their use. As a first step in creating such libraries, long dsRNA was expressed in mammalian cells. The 250 nt dsRNAs were capable of efficiently silencing a luciferase reporter gene that was stably transfected in MDA-MB-231 cells without inducing the interferon response or off-target effects any more than reported for siRNAs. In addition, a long dsRNA expressed in the same cell line was capable of silencing endogenous c-met expression and inhibited cell migration, whereas the dsRNA against luciferase had no effect on c-met or cell migration. The studies suggest that large dsRNA libraries are feasible and that functional selection of genes will be possible.
INTRODUCTION
A number of different approaches have been advanced to validate the function of genes, including the function of a number of genes involved in cancer cell migration and invasion. The use of RNAi is generally regarded to be one of the most promising approaches since, at least in non-mammalian systems, it acts systemically (1), thus providing the potential to carry out both in vitro and in vivo target validation. RNAi was initially described in plants, where it was believed to play a key role in protection against viral pathogens (2). The pathway involves a dsRNA of >21 bp triggering an RNaseIII-like enzyme now called Dicer (3–5). Dicer cleaves long dsRNAs, into small interfering RNAs (siRNAs) of 21–25 bp. These siRNAs are then incorporated into a multi-subunit RNA-induced silencing complex (RISC), which acts catalytically to target degradation of cellular mRNA in a sequence dependent manner (6,7). Given the systemic and catalytic nature of RNAi, this class of molecules has been proposed for use both in target identification/validation and the development of therapeutics (8,9). While the great majority of the target validation studies using RNAi have been focused on a specific target, the efficiency of RNAi has led to the recognition that this technology may represent an exceptionally strong approach to functional genomics through the creation of RNAi libraries.
Libraries of siRNA molecules targeting a specific gene can be produced synthetically, although new enzymatic approaches (10) have reduced the time and costs associated with producing libraries, and allow for the selection of the siRNA with the highest level of activity against the target of interest. Functional RNAi libraries have been designed and synthesized to study cytoskeleton organization in Drosophila (11) and are being used extensively to study gene function in Caenorhabditis elegans, including the functional analysis of entire chromosomes and the full genome (12,13), but have only recently been described for use in mammalian systems (14). While the above laboratories reported the ability to generate functional libraries of siRNAs against specific targets and the ability to generate a complex, 415 000 member siRNA library from an existing cDNA library, they did not report that this latter library was functional. The authors go on to report that they generate on average 34 unique siRNA clones per kilobase of sequence, a distinct advantage since not all siRNAs are active. However, this also points out the limitations of generating siRNA libraries. Significantly larger libraries will be required to ensure that active siRNAs are present for each transcript. Other approaches have recently been undertaken to develop functional siRNA libraries ranging from ∼500 to 8000 known genes (15,16). Plasmids and retroviral vectors have also been used to generate libraries of short hairpin RNAs (shRNAs) of similar size to the larger siRNA library (17,18), with which genetic screens have been carried out to identify tumor suppressor genes (18–20). Recently, libraries corresponding to most known human and mouse genes have been developed (21). Many of these si-and shRNA libraries are commercially available and a database, RNAi Codex, has been developed to annotate the data generated in the various studies (22).
While the above strategies have demonstrated promise for functional genomic screening, most are based on developing libraries using known sequences and when a random library was generated, the complexity was significant. An alternative approach to that taken in generating the random siRNA library (14), would be to restriction digest cDNA to generate a library of long dsRNAs. Long dsRNA libraries have been used to analyze gene function in Drosophila (23,24) but potential off-target effects and induction of the stress response are issues for mammalian gene silencing. Thus, the focus of this study was to analyze the efficiency, selectivity and toxicity of long dsRNAs in mammalian cells and to determine whether long dsRNAs lead to the desired biological response in these cells, as a first step to generate long dsRNA libraries.
MATERIALS AND METHODS
Plasmid construction, primers and probes
For the construction of the vectors containing dual, convergent promoters for expressing dsRNA molecules, two different polIII human promoters, U6 from psiSTRIKE-U6 (Promega) and H1 from pSilencer 3.0-H1 (Ambion), were cloned into pGEM3Z (Promega) (see Supplemental Figure 1). The primers (obtained from Invitrogen) used to amplify these promoters by PCR were designed to contain unique restriction sites at the 5′ and 3′ ends for cloning (see Supplemental Table 1) and a TTTTT stretch in the reverse primers to create the polIII termination signal.
The firefly luciferase fragments were generated by PCR amplification from pMS110 (Message Pharmaceuticals) using primer pairs (Invitrogen) each containing a KpnI restriction site (see Supplemental Table 1). Following amplification, the luciferase fragments were cloned into the KpnI site in pGEM/U6/H1.
The c-met gene (gb accession number: NM_000245) was targeted with a long dsRNA generated by cloning the region between 3209 and 3330 into the KpnI site of pGEM/U6/H1 vector using the primer pair (Invitrogen) described in Supplemental Table 1.
