Abstract
Rickettsia prowazekii, the causative agent of epidemic typhus, is an obligate, intracellular, parasitic bacterium that grows within the cytoplasm of eucaryotic host cells. Rickettsiae exploit this intracellular environment by using transport systems for the compounds available in the host cell's cytoplasm. Analysis of the R. prowazekii Madrid E genome sequence revealed the presence of a mutation in the rickettsial metK gene, the gene encoding the enzyme responsible for the synthesis of S-adenosylmethionine (AdoMet). Since AdoMet is required for rickettsial processes, the apparent inability of this strain to synthesize AdoMet suggested the presence of a rickettsial AdoMet transporter. We have confirmed the presence of an AdoMet transporter in the rickettsiae which, to our knowledge, is the first bacterial AdoMet transporter identified. The influx of AdoMet into rickettsiae was a saturable process with a KT of 2.3 μM. Transport was inhibited by S-adenosylethionine and S-adenosylhomocysteine but not by sinfungin or methionine. Transport was also inhibited by 2,4-dinitrophenol, suggesting an energy-linked transport mechanism, and by N-ethylmaleimide. AdoMet transporters with similar properties were also identified in the Breinl strain of R. prowazekii and in Rickettsia typhi. By screening Escherichia coli clone banks for AdoMet transport, the R. prowazekii gene coding for a transporter, RP076 (sam), was identified. AdoMet transport in E. coli containing the R. prowazekii sam gene exhibited kinetics similar to that seen in rickettsiae. The existence of a rickettsial transporter for AdoMet raises intriguing questions concerning the evolutionary relationship between the synthesis and transport of this essential metabolite.
Rickettsia prowazekii, the etiologic agent of epidemic typhus, is an obligate, intracellular, parasitic bacterium that grows within the cytoplasm of the eucaryotic host cell rather than within an intracytoplasmic vesicle. The rickettsiae exploit this environment by expressing distinctive transport systems for the high-energy intermediates available in the host cell cytoplasm. For example, R. prowazekii can transport such highly charged molecules as ATP/ADP, AMP, GMP, NAD, UMP, and UDPG (5, 6, 32-34). Due to the importance of transport systems in rickettsial intracellular parasitism, the identification and characterization of rickettsial transporters is critical to our understanding of how these unusual bacteria grow and cause disease.
The search for rickettsial transporters has benefited from the publication of the R. prowazekii Madrid E genome sequence (4). Many genes coding for putative membrane proteins can be identified and are now available for further study. In addition, the genome sequence identified mutations that point to the possible existence of specific transporters. For example, analysis of the R. prowazekii Madrid E genome sequence revealed the presence of numerous pseudogenes. These genes code for proteins with high homologies to characterized gene products of other organisms but contain mutations that would presumably preclude expression of functional products (4). One such gene is metK, coding for methionine adenosyltransferase (MAT). In the Madrid E strain of R. prowazekii, this gene contains a stop codon in the middle of the MAT coding sequence (1, 4). Interestingly, the R. prowazekii Breinl strain and a strain of Rickettsia typhi exhibit complete metK open reading frames, while the metK genes of the spotted fever group rickettsiae possess numerous stop codons and frameshifts (2-4). MAT, the enzyme that catalyzes the synthesis of S-adenosylmethionine (AdoMet), has been identified in a wide variety of species of the bacterial, eucaryotic, and archaeal lineages (9, 27). AdoMet is an essential metabolite in both procaryotes and eucaryotes, where it serves as the primary methyl donor in a variety of methylation reactions (18, 27). In addition, the aminopropyl group of AdoMet serves as a substrate in the polyamine biosynthetic pathway (7, 18). Based on the facts that R. prowazekii has been shown to synthesize but not transport polyamines and that the R. prowazekii Madrid E strain contains a nonsense mutation within the coding sequence of the metK gene, a rickettsial transport system for AdoMet should be present (3, 26).
