Abstract
In the group A streptococcus (GAS; Streptococcus pyogenes), a two-component system known as CovRS (or CsrRS) regulates about 15% of the genes, including several important virulence factors like the hyaluronic acid capsule. Most of these genes, including covR itself, are negatively regulated by CovR. We have isolated two independent ISS1 insertions in an open reading frame (ORF) that increases CovR expression as measured by a Pcov-gusA reporter fusion in single copy in the GAS chromosome. This ORF, named rocA for “regulator of Cov,” activates covR transcription about threefold. As expected, a rocA mutant is mucoid and produces more transcript from the has promoter since this promoter is repressed by CovR. This effect is dependent on the presence of a wild-type covR gene. In contrast to its activation of Pcov, RocA negatively regulates its own expression. This autoregulation is not dependent on the presence of the covR gene. All the phenotypes of the rocA mutant were complemented by the presence of the rocA gene on a plasmid. The rocA gene is present in strains of all nine M serotypes of GAS tested and is absent from strains representing 11 other groups of streptococci and related bacteria, including strains of the closely related group C and G streptococci. It seems likely that rocA plays an important role in the pathogenesis of GAS since it affects expression of the global regulator CovR.
The human pathogen Streptococcus pyogenes (group A streptococcus [GAS]) is a gram-positive bacterium that causes a wide variety of diseases, including relatively mild and self-limiting infections of the throat and skin, such as pharyngitis and pyoderma, as well as life-threatening invasive diseases like septicemia, myositis, necrotizing fasciitis, and streptococcal toxic shock syndrome (for a recent review, see reference 8). The primary infection may also lead to serious sequelae like rheumatic fever, glomerulonephritis, and reactive arthritis (3, 5). In many cases, it appears that single GAS strains can cause all or most of these diseases.
Many virulence factors of GAS strains appear to be involved in the survival, spread, and persistence of the organism inside the human host. These are (i) factors that damage or degrade the host tissues, such as cysteine protease, hyaluronidase, pyrogenic exotoxins, streptokinase, and streptolysins; (ii) factors that are required for resistance to phagocytosis by the host immune system, such as M protein, C5a peptidase, and hyaluronic acid capsule; and (iii) proteins that promote adherence and colonization, such as M protein and protein F (for reviews, see references 1, 8, 48, and 55). GAS encounters distinct microenvironments during infection and probably responds to them by expressing genes that produce proteins necessary for infection at that particular site. The ability to differentially regulate a wide range of virulence factors is therefore likely to be essential for the success of GAS infections.
Two-component signal transduction systems, consisting of sensor kinases and DNA-binding response regulators, allow bacteria to respond differently to diverse environmental stimuli (27, 46, 57). In the sequenced M1, M3, and M18 GAS genomes, 13 two-component systems have been identified (19) and several of them have been studied in various GAS strains (17, 22, 26, 31). Among them, CovRS (also called CsrRS) is a major regulator, repressing at least seven known or presumed virulence factors, including the hyaluronic acid capsule synthesis operon (hasABC), pyrogenic exotoxin B (speB), the streptolysin S-associated operon (sag), streptokinase (ska), mitogenic factor/streptodornase (speMF/sda), and inhibitor of innate immunity (mac) (2, 17, 25, 33, 34). In addition, CovR represses its own expression (17). A recent analysis by microarray and real-time PCR indicates that CovR controls as many as 15% of the GAS genes, either directly or indirectly (22). Because this is such a large number of genes and because some genes controlled by CovR are themselves known or suspected regulators, it seems likely that CovR is involved in a regulatory cascade.
Response regulators usually act by binding to DNA in the vicinity of promoter sequences. This is true for CovR as well, since recent in vitro studies have shown that CovR binds near several covR-regulated promoters (18, 41). For example, CovR binds to the promoter region of hasA, sagA, ska, speB, and speMF (18, 41). In vitro binding studies with the Phas region identified five CovR binding sites, all of which are required for complete CovR-mediated repression in vivo (18). Thus, CovR appears to regulate the expression of some genes through direct interactions with their promoter regions.
Although we have begun to understand how CovR regulates the expression of different virulence genes, regulation of covRS expression is not well understood. It is known that CovR negatively regulates its own transcription (17) and that Rgg, a transcriptional regulator, activates covRS transcription (7). In addition, covR expression is also regulated by growth conditions. For example, covRS transcription reaches its maximum during the exponential growth phase and declines as the stationary phase begins (17, 41). Nutritional conditions, such as amino acid starvation, also affect covRS transcription by some unknown mechanism (54). Therefore, it appears that expression of the covRS operon may be controlled by a complex regulatory network.
The goal of this investigation was to isolate a regulatory factor(s) that modulates the expression of the covRS operon. In the present work, we report the identification and characterization of a gene that activates covRS expression. Because it activates covR, we named this gene rocA (for “regulator of CovR”). Inactivation of rocA in GAS results in decreased covR expression and derepression of has transcription.