The following customized primers and probe sets for qRT–PCR and qPCR plasmid quantification were purchased from Applied Biosystems:
Primers Forward (U6): 5′-GCTTACCGTAACTTGAAAGTA-3′; Reverse (H1): 5′-CTGGGAAATCACCATAAACGT-3′; Luc probe:5′-CCGGCGCCATTCTATCCGCTGGAAGATGGAACCGCTGGAGAGCAACTGCA-3′; c-met probe: 5′-CAGAAGATCAGTTTCCTAATTCATCTCAGAACGGTTCATGCCGACAAGTG-3′.
Cell culture and transfection
MDA-MB-231 cells were maintained in DMEM/F12 (1:1), supplemented with 10% FBS, 100 μg/ml penicillin, 100 μg/mlstreptomycin, 40 μg/ml gentamycin, 0.4 mM sodium pyruvate and 2 mM l-glutamine. MCF-7 cells permanently transfected with pMS110 (a generous gift from Message Pharmaceutics) were maintained in DMEM and 10% FBS, containing the supplements described above. Twenty-four hours after plating, cells were transfected with Lipofectamine 2000 according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA) in 96-well, 6-well or T75 flasks as necessary for the desired number of cells.
Luciferase experiments
MDA-MB-231 cells permanently expressing the pMS110 plasmid containing firefly luciferase (MDA-MB-231/FFLuc) or the permanently transfected MCF-7 cell line (Message Pharmaceuticals) were seeded in 96-well culture plates at 104 cells/well and transfected with Lipofectamine 2000 according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). Each well received 45 ng pGEM/U6/H1/dsRNALuc expression vector. All samples were transfected in triplicate and the experiment was performed a minimum of three times. For control transfections, empty vector pGEM/U6/H1 was included. The luciferase assays were performed 24 h after transfection using the Bright Glo Luciferase Assay system according to the manufacturer's instructions (Promega, Madison, WI) with activity quantified on a Lucy II (Rosys Anthos) luminometer.
Interferon β analysis
To determine whether expression of long dsRNA induced the interferon response in mammalian cells, 2 × 105 of MDA-MB-231 or MCF-7 cells were plated on 6-well plates, then 24 h later pGEM/U6/H1 or one of the four pGEM/U6/H1dsRNAFFLuc were transfected using Lipofectamine 2000. Forty-eight hours after transfection interferon β levels in the media were analyzed. To confirm that each cell line retained an inducible interferon-β pathway, the interferon response was assessed against an adenovirus infection. As above, 2 × 105 of MDA-MB-231 and MCF-7 cells were plated on 6-well plates then 24 h later, adenovirus was introduced at a Multiplicity of Infection (MOI) of 0, 5, 50 and 500 for 4 h. Thereafter, the media was replaced without adenovirus and incubated for an additional 48 h. At the end of each experiment, 220 μl media was collected and the level of secreted interferon-β was determined using the Human INF-beta ELISA kit (Invitrogen) following the manufacturer's instructions.
Transwell migration assay
MDA-MB-231/FFLuc cells were grown in T75 flasks and transfected with the desired dsRNA expression vector. Twenty-four hours following transfection, cells were harvested for the migration assay by trypsinization, suspended in DMEM/F12 without serum and seeded at densities of 2 × 104 or 106 cells per well in 24 or 6 well transwell inserts, respectively. The lower chamber was supplemented with DMEM/F12 medium with 10% serum. The upper and lower chambers were separated by an 8 μm pore polycarbonate membrane (Costar, Corning, NY) that was coated with 50 or 500 μL of 0.5 μg/μL Matrigel (Becton Dickinson). After 48 h of incubation at 37°C, the cells in the bottom well were harvested with trypsin and counted for invasion index evaluation (24-well) and RNA or DNA were extracted (6-well) for qRT–PCR and microarray hybridization or plasmid partition quantification, respectively. The number of migrating cells was determined by counting a 10 μl aliquot using a haemocytometer (Neubauer). For each experiment of invasion index evaluation (invasion index = percentage of cells that migrated from the total number of plated cells), values were normalized to the empty vector transfected cells set at 100%.
RNA isolation and quantitative real-time PCR
Total RNA was extracted with Trizol (Invitrogen) from MDA-MB-231 FFLuc cells 48 h after transfection with either pGEM/U6/H1 or pGEM/U6/H1dsRNAFFLuc. The estimated transcript concentrations of the human c-met, GAPDH and firefly luciferase genes were based on the comparison of the target transcript PCR signal in dsRNA expression vector transfected cells to the signal measured in an empty vector transfected control. Analysis was done using the 2−ΔΔCT method, which first normalizes the RNA levels of the gene of interest to a housekeeping gene then normalizes this relative RNA level to that of the control treatment group, as described by Livak and Schmittgen (25) for target gene transcripts and interpolation from standard curves for absolute quantification of plasmid partitioning. The TaqMan PCR system (Applied Biosystems) with a 7700 ABI Detector (Applied Biosystems) was used for analysis. cDNA was synthesized from 1 μg of the total RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems) for use in the microarray experiments. Pre-designed, gene-specific TaqMan probe and primer sets (Applied Biosystems), consisting of a specific fluorogenic probe and a pair of oligonucleotides, were used to run standard qPCRs for human c-met,GAPDH and firefly luciferase genes. cDNA (0.1 ng) was used for all genes and 5 μg DNA for plasmid quantification. The reactions were carried out in triplicate in a 50 μl reaction volume and a 96-well plate format.