While AdoMet synthesis has been found in a wide variety of cells, AdoMet transport has been identified in only a few eucaryotes, including Saccharomyces cerevisiae, Pneumocystis carinii, Trypanosoma brucei, Leishmania donovani, and rat liver mitochondria (8, 11, 17, 19, 20, 22, 25). In this paper we identify and characterize a rickettsial AdoMet transporter, the first reported bacterial transporter for this essential metabolite.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The rickettsial strains used included the R. prowazekii Madrid E and Breinl strains and the R. typhi Wilmington strain. Rickettsiae were purified from the yolk sacs of embryonated hen eggs as described previously (32) and were suspended in a sucrose-phosphate-glutamate-magnesium solution (SPG-Mg; 0.218 M sucrose, 3.76 mM KH2PO4, 7.1 mM K2HPO4, 4.9 mM potassium glutamate, and 10 mM MgCl2). For uptake assays, rickettsial suspensions were concentrated so that rickettsiae derived from 8 g of infected yolk sac were present in 1 ml. The concentrated suspensions ranged from 4 to 15 mg of protein per ml.
E. coli strain XL1-Blue (Stratagene, La Jolla, Calif.) was used as the standard recipient in these studies. An R. prowazekii cosmid clone bank constructed in E. coli DH1 was used in transport screening experiments (14, 15). E. coli strains were cultured in Luria-Bertani (LB) medium at 37°C. Where appropriate for selection of E. coli transformants, ampicillin was added to a final concentration of 50 μg/ml.
Transport assays.
Rickettsial AdoMet uptake assays were initiated by adding 1/10 the final volume of the concentrated rickettsial suspension to SPG-Mg containing S-adenosyl-l-[methyl-14C]methionine (Amersham Biosciences Corp., Piscataway, N.J.) at concentrations ranging from 0.5 to 20 μM. For measuring the effect of substrate concentration on AdoMet uptake, solutions were incubated at 34°C for 15 s and 0.1 ml aliquots were placed on prewetted membrane filters (25 mm; Durapore PVDF; Millipore Corp.) and were washed with 5 ml of 0.25 M sucrose. Filters were dried, and radioactivity was assayed by liquid scintillation. Intracellular accumulation of AdoMet was measured by using a microspace technique as previously described (31). To determine specificity, unlabeled, putative competitive inhibitors, at a final concentration of 25 μM, were added to reaction mixtures containing 10 μM labeled AdoMet. Sensitivity of transport to metabolic inhibitors was assayed similarly but at a final concentration of 1 mM, and they were preincubated with rickettsiae for 10 min. Assays were initiated by the addition of labeled AdoMet. All inhibitors were purchased from Sigma (St. Louis, Mo.). For E. coli assays, an overnight culture was used to inoculate fresh LB medium and the culture was grown to exponential phase. Bacteria were harvested by centrifugation, washed with 5 ml of 50 mM potassium phosphate buffer (pH 7.0), and finally suspended in SPG-Mg to a calculated optical density at 600 nm of 2.0 and assayed for AdoMet transport as described above for the rickettsiae. Kinetic parameters for all uptake experiments were calculated by nonlinear regression and were plotted by using GraphPad Prism software (GraphPad Software, Inc., San Diego, Calif.).
Clone bank screening and subcloning.
A previously established cosmid clone bank was replicated to 96-well microtiter plates containing 100 μl of LB medium plus ampicillin. After overnight incubation at 37°C, 100 μl of LB medium plus ampicillin containing 20 μM [14C]AdoMet was added to each well. After a 4-h incubation the cells were placed in a 96-well vacuum manifold containing a nitrocellulose filter, the medium was removed, and the cells were washed with two 500-μl aliquots of potassium phosphate buffer. The filters were dried, and the radioactivity was visualized by using a Cyclone Storage Phosphor System (PerkinElmer Life Sciences Inc., Boston, Mass.). Plasmid vectors used in the cloning of specific rickettsial fragments included pBluescript SKII(+) (Stratagene) and pSMART HCAmp (Lucigen Corp., Middleton, Wis.).