MATERIALS AND METHODS
Bacterial strains and media.
GAS (S. pyogenes) strain JRS4 is a streptomycin-resistant derivative of serotype M6 strain D471 (50). The GAS strain RTG229 was derived from JRS4 and contains a copy of the Tn916-J4 transposon on the chromosome, which provides homology for recombination of the pVIT plasmids as described previously (6, 20). All GAS strains described here are derivatives of JRS4 and RTG229. Escherichia coli XL1-Blue (Stratagene) and TG1 Rep (47) were used as the hosts for plasmid construction and were grown in Luria-Bertani broth with agitation (49). GAS strains were grown at 37°C without agitation in Todd-Hewitt broth supplemented with 0.2% yeast extract (THY). Antibiotics were used at the following concentrations: ampicillin at 100 μg/ml for E. coli, chloramphenicol at 30 μg/ml for E. coli and 10 μg/ml for GAS, erythromycin at 500 μg/ml for E. coli and 1 μg/ml for GAS, kanamycin at 50 μg/ml and 200 μg/ml for GAS, and spectinomycin at 100 μg/ml for both E. coli and GAS.
Construction of the pVIT GAS strain used to measure Pcov expression.
The VIT system was used to deliver the Pcov-gusA reporter fusion in single copy to an ectopic chromosomal location to study Pcov expression (6). The pVIT plasmid contains regions of homology to the resident Tn916-J4 transposon on the RTG229 chromosome. A DNA fragment containing the Pcov-gusA reporter was cloned into the homologous region of pVIT for integration into the chromosome.
The covR promoter region used for translational fusions was amplified from the JRS4 chromosome by using primers R1-CovOrf1 (gaattcTGGTATTAGTTTTAGACAAAGACGC) and Cov-GusAS (tctagaTGTCATTTATACCAACCCTTATCCTCTTA). The restriction site at the 5′ end of each primer is indicated by lowercase letters, the sequence homologous with the chromosome is indicated by uppercase letters, and the first two codons of the covR open reading frame (ORF) are in boldface. The 504-bp amplified fragment, which contains the covR promoter, untranslated sequence, and two codons, was blunt end ligated into SmaI-digested pBluescript II KS (Stratagene) to generate pEU7415. To clone the gusA reporter gene in frame with covR, we used the XbaI site that was inserted into the primer.
The gusA gene (1.8 kb) was amplified from plasmid pNZ8008 (9) by using primers GusL (gctctagaATGTTACGTCCTGTAGAA) and GusR (gctctagaTCATTGTTTGCCTCCCTG) and was restricted with XbaI (site synthesized into the primer; underlined). The XbaI fragment containing the gusA gene was ligated to XbaI-restricted pEU7415 to create pEU7416. The orientation of gusA and the in-frame fusion of the Pcov-gusA gene were verified by sequencing.
To insert the Pcov-gusA reporter fusion at the pVIT locus, plasmid pVIT164 (20) was used. A PstI-SacI fragment from pEU7416 carrying Pcov-gusA was cloned into PstI-SacI-digested pVIT164 to generate pJRS2227. Plasmid pJRS2227 was linearized with PvuII and transformed into RTG229 to produce JRS2227 as described previously (6). JRS2227 is kanamycin resistant and erythromycin sensitive, and the presence of both junctions of the Pcov-gusA fusion at the pVIT locus was verified by PCR.
Isolation of rocA mutants.
The procedure described by Maguin et al. (39) was used to generate insertion mutants of GAS. Briefly, JRS2227 was transformed with pGhost9::ISS1 and transformants were selected on THY agar containing kanamycin and erythromycin at 30°C. An overnight culture was made from a single transformed colony at 30°C with erythromycin. Cultures were diluted 100-fold in the same medium, grown for 2 h at 30°C, and then shifted to 37°C for 2.5 h to select for transposition. Insertion mutants were selected on THY agar containing kanamycin and erythromycin at 37°C.
The location of the inserted ISS1 element was identified by one of two methods. A template generated by self-ligation of HindIII-digested chromosomal DNA was subjected to inverse PCR by using primers GATTGTAACGTAGATAATAACCAACAGC (ISS1Rout1) and GCAAGAACCGAAGAAATGGAACG (ISS1Fout1). The PCR product was sequenced with primer AATAGTTCATTGATATATCCTCGCTGTCA (ISS1-Rout2) to identify the flanking sequences. Alternatively, the self-ligated product was used to transform E. coli TG1 and erythromycin-resistant transformants were selected. Plasmid DNA containing flanking sequence was isolated and sequenced with primer GGTCTTAATGGGAATATTAGC (ISS1-For4) as previously described (13). The flanking sequences were identified by comparison to the M1 and M5 genome sequences.
Colonies in which the plasmid vector sequence had been excised from the chromosome were obtained by growth at 30°C without antibiotic selection, which permits plasmid replication (39). Erythromycin-sensitive (Ems) colonies, indicating that the plasmid had been excised, were selected by plating on solid medium at 37°C. Ems colonies were confirmed for loss of the plasmid sequence by PCR with primers homologous to the flanking regions.