Microarray hybridization
Total RNA was extracted as described above. Quality of each sample was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Biotinylated cRNA was synthesized from total RNA (Enzo, Farmingdale, NY). Following processing according to the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA), the labeled cRNA was hybridized to Affymetrix U133A GeneChips and the resulting expression levels were quantified. Comparative analysis of the two samples was performed with the statistical algorithm of MAS 5.0 software (Affymetrix) by scaling at fixed target intensity of 150, a fold change of 1.5 (approximate signal log ratio of 0.6) and P-value < 0.01. Comparison results were expressed as percentage change in the study, by measuring the fluorescence signal of the targets on the array.
Statistical analysis
With the exception of the microarray data, all data were analyzed using the Sigma Plot 8.02 statistical software package (SPSS Inc. 2002). All quantitative values were expressed and plotted as mean ± SEM. Comparisons were evaluated by the unpaired Student's t-test. A value of P < 0.01 was considered statistically significant.
RESULTS
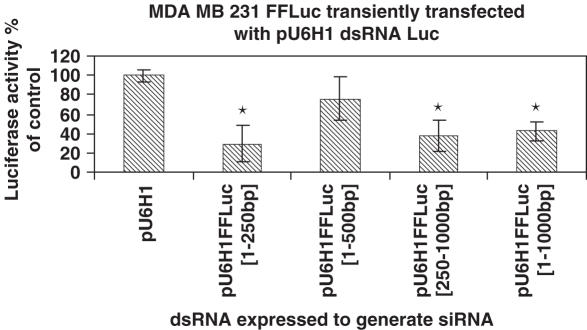
To create vectors containing dual, convergent promoters for expressing dsRNA molecules two different polIII promoters, U6 and H1, were cloned into pGEM3Z. The primers used to amplify these promoters were designed to contain unique restriction sites at the 5′ and 3′ ends for cloning and a TTTTT stretch in the reverse primers to create a strong polIII termination signal. To test various long dsRNAs, regions of the firefly luciferase gene were amplified by PCR and cloned into a KpnI site between the two promoters. Sequences of 250, 500, 750 and 1000 bp (see Supplemental Figure 2) were tested for their ability to inhibit firefly luciferase activity in a MDA-MB-231 cell line that stably expressed firefly luciferase. Each of the constructs were capable of silencing luciferase activity 48 h after transfection with inhibition ranging from 20% for the 500 bp sequence to 80% for the 250 bp sequence (Figure 1). A second cell line, MCF-7, was also transfected with the plasmid generating the 250 bp luciferase dsRNA. Similar to the MDA-MB-231 transfected cells, the dsRNA resulted in a 60% reduction of luciferase activity. For both the MDA-MB-231 and MCF-7 cell lines, silencing of luciferase expression was consistent with transfection efficiency. A plasmid expressing GFP was transfected as described above, 48 h later cells were sorted by FACS and the transfection efficiencies were determined to be 55 and 41%, respectively.
Figure 1.
Firefly luciferase gene silencing in transiently transfected MDA-MB-231 cells permanently expressing firefly luciferase, with long dsRNA expressed from the pGEM/U6/H1 vector. Luciferase activity was analyzed 48 h following transfection of the vectors expressing dsRNA. Firefly luciferase activity was used as a read out and normalized to vector control. *P < 0.01.
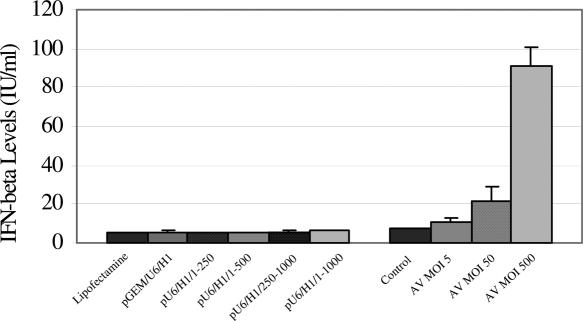
To analyze whether expression of long dsRNAs had adverse effects on the cell, the stress response and off-target effects were monitored. The four constructs were analyzed to determine if expression of long dsRNAs led to the induction of interferon β in order to assess their capability to induce the stress response. The MDA-MB-231 cells, which had previously been shown to possess a functional interferon pathway (26), were demonstrated in this study to be β-interferon competent by monitoring secreted interferon β levels following infection with adenovirus. At 48 h post-transfection or infection, neither the lipofectamine reagent, the pGEM/U6/H1 plasmid nor any of the four constructs led to detection of interferon β whereas adenovirus infection demonstrated at least a 10-fold induction of interferon β secreted in the media (Figure 2). Similar results were observed using MCF-7 cells, with non-transfected and cells transfected with the 250 bp dsRNA producing essentially undetectable levels of interferon and adenoviral infected cells secreting ∼90 IU/ml, which was ∼10-fold higher than non-infected MCF-7 cells. Since, the 250 bp dsRNA resulted in maximal silencing of luciferase without inducing interferon β in the MDA-MB-231 cells, this construct was used to analyze off-target effects by expression profiling. Of 22 283 probe sets on the array, 68 were increased by 1.5-fold and 189 decreased by the same level 48 h after transfection of this construct (Table 1 and Supplemental Table 2). Interferon β, 2′-5′ OAS and PKR were found absent, present and present with no change in the expression level when compared with vector alone transfected cells, respectively.