RESULTS
Transport of radiolabeled AdoMet in R. prowazekii Madrid E.
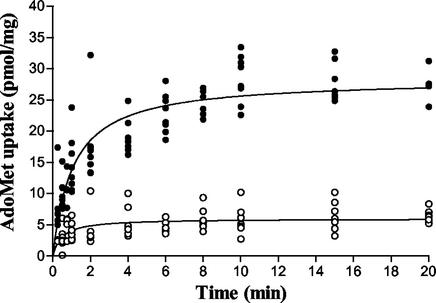
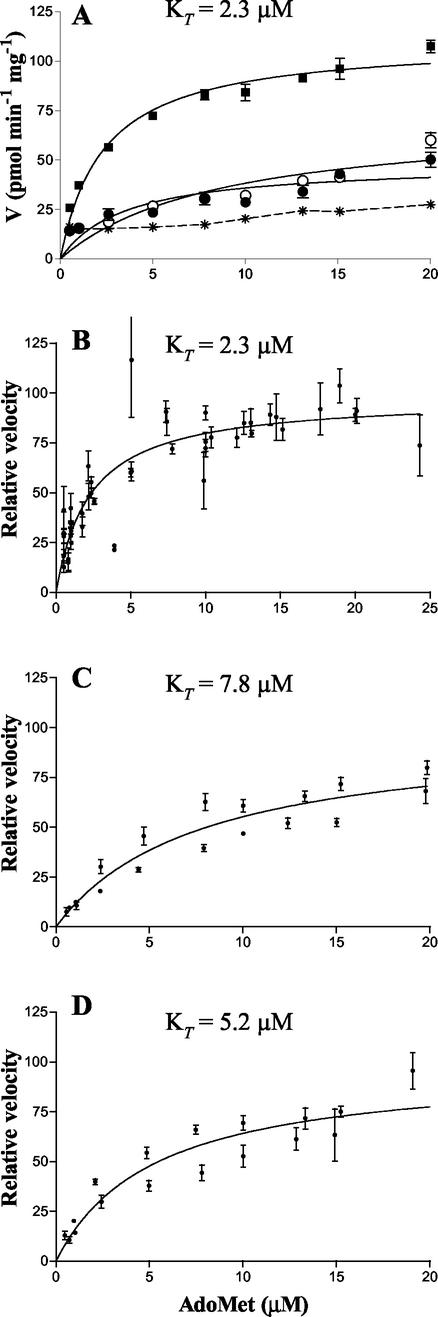
Figure 1 shows the kinetics of radioactive AdoMet (10 μM) uptake as a function of time. Uptake was linear for the first minute and reached a steady state within 8 min. A microspace assay was used to examine the accumulation of AdoMet by the rickettsiae at steady state (31). In two independent experiments, accumulation ratios (the ratio of the concentration inside to that outside) of 13 and 28 were obtained. Since uptake was linear for the first minute, subsequent uptake experiments were sampled at periods of less than 1 min to minimize the possible effect of downstream processes on transport kinetics. The effect of substrate concentration on AdoMet uptake can be seen in Fig. 2. In contrast to nonspecific interactions that occur in the presence of 2,4-dinitrophenol (DNP) or when the rickettsiae are assayed at 4°C, accumulation of AdoMet by R. prowazekii at 34°C was a saturable process (Fig. 2A). A composite of R. prowazekii AdoMet uptake experiments that used four independent R. prowazekii preparations and that were normalized to a standard Vmax revealed a KT of AdoMet transport of 2.3 μM (range of 0.9 to 4.2 μM) (Fig. 2B). Due to the variability between rickettsial preparations it was impossible to determine the Vmax for rickettsial AdoMet transport. In the series of experiments described above, the Vmax spanned a range of 11 to 95 pmol mg−1 min−1 after background subtraction.
FIG. 1.