Insertional inactivation of rocA.
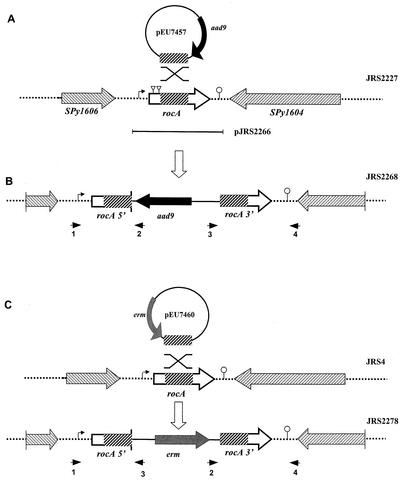
To create plasmids for inactivation of the rocA gene, a fragment internal to the coding region was amplified from JRS4 with high-fidelity Herculase polymerase (Stratagene) with primers AGCATTCAAGATACCTTCATAAGTAAGTCT and TCCACTAGTGTCTATTAGTTTGGTTAAGCG. The amplified fragments were blunt end ligated to SmaI-restricted pUC-Spec (28), a suicide vector unable to replicate in GAS, to create pEU7457 (Fig. 2). The amplified internal rocA fragment was also blunt end ligated to SmaI-restricted pSK-Erm (a pBluescript SK derivative in which the ampicillin resistance gene has been replaced with the erythromycin resistance gene from Tn1545; I. Stojiljkovic, unpublished data) to generate pEU7460 (Fig. 2). Plasmid pEU7457 was introduced into JRS2227 by electroporation, and transformants were selected on THY agar with spectinomycin. A spectinomycin-resistant transformant resulting from homologous single-crossover integration of pEU7457 into the GAS chromosome and inactivation of the rocA gene was named JRS2268. The plasmid-chromosome junctions of JRS2268 were verified with the following primer pairs: 5′ junction, ccgaattcCTGATTTAATCTTTTAAGCTGT (RocAS1; see Fig. 2, arrowhead 1) and ggaaacagctatgaccatg (M13Rev; see Fig. 2, arrowhead 2); 3′ junction, ccgaattcATTAGTAATAGATTAACATATT (RocAA1; see Fig. 2, arrowhead 4) and tgtaaaacgacggccagtg (M13For; see Fig. 2, arrowhead 3). The rocA gene was also inactivated in strains JRS4, JRS948 (17), JRS 964, JRS965, and JRS966 (11, 18) by transformation with pEU7460 and selection for erythromycin-resistant transformants. This generated JRS2278, JRS2279, JRS2351, JRS2285, and JRS2286, respectively. These strains, in which rocA was inactivated, were verified by PCR across the plasmid-chromosome junctions by using primer pairs RocAS1 and M13For for the 5′ junction and RocAA1 and M13Rev for the 3′ junction.
FIG. 2.
Construction of a rocA null mutant. (A) Region surrounding rocA in the chromosome of GAS. Plasmid pEU7457 (circle), which was used for insertional inactivation, contains a region internal to the rocA ORF (thick hatched box) and contains aad9, which encodes spectinomycin resistance (thick black arrow). The direction of transcription of the ORFs flanking rocA is indicated by striped arrowheads. The region of rocA that was cloned in complementing plasmid pJRS2266 is indicated by a bar below the chromosome. (B) JRS2268 was produced by homologous recombination (indicated by the X above the representation of the chromosome in panel A), which inserted pEU7457 into the wild-type rocA gene in the JRS2227 chromosome. (C) JRS2278 was produced by a similar targeted insertion in JRS4 by using integrational plasmid pEU7460, which contains erm, encoding erythromycin resistance (gray box), and a region internal to the rocA ORF (thick hatched box) as shown. Symbols: dotted lines, chromosome; striped boxes, coding regions with the directions of transcription indicated by the arrowheads; bent arrow, putative promoter; triangles, ISS1 insertion sites; lollipop, putative rho-independent transcription terminator. Small arrowheads below the chromosome represent primers used to confirm plasmid insertion into the chromosome (see Materials and Methods). The figure is not drawn to scale.
Construction of the rocA-complementing plasmid pJRS2266.
A PCR fragment containing all of rocA and 330 bp upstream of its start codon was amplified from JRS4 by using primers RocAS1 and RocAA1, which introduced unique EcoRI sites at both ends. The resulting 1.78-kb fragment was digested with EcoRI and ligated into EcoRI-digested pLZ12 (11), a chloramphenicol resistance-encoding natural shuttle vector able to replicate in GAS, to create pJRS2266.
Deletion of the covR gene.
Plasmid pEU7529 was used to delete covR from the GAS strains (A. Gusa and J. R. Scott, unpublished). This plasmid was constructed from pJRS943 (17), which contains, in addition to the covR ORF, the upstream and downstream