Figure 2.
Interferon β production at 48 h after transfection in the supernatant of MDA-MB-231 FFLuc cells transfected with lipofectamine or various dsRNA expression vectors or infected with adenovirus for 48 h. Interferon β levels in the supernatant were measured by ELISA and the IU/ml determined from a standard curve using recombinant interferon β.
Table 1.
Pairwise analysis of off-target effects of dsRNA
| −5.9 | Moderately similar to RAS-RELATED PROTEIN RAB-7 |
| −5.4 | Delta-catenin mRNA, complete cds. |
| −4.9 | Plexin C1 (PLXNC1) |
| −4.7 | Phospholipase A2, group VII |
| −4.6 | Myeloid cell nuclear differentiation antigen |
| −4.6 | Dsarcosine dehydrogenase (SARDH) |
| −4.6 | UDP glycosyltransferase 2 family, polypeptide A1 |
| −3.9 | Fasciculation and elongation protein zeta 1 (zygin I) |
| −3.9 | ATP-binding cassette, sub-family C (CFTRMRP) |
| −3.8 | Ninein (LOC51199) |
| −3.8 | MIP-1 delta mRNA |
| −3.6 | Protein phosphatase 1, regulatory (inhibitor) |
| −3.6 | Growth factor independent 1B |
| −3.5 | Wilms tumor suppressor protein human |
| −3 | Transcription factor 15 (basic helix-loop-helix) (TCF15) |
| −3 | Protein kinase C, zeta (PRKCZ) |
| −2.9 | G protein-coupled receptor kinase 5 |
| −2.7 | Protein kinase, cAMP-dependent, regulatory, type I, beta |
| −2.7 | Ribosomal protein S6 kinase, 90kD, polypeptide 6 (RPS6KA6) |
| −2.6 | TNF superfamily, member 8 (TNFRSF8) |
| −2.5 | Zinc finger protein (ZNF154) |
| −1.9 | Platelet-activating factor receptor |
| −1.9 | Receptor-associated coactivator 3 (RAC3) |
| −1.8 | Hepatocellular carcinoma-associated gene TD26 (LOC55908) |
| −1.3 | Protein kinase, AMP-activated, beta 1 (PRKAB1) |
| −1.3 | Small nuclear RNA activating complex (SNAPC2) |
| −1.3 | Guanine nucleotide binding protein (G protein) (GNB3) |
| −1 | Calciumcalmodulin-dependent protein kinase II beta 6 (CAMKB) |
| −0.9 | Protein kinase, cAMP-dependent, regulatory, type II, _ |
| −0.9 | Interleukin-1 Superfamily z (FIL1(ZETA)) |
| −0.8 | N6-DNA-methyltransferase (N6AMT1) |
| −0.8 | GTPase activating protein-like (GAPL) |
| −0.8 | Cell-line MDA-MB-453 androgen receptor |
Interferon β, 2′-5′ OAS and PKR were found absent, present and present with no change in the expression level when compared with vector alone transfected cells, respectively. Selected changes and the log change following dsRNA treatment are listed below. The complete list of changes in expression pattern can be found in Supplemental Table 2.
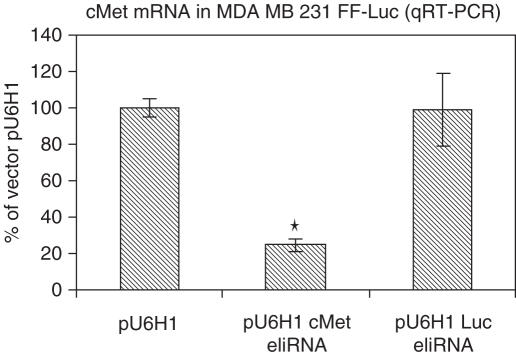
While the above results demonstrate that long dsRNA can effectively inhibit the expression of a transgene in MDA-MB-231 cells without inducing the interferon response or strong off-target effects, the ability to silence an endogenous gene and generate the expected phenotypic change with long dsRNA must be assessed. To this end, the ability to silence c-met and prevent cell migration was analyzed. A dsRNA corresponding to the region 3209–3300 of the c-met sequence was cloned into the vector described above and transfected into MDA-MB-231 cells. The 250 bp luciferase silencing construct was independently transfected into this cell line and 48 h after transfection, c-met RNA levels were determined using quantitative RT–PCR. The c-met dsRNA construct led to a 60% decrease in c-met RNA levels, whereas the luciferase dsRNA construct did not alter c-met RNA levels (Figure 3).
Figure 3.