Time course of AdoMet transport in R. prowazekii Madrid E. The uptake of AdoMet (10 μM) was measured at 34°C (filled circles) or at 4°C (open circles).
FIG. 2.
Effect of substrate concentration on AdoMet transport in rickettsiae. KT values were obtained by nonlinear regression analysis with GraphPad Prism software. (A) Transport of AdoMet by R. prowazekii Madrid E at 34°C (▪), at 4°C (○), and in the presence of DNP (•). Nonspecific background binding of labeled AdoMet to filters is also shown (∗) and is subtracted from experimental values in panels B to D. (B) A composite of four independent R. prowazekii Madrid E AdoMet transport assays normalized to a Vmax of 100 pmol min−1 mg−1. (C) A composite of two independent R. prowazekii Breinl AdoMet transport assays normalized to a Vmax of 100 pmol min−1 mg−1. (D) A composite of two independent R. typhi Wilmington AdoMet transport assays normalized to a Vmax of 100 pmol min−1 mg−1.
The specificity of transport was examined by measuring rickettsial AdoMet transport in the presence of various AdoMet analogues (Table 1). The most effective inhibitor, comparable to that of unlabeled AdoMet, was S-adenosylethionine, which differs from AdoMet in the substitution of an ethyl group for the donor methyl group. S-adenosylhomocysteine, a byproduct of AdoMet methylation reactions, also inhibited uptake to a lesser extent, while sinfungin, an effective inhibitor of some eucaryotic AdoMet transporters (8, 11, 17), exhibited no significant inhibition. With inhibitor concentrations of 12.5, 25, 50, and 100 μM, the calculated Ki values for S-adenosylethionine and S-adenosylhomocysteine were 6.4 ± 1.0 and 14.3 ± 6.9 μM, respectively. Additional potential inhibitors (methionine, ethionine, adenosine, and methylthioadenosine) did not inhibit rickettsial AdoMet transport (data not shown).
TABLE 1.
Effect of inhibitors on AdoMet uptake by R. prowazekii Madrid E and E. coli MOB 1402a
| AdoMet analogues | % Inhibition ± SEM for
|
|
|---|---|---|
| R. prowazekii Madrid E | E. coli MOB 1402 | |
| Unlabeled metabolitesb | ||
| AdoMet | 62 ± 2.8 | 36 ± 1.8 |
| SAE | 50 ± 3.2 | 36 ± 2.2 |
| SHC | 34 ± 4.0 | 31 ± 4.2 |
| Sinfungin | 7 ± 3.1 | 0 |
| Methionine | 6 ± 5.6 | 25 ± 3.8 |
| No inhibitor at 4°C | 64 ± 2.8 | 66 ± 3.7 |
| Metabolic inhibitorsc | ||
| NEM | 42 ± 6.5 | 34 ± 8.0 |
| DNP | 72 ± 3.6 | 82 ± 3.6 |
Two independent bacterial preparations were assayed three to five times for each inhibitor tested.
Assays contained 10 μM labeled AdoMet and 25 μM unlabeled inhibitor. SAE, S-adenosylethionine; SHC, S-adenosylhomocysteine.
Assays contained 10 μM labeled AdoMet and inhibitors at a concentration of 1 mM added 10 min prior to assay.
The sensitivity of the R. prowazekii AdoMet transporter to metabolic inhibitors was also assessed (Table 1). DNP significantly inhibited accumulation of AdoMet, suggesting that AdoMet transport is an energy-dependent process. Rickettsial AdoMet transport was also sensitive to N-ethylmaleimide (NEM), a sulfhydryl group blocking agent.
AdoMet transport by other rickettsiae.
The Breinl strain of R. prowazekii and the Wilmington strain of R. typhi do not have stop codons within their metK genes and thus may be able to synthesize AdoMet (2). In order to evaluate their transport capabilities, the kinetics of AdoMet transport were determined. The kinetics of uptake were found to be similar to those of the R. prowazekii Madrid E strain (Fig. 2C and D). The Breinl strain exhibited a KT of 7.8 μM while the R. typhi KT was 5.2 μM. Both substrate specificity and the sensitivity of these transporters to poisons were comparable to those of the R. prowazekii Madrid E strain (data not shown).
Identification of the AdoMet transporter gene.
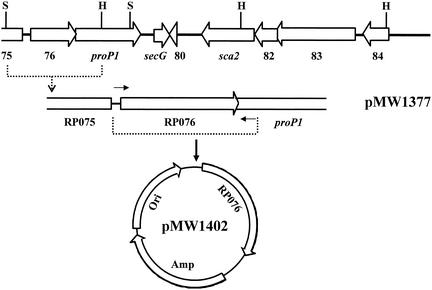
Since none of the genes contained in the R. prowazekii genome exhibited homology to any of the known eucaryotic AdoMet transporters, a genome screening method was used to identify the rickettsial transporter. A previously established cosmid clone bank (14, 15) was screened for clones that demonstrated uptake of radiolabeled AdoMet. One such clone that was identified contained a rickettsial Sau3A fragment encompassing the RP075-RP084 gene region (Fig. 3). A SnaBI-HpaI fragment containing the complete gene sequence of the RP076 gene and partial sequences of the RP075 and proP1 genes was subsequently identified as the region imparting AdoMet transport to E. coli clones (Fig. 3).
FIG. 3.
Schematic maps of the cosmid insert containing the R. prowazekii RP075-RP084 gene region and subsequent recombinants. The orientation of the genes is indicated by open arrows. Dashed lines identify the regions cloned to generate specific recombinants. The indicated SnaB1-HpaI fragment was cloned into pBluescript SKII(+) to generate pMW1377. Small arrows represent primers used to amplify the RP076 gene. The amplified gene was cloned into pSMART to generate pMW1402. All the constructs shown transport AdoMet in E. coli. H, HpaI; S, SnaBI; Amp, ampicillin resistance gene; Ori, origin of replication.
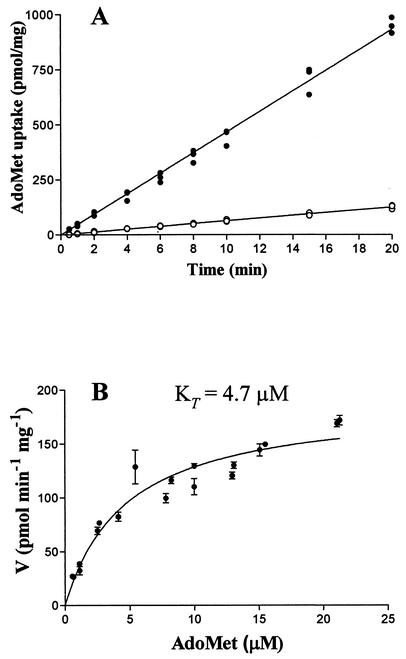
In order to conclusively prove that only the RP076 gene is necessary for AdoMet transport, the coding region of RP076, flanked by 56 bp upstream and 32 bp downstream, was PCR amplified and ligated into the blunt cloning vector pSMART, generating plasmid pMW1402. Originally, the pSMART vector was chosen to eliminate possible toxicity problems during the initial cloning of the RP076 PCR product. However, it was discovered that insertion of the RP076 coding region into pSMART in one orientation (pMW1402) resulted in constitutive AdoMet transport. Thus, this construction was used to investigate transport kinetics in E. coli (Fig. 4). The fragment inserted into the vector in the opposite orientation (pMW1410) served as a negative control. AdoMet uptake in E. coli cells containing pMW1402 remained linear over the 20-min assay, presumably due to the rapid metabolism of AdoMet within E. coli. To minimize the effect of downstream processes on the kinetics of transport, samples for determining the effect of substrate concentration on transport were taken at 30 s. A KT of 4.7 μM, similar to that found for rickettsial transport, was found under these conditions. In addition, the sensitivity of the cloned transporter to the inhibitors S-adenosylethionine and S-adenosylhomocysteine and to the metabolic inhibitors DNP and NEM was similar to that found for rickettsial transport (Table 1). Based on this data, we have assigned the gene designation sam to RP076.