Cells were transiently transfected with vector alone, a vector expressing 250 bp of luciferase dsRNA or a vector expressing 121 bp of c-met dsRNA using Lipofectamine 2000. Forty-eight hours after transfection, total RNA was isolated using Trizol and levels of c-met were quantified. The levels of c-met RNA were expressed relative to those found in vector alone transfected cells. *P < 0.01.
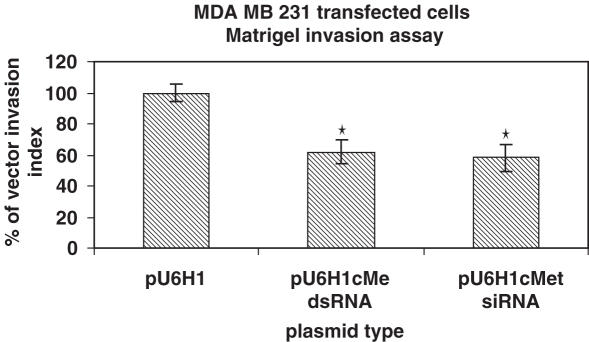
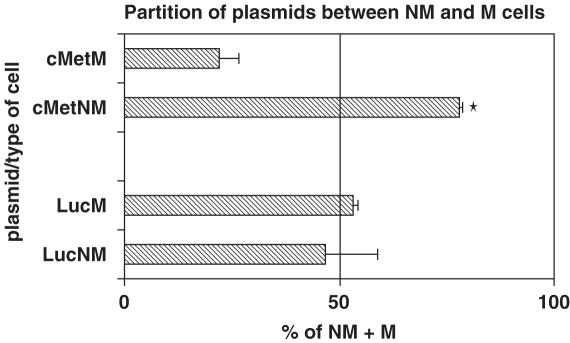
The transiently transfected cells were then tested for their ability to inhibit cell migration using a transwell assay. The ability of expressed long c-met dsRNA to inhibit migration was compared with that of an siRNA that had been previously demonstrated to efficiently knock down c-met (27). As can be seen in Figure 4, both the expressed long dsRNA and the siRNA inhibited cell migration by ∼40%. The inability to inhibit cell migration to higher levels is probably owing to the inefficiency of transfection, such that the cells that still migrate may lack plasmid or have significantly less plasmid expressing the dsRNA. To address this possibility, the partitioning of the plasmids expressing long dsRNA between the migrated and non-migrated cells was quantified using real time PCR to determine the abundance of plasmid DNA per unit of total DNA. Standard curves were generated using the plasmids encoding c-met and luciferase dsRNAs to quantify actual plasmid levels from migrating and non-migrating cells. As expected, the luciferase plasmid was found at relatively similar levels in both the migrating and non-migrating cell populations, whereas the c-met plasmid strongly partitioned with the non-migrating population (Figure 5).
Figure 4.
MDA-MB-231 cells were transiently transfected with either a plasmid encoding c-met long dsRNA or c-met siRNA by Lipofectamine 2000. Twenty-four hours after transfection, cells were added to a transwell chamber coated with matrigel, allowed to migrate for 48 h and the number of cells from the lower compartment of the chamber was compared with those transfected with vector alone. *P < 0.01.
Figure 5.
MDA-MB-231 cells were transiently transfected with a plasmid capable of generating either c-met or luciferase long dsRNA. Following a transient transfection, the cells were placed in a transwell chamber coated with matrigel and 48 h later the migrated and non-migrated populations of cells were isolated. DNA was recovered from each population and the levels of plasmid were analyzed using primers derived from the U6 and H1 promoters by qPCR using the ABI7700. *P < 0.01.
In total, these experiments present data that demonstrate (i) the ability to produce long dsRNA and efficiently silence gene expression when expressed from a plasmid; (ii) the lack of interferon induction following transfection and efficient silencing; (iii) the ability of a long dsRNA (c-met) to inhibit migration and (iv) the increased partitioning of this plasmid (c-met) with the non-migrating cells since silencing of c-met would be expected to lead to prevention of migration. Similar expression of long dsRNA targeting Arc and NR1 in a neuronal cell line has also been carried out and confirms the above observations, both for efficacy of silencing and lack of interferon induction (data not shown).
DISCUSSION
There are a number of types of dsRNAs that have been used to silence gene expression. By far the most common of these is synthetic siRNA. These 21–23 nt RNA molecules are commonly used both in vitro and in vivo. Another form of siRNA commonly used is shRNA, which is generally expressed intracellularly from a vector. The use of long dsRNAs have been very limited owing to the belief that these longer regions of double stranded RNA will result in the induction of the interferon response (7). However, the above studies demonstrate that long dsRNAs can be used to silence gene expression without inducing the stress response or causing significant off-target effects. The use of long dsRNAs can provide numerous advantages in that the cell can select the optimal silencing sequence alleviating the need to test numerous siRNAs; long dsRNAs will allow for silencing libraries to have less complexity than would be necessary for siRNAs; and, perhaps most importantly, long dsRNA could prevent viral escape mutations when used as therapeutics.