FIG. 4.
Kinetics of AdoMet transport in MOB1402. (A) Time course of AdoMet uptake in E. coli MOB1402 and MOB1410. (B) Effect of substrate concentration on AdoMet transport in MOB1402. The data represent a composite of two independent bacterial preparations. The KT value was obtained by nonlinear regression analysis with GraphPad Prism software.
The R. prowazekii sam gene exhibits a G+C content (32.4%) typical of R. prowazekii protein-coding genes (4). The transporter encoded by sam possesses characteristics of an integral membrane protein. The 294-amino-acid deduced protein has a calculated Mr of 33,092 and a pI value of 9.98. A Kyte-Doolittle hydrophobicity analysis (16) revealed a hydrophobic protein with 10 potential membrane-spanning regions (data not shown). Several topology models for transmembrane proteins also predicted 10 membrane-spanning regions (10, 13, 30). A homolog to sam (RC0106) with 90.5% identity at the amino acid sequence level can be identified in the Rickettsia conorii genome sequence (21). A BLAST search revealed that the rickettsial transporter exhibits a small but significant relationship (23 to 28% identity) to hypothetical proteins from a wide range of bacterial genera. The only annotated genes identified in the BLAST search, from Brucella melitensis and Vibrio vulnificus, are members of the DMT superfamily of drug/metabolite transporters. The Brucella transporter is also annotated as a member of the DME family, a subgroup of the DMT superfamily that contains integral membrane proteins with sizes ranging from 246 to 353 amino acids and having 10 membrane-spanning regions (12).
DISCUSSION
As obligate intracellular bacteria growing within the cytoplasm of the eucaryotic host cell, the rickettsiae are immersed in pools of metabolic intermediates. The rickettsiae have evolved to exploit this rich environment by expressing transport proteins specific for these metabolites. Consequently, the capability of rickettsiae to synthesize many of these compounds has been lost. Upon publication of the R. prowazekii genome sequence, an example of this reductive evolution process was encountered with the identification of a translational stop codon within the metK gene of R. prowazekii (1, 4). Since this gene codes for the enzyme responsible for synthesizing the essential metabolite AdoMet, the identification of a translational stop within this gene led to the hypothesis that R. prowazekii Madrid E should transport AdoMet (3).
Our data demonstrate that rickettsiae transport AdoMet via a high-affinity system. This is the first bacterial AdoMet transporter identified, and the 2 to 8 μM KT values for the rickettsial transporters are comparable to the values for the S. cerevisiae transporter (3.3 μM), the high-affinity transporter of P. carinii (4.5 μM), and the transporter found in rat liver mitochondria (8.9 μM) (11, 19, 20). In sensitivity to inhibitors, the rickettsial transporter is comparable to that of S. cerevisiae. However, there is no significant homology between any eucaryotic AdoMet transporter and the rickettsial transporter identified in this study. Thus, the rickettsial transporter offers a unique model for examining the transport of this essential metabolite.
Uptake of AdoMet by E. coli expressing the cloned transporter remained linear over 20 min, while rickettsial uptake reached a steady state within 8 min. This difference is likely due to a much lower metabolic demand for AdoMet in the rickettsiae. In rickettsiae, the only known use for AdoMet is in the synthesis of polyamines (26). AdoMet-dependent rickettsial methylation reactions have not been characterized. In addition, preliminary experiments examining the intracellular presence of AdoMet in rickettsiae by thin-layer chromatography identified AdoMet as the major labeled compound, suggesting a slower rate of AdoMet metabolism within the time frame of the experiments.