One concern with using long dsRNAs randomly generated from cDNA libraries and expressed from promoters constructed of two convergent polIII promoters is premature termination of the transcript owing to the presence of polIII terminators. A long stretch of thymidine nucleotides leads to polIII termination, with a minimum of three necessary but five being optimal (28). Thus, if a terminator was present in either the sense or antisense strand very near to the promoter or if terminators for both strands were present adjacent to one another, it would be possible to generate no dsRNA. It was interesting to note that the optimal long dsRNA for silencing luciferase expression was in fact, the shortest of the sequences. However, when the sequence was analyzed for stretches of five thymidine nucleotides in either the forward or reverse orientation, the 250 bp sequence generates a region of ∼235 bp of dsRNA, whereas the 750 and 1000 bp sequences generate identical ∼330 bp of dsRNA and the 500 bp sequence results in a dsRNA of ∼80 bp, perhaps explaining its poor silencing activity. These results demostrate that while the existance of polIII terminators may prevent some dsRNAs from forming, a stretch of five thymidines should occur on average once every 1000 bp so the likelihood that such stretches will occur on opposite strands of a 250 nt base pair digestion product is remote. In addition, it is extremely unlikely that if a cDNA was digested to numerous 250 bp fragments, every fragment would contain terminators preventing formation of dsRNAs. Thus, use of random 250 bp regions should result in effective silencing.
Long dsRNAs are capable of undergoing dicer-mediated processing to siRNAs in mammalian cells resulting in the production of numerous siRNAs of 22–23 nt (29). The presence of numerous fragments against a single target has the advantage of allowing the cell to select the most efficient siRNA for target accessibility and by producing numerous dsRNAs, can offer better potential therapeutic protection against viral infections by making resistance difficult (9). However, there has been a long standing belief that long dsRNAs induce an interferon response, beginning with dsRNA viruses induction of a stress response (30–32) and further supported when early work comparing long dsRNA with siRNAs was carried out (7). These studies are often cited as rationale for working with siRNAs but other studies have reported that siRNA can also induce an interferon response in mammalian cells (33,34). Although siRNAs are still the method of choice for silencing gene expression, there have been a number of reports in which long dsRNAs have been used in mammalian cells and in mammals without signs of toxicity or adverse effects on the animal (35–38). The lack of interferon response associated with long dsRNA in this study was an encouraging first step in the development of libraries since long dsRNA would reduce the required complexity and allow processing to numerous siRNAs intracellularly, enhancing the probability of target acessibility.
A second important consideration when using long dsRNA is the potential for significant off-target silencing. One of the proposed strengths of an siRNA is that the 21–23 bp sequence can be analyzed against the genomic database and identical matches can be avoided. This becomes difficult, if not impossible, when using a 250 bp dsRNA that will be processed into a number of potential overlapping 21–23 bp siRNAs (29). One of the real strengths of siRNAs was originally thought to be the exquisite specificity of the interaction. When a reporter gene, GFP, was used as a target to assess silencing ot the reporter expression in HEK293 cells, no detectable secondary effects were noted by genomewide expression profiling (39) with a similar profiling demonstrating high specificity when the profiles of three endogenous genes were compared (40). The tremendous specificity of the response was also inferred from data generated using siRNAs as antiviral treatments—resistant viruses were shown to contain a single nucleotide substitution in the region corresponding to the sequence of the siRNA (41). More recently, this specificity has come into question. When similar expression profiling was carried out using multiple siRNAs against a MAPK14 or IGF1R, there were reproducible expression patterns for each siRNA across three experiments but a set of siRNAs against a given target found only a few genes regulated similarly for the given target (42). This was unexpected since siRNAs targeting the same gene should result in the same changes in an expression profile. They also reported that the observed off-target effects could not be titrated out using lower concentrations of siRNA and that an siRNA targeting luciferase reproducibly silenced other genes in the absence of any sequence similarity. In another study, a 21 bp siRNA against luciferase was also found to elicit significant off-target effects, stimulating or repressing >1000 genes (43). They explained this, at least in part, through the induction of the stress response through the PKR pathway. In light of these latter studies, the several hundred genes identified by expression analysis in this study that exhibit an increase or decrease following silencing of luciferase with long dsRNA were not unexpected for silencing in general. Although the effectiveness of the luciferase could not be monitored on the array since its sequence was not present on the array, analysis of luciferase expression in a popultion of cells transfected at the same time and in the same way demonstrated the on-target efficacy of the dsRNA (data not shown). From these results, long dsRNAs do not appear to be toxic and the non-specific effects are modest when compared with those of siRNAs.
We next tested whether a long dsRNA expressed from a plasmid would not only silence gene expression but would also result in the desired phenotypic change. To analyze this activity we choose to attempt to inhibit cell migration as a strategy for targeting cancer metastasis. The understanding of how a primary tumor cell migrates from the initial tumor, disperses throughout the body in the vascular/lymphatic system, invades a new site and begins to proliferate and form secondary tumors in not well understood (44), although it is well accepted that the mechanisms leading to metastases will represent attractive therapeutic targets (45). Since migration and invasion are key components of cellular metastasis (45,46), these processes have been well studied in cell culture systems and represent an attractive model to look at the functional effects of large dsRNAs.