The existence of an AdoMet transporter in rickettsiae raises questions about the contribution of AdoMet synthesis in those rickettsiae containing an intact metK gene. Since AdoMet is an essential metabolite, it is obvious that a transporter must exist before the ability to synthesize this compound is lost completely. Thus, the hypothesis that the R. prowazkeii Madrid E strain must have a transporter was logical when coupled to the fact that the Madrid E metK gene has a nonsense mutation within the coding sequence. However, the Breinl strain of R. prowazekii and the Wilmington strain of R. typhi possess complete open reading frames of the metK gene. While this does not preclude the existence of missense mutations within these metK genes, the presence of complete open reading frames raises the question of whether these strains would need to transport AdoMet in a comparable manner. Our data demonstrated that both strains transported AdoMet with kinetics similar to that seen for the Madrid E strain. While the KT values of these strains were found to be slightly higher than that of Madrid E, this is insignificant considering the variability of rickettsial preparations. Obviously, the next step is to determine whether these rickettsial strains possess active MAT enzymes and how these rickettsial enzymes compare in activity with the well-studied MAT enzyme of E. coli. Preliminary data from whole-cell assays suggest that both the Breinl and Wilmington strains are expressing active enzyme, and analysis of the protein sequences reveals that both exhibit conservation of the active-site residues identified in E. coli (23, 24, 28, 29). Studies are under way to purify the MAT enzymes from the Breinl and Wilmington strains in order to directly compare and contrast the activities of these enzymes with those of the E. coli standard.
Acknowledgments
This work was supported by National Institutes of Health grant AI44997.
We thank Robin Daugherty and Rose Robertson for assistance in developing the transport assay and in rickettsial isolations.
REFERENCES
- 1.Andersson, J. O., and S. G. E. Andersson. 1997. Genomic rearrangements during evolution of the obligate intracellular parasite Rickettsia prowazekii as inferred from an analysis of 52015 bp nucleotide sequence. Microbiology 143:2783-2795. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, J. O., and S. G. E. Andersson. 1999. Genome degradation is an ongoing process in Rickettsia. Mol. Biol. Evol. 16:1178-1191. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, J. O., and S. G. E. Andersson. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664-671. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Pontén, U. C. M. Alsmark, R. M. Podowdki, A. K. Näslund, A.-S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-143. [DOI] [PubMed] [Google Scholar]
- 5.Atkinson, W. H., and H. H. Winkler. 1985. Transport of AMP by Rickettsia prowazekii. J. Bacteriol. 161:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson, W. H., and H. H. Winkler. 1989. Permeability of Rickettsia prowazekii to NAD. J. Bacteriol. 171:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman, W. H., C. W. Tabor, and H. Tabor. 1973. Spermidine biosynthesis: purification and properties of propylamine transferase from Escherichia coli. J. Biol. Chem. 248:2480-2486. [PubMed] [Google Scholar]
- 8.Goldberg, B., D. Rattendi, D. Lloyd, J. R. Sufrin, and C. J. Bacchi. 1998. Effects of intermediates of methionine metabolism and nucleoside analogs on S-adenosylmethionine transport by Trypanasoma brucei brucei and a drug-resistant Trypanasoma brucei rhodesiense. Biochem. Pharmacol. 56:95-103. [DOI] [PubMed] [Google Scholar]
- 9.Graham, D. E., C. L. Bock, C. Schalk-Hihi, Z. J. Lu, and G. D. Markham. 2000. Identification of a highly diverged class of S-adenosylmethionine synthetases in the archaea. J. Biol. Chem. 275:4055-4059. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann, K., and W. Stoffel. 1993. TMBASE-a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 11.Horne, D. W., R. S. Holloway, and C. Wagner. 1997. Transport of S-adenosylmethionine in isolated rat liver mitochondria. Arch. Biochem. Biophys. 343:201-206. [DOI] [PubMed] [Google Scholar]