Recently, an siRNA to c-met has been identified and shown to lead to apoptosis and reduction of tumor growth (27). In addition, when c-met was knocked-down using ribozymes, cell invasion and metastasis were inhibited both in vitro and in mice (47,48). A number of groups are targeting c-met for therapeutic intervention with small molecules (49,50) given its important role in tumor metastasis (51). For these reasons, c-met was used in the present studies as a positive control dsRNA for cell invasion. When comparing the dsRNA with the previously reported siRNA, both inhibited cell migration to an identical level. To determine whether this inhibition was owing to the transfection of the plasmid containing the dsRNA for c-met, the migrating and non-migrating populations of cells were compared. While a plasmid expressing dsRNA against luciferase was found in equal abundance in the two populations of cells, the plasmid expressing dsRNA against c-met was enriched almost 4-fold in the non-migrating cell population. This enrichment is consistent with the loss of c-met function, which should result in inhibition of migration. Thus, these studies demonstrate the ability to silence an endogenous gene and obtain the desired phenotype. The presence of c-met dsRNA plasmid in the migrating population of cells may represent low number of copies per cell resulting in low levels of dsRNA that are insufficient to elicit the change in migration or may represent a population of the MDA-MD-231 cells whose migration is not dependent on c-met expression. A further analysis of this population would be required to better understand the biological significance of this phenomenon.
Results from these studies have demonstrated the ability of dsRNA to silence endogenous gene expression, leading to a biological response, without inducing an interferon response or significant off-target effects. This is the first step in creating functional dsRNA libraries. Using long dsRNAs, libraries can be generated from cells expressing a phenotype of interest and transfected back into the same type of cells to look for loss of function. The dsRNA sequence resulting in loss of function can then be rescued, sequenced and the isolated sequence further tested for function. The use of long dsRNAs also offer promise for development of therapeutics, particularly since off-target effects are modest when compared with those found with siRNAs.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
Acknowledgments
The authors are also grateful to Message Pharmaceuticals for the gift of pMS110. This work was supported by Department of Defense Grant No. W81XWH-04-1-0639. Funding to pay the Open Access publication charges for this article was provided by Funds to TG from the Feist-Weiller Cancer Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Winston W.M., Molodowitch C., Hunter C.P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 3.Tuschl T., Zamore P.D., Lehmann R., Bartel D.P., Sharp P.A. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 5.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 6.Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 8.Chopra M., Pachuck C., Satishchandran C., Giordano T. Using RNA interference to modulate gene expression. Targets. 2002;1:102–108. [Google Scholar]
- 9.Shuey D., McCallus D.E., Giordano T. RNAi: gene-silencing in therapeutic intervention. Drug Discov. Today. 2002;7:1040–1046. doi: 10.1016/s1359-6446(02)02474-1. [DOI] [PubMed] [Google Scholar]
- 10.Shirane D., Sugao K., Namiki S., Tanabe M., Iino M., Hirose K. Enzymatic production of RNAi libraries from cDNAs. Nature Genet. 2004;36:190–196. doi: 10.1038/ng1290. [DOI] [PubMed] [Google Scholar]
- 11.Kiger A., Baum B., Jones S., Jones M., Coulson A., Echeverri C., Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 13.Sonnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.-M., Artelt J., Bettencourt P., Cassin E., et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 14.Sen G., Wehrman T.S., Myers J.W., Blau H.M. Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nature Genet. 2004;36:183–189. doi: 10.1038/ng1288. [DOI] [PubMed] [Google Scholar]
- 15.Aza-Blanc P., Cooper C.L., Wagner K., Batalov S., Deveraux Q.L., Cooke M.P. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell. 2003;12:627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- 16.Zheng L., Liu J., Batalov S., Zhou D., Orth A., Ding S., Schultz P.G. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paddison P.J., Silva J.M., Conklin D.S., Schlabach M., Li M., Aruleba S., Balija V., O'Shaughnessy A., Gnoj L., Scobie K., et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 18.Berns K., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kekhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 19.Kolfschoten I.G.M., van Leeuwen B., Berns K., Mullenders J., Beijersbergen R.L., Bernards R., Voorhoeve P.M., Agami R. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 20.Westbrook T.F., Martin E.S., Schlabach M.R., Leng Y., Liang A.C., Feng B., Zhao J.J., Roberts T.M., Mandel G., Hannon G.J. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 21.Silva J., Li M.Z., Chang K., Ge W., Golding M.C., Rickles R.J., Siolas D., Hu G., Paddison P.J., Schlabach M.R., et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 22.Olson A., Sheth N., Lee J.S., Hannon G., Sachidanandam R. RNAi Codex: a portal/database for short-hairpin RNA (shRNA) gene-silencing constructs. Nucl. Acids Res. 2006;34:D153–157. doi: 10.1093/nar/gkj051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov A.I., Rovescalli A.C., Pozzi P., Yoo S., Mozer B., Li H.-P., Yu S.-H., Higashida H., Guo V., Spencer M., et al. Genes required for Drosophila nervous system development identified by RNA interference. Proc. Natl Acad. Sci. USA. 2004;101:16216–16221. doi: 10.1073/pnas.0407188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y.-O., Park S.-J., Balaban R.S., Nirenberg M., Kim Y. A functional genomic screen for cardiogenic genes using RNA interference in developing Drosophila embryos. Proc. Natl Acad. Sci. USA. 2004;101:159–164. doi: 10.1073/pnas.0307205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Vidalain P.-O., Laine D., Zaffran Y., Azocar O., Servet-Delprat C., Wild F., Rabourdin-Combe C., Valentin H. Interferons mediate terminal differentiation of human cortical thymic epithelial cells. J. Virol. 2002;76:6415–6424. doi: 10.1128/JVI.76.13.6415-6424.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinomiya N., Gao C.F., Xie Q., Gustafson M., Waters D.J., Zhang Y.-W., Vande Woude G.F. RNA interference reveals that ligand-independent met activity is required for tumor cell signaling and survival. Cancer Res. 2004;64:7962–7970. doi: 10.1158/0008-5472.CAN-04-1043. [DOI] [PubMed] [Google Scholar]
- 28.Bakken A., Morgan G., Sollner-Webb B., Roan J., Busby S., Reeder R.H. Mapping of transcription initiation and termination signals on Xenopus laevis ribosomal DNA. Proc. Natl Acad. Sci. USA. 1982;79:56–60. doi: 10.1073/pnas.79.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Billy E., Brondani V., Zhang H., Muller U., Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clemens M., Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J. Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 31.Gil J.S., Esteban M. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis. 2000;5:107–114. doi: 10.1023/a:1009664109241. [DOI] [PubMed] [Google Scholar]
- 32.Manche L., Green S.R., Schmedt C., Mathews M.B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R.G. Activation of the interferon system by short-interfering RNAs. Nature Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 34.Bridge A.J., Pebernard S., Ducraux A., Nicoulaz A.-L., Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genetics. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 35.Bhargava A., Dallman M.F., Pearce D., Choi S. Long double-stranded RNA-mediated RNA interference as a tool to achieve site-specific silencing of hypothalamic neuropeptides. Brain Res. Protoc. 2004;13:115–125. doi: 10.1016/j.brainresprot.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Diallo M., Arenz C., Schmitz K., Sandhoff K., Schepers U. Long endogenous dsRNAs can induce complete gene silencing in mammalian cells and primary cultures. Oligonucleotides. 2003;13:381–392. doi: 10.1089/154545703322617069. [DOI] [PubMed] [Google Scholar]
- 37.Paddison P.J., Caudy A.A., Hannon G.J. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran N., Raponi M., Dawes I.W., Arndt G.M. Control of specific gene expression in mammalian cells by co-expression of long complementary RNAs. FEBS Lett. 2004;573:127–134. doi: 10.1016/j.febslet.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 39.Chi J.-T., Chang H.Y., Wang N.N., Chang D.S., Dunphy N., Brown P.O. Genomewide view of gene silencing by small interfering RNAs. Proc. Natl Acad. Sci. USA. 2003;100:6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semizarov D., Frost L., Sarthy A., Kroeger P., Halbert D.N., Fesik S.W. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl Acad. Sci. USA. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gitlin L., Karelsky S., Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 42.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 43.Persengiev S.P., Zhu X., Green M.R. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch D.R., Steeg P.S., Rinker-Schaeffer C.W. Molecular biology of breast metastasis: Genetic regulation of human breast carcinoma metastasis. Breast Cancer Res. 2000;2:408–416. doi: 10.1186/bcr87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sledge G.W., Miller K.D. Exploiting the hallmarks of cancer. Eur. J. Cancer. 2003;39:1668–1675. doi: 10.1016/s0959-8049(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 46.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 47.Kim S.J., Johnson M., Koterba K., Herynk M.H., Uehara H., Gallick G.E. Reduced c-Met expression by an adenovirus expressing a c-Met ribozyme inhibits tumorigenic growth and lymph node metastases of PC3-LN4 prostate tumor cells in an orthotopic nude mouse model. Clin. Cancer Res. 2003;9:5161–5170. [PubMed] [Google Scholar]
- 48.Davies G., Watkins G., Mason M.D., Jiang W.G. Targeting the HGF/SF receptor c-met using a hammerhead ribozyme transgene reduces in vitro invasion and migration in prostate cancer cells. Prostate. 2004;60:317–324. doi: 10.1002/pros.20068. [DOI] [PubMed] [Google Scholar]
- 49.Christensen J.G., Burrows J., Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Letters. 2005;225:1–26. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 50.Wang X., Le P., Liang C., Chan J., Kiewlich D., Miller T., Harris D., Sun L., Rice A., Vasile S., et al. Potent and selective inhibitors of the Met [hepatocyte growth factor/scatter factor (HGF/SF) receptor] tyrosine kinase block HGF/SF-induced tumor cell growth and invasion. Mol. Cancer Ther. 2003;2:1085–1092. [PubMed] [Google Scholar]
- 51.Corso S., Comoglio P.M., Giordano S. Cancer therapy: can the challenge be MET? Trends Mol. Med. 2005;11:284–292. doi: 10.1016/j.molmed.2005.04.005. [DOI] [PubMed] [Google Scholar]