- 12.Jack, D. L., N. M. Yang, and M. H. Saier, Jr. 2001. The drug/metabolite transporter superfamily. Eur. J. Biochem. 268:3620-3639. [DOI] [PubMed] [Google Scholar]
- 13.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 14.Krause, D. C., H. H. Winkler, and D. O. Wood. 1985. Cloning and expression of the Rickettsia prowazekii ADP/ATP translocator in Escherichia coli. Proc. Natl. Acad. Sci. USA 82:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krause, D. C., H. H. Winkler, and D. O. Wood. 1985. Cosmid cloning of Rickettsia prowazekii antigens in Escherichia coli K-12. Infect. Immun. 47:157-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence, F., T. Derbécourt, and M. Robert-Gero. 1998. Proton-ATPase activities involved in the uptake of an S-adenosylmethionine analogue. Mol. Biochem. Parasitol. 92:99-109. [DOI] [PubMed] [Google Scholar]
- 18.Mato, J. M., L. Alvarez, P. Ortiz, and M. A. Pajares. 1997. S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacol. Ther. 73:265-280. [DOI] [PubMed] [Google Scholar]
- 19.Merali, S., D. Vargas, M. Franklin, and A. B. Clarkson, Jr. 2000. S-adenosylmethionine and Pneumocystis carinii. J. Biol. Chem. 275:14958-14963. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, J. T., and K. D. Spence. 1972. Transport of S-adenosylmethionine in Saccharomyces cerevisiae. J. Bacteriol. 109:499-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogata, H., S. Audic, P. Renesto-Audiffren, P.-E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J.-M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 22.Petrotta-Simpson, T. F., J. E. Talmadge, and K. D. Spence. 1975. Specificity and genetics of S-adenosylmethionine transport in Saccharomyces cerevisiae. J. Bacteriol. 123:516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reczkowski, R. S., and G. D. Markham. 1995. Structural and functional roles of cysteine 90 and cysteine 240 in S-adenosylmethionine synthetase. J. Biol. Chem. 270:18484-18490. [DOI] [PubMed] [Google Scholar]
- 24.Reczkowski, R. S., J. C. Taylor, and G. D. Markham. 1998. The active-site arginine of S-adenosylmethionine synthetase orients the reaction intermediate. Biochemistry 37:13499-13506. [DOI] [PubMed] [Google Scholar]
- 25.Rouillon, A., Y. Surdin-Kerjan, and D. Thomas. 1999. Transport of sulfonium compounds. J. Biol. Chem. 274:28096-28105. [DOI] [PubMed] [Google Scholar]
- 26.Speed, R. R., and H. H. Winkler. 1990. Acquisition of polyamines by the obligate intracytoplasmic bacterium Rickettsia prowazekii. J. Bacteriol. 172:5690-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabor, C. W., and H. Tabor. 1984. Methionine adenosyltransferase (S-adenosylmethionine synthetase) and S-adenosylmethionine decarboxylase. Adv. Enzymol. 56:251-282. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, J. C., and G. D. Markham. 1999. The bifunctional active site of S-adenosylmethionine synthetase: roles of the active site aspartates. J. Biol. Chem. 274:32909-32914. [DOI] [PubMed] [Google Scholar]
- 29.Taylor, J. C., and G. D. Markham. 2000. The bifunctional site of S-adenosylmethionine synthetase: roles of the basic residues. J. Biol. Chem. 275:4060-4065. [DOI] [PubMed] [Google Scholar]
- 30.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 31.Winkler, H. H. 1975. Rickettsial cell water and membrane permeability determined by a microspace technique. Appl. Environ. Microbiol. 31:146-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler, H. H. 1976. Rickettsial permeability: an ADP-ATP transport system. J. Biol. Chem. 251:389-396. [PubMed] [Google Scholar]
- 33.Winkler, H. H., and R. M. Daugherty. 1986. Acquisition of glucose by Rickettsia prowazekii through the nucleotide intermediate uridine 5′-diphosphoglucose. J. Bacteriol. 167:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkler, H. H., R. Daugherty, and F. Hu. 1999. Rickettsia prowazekii transports UMP and GMP, but not CMP, as building blocks for RNA synthesis. J. Bacteriol. 181:3238-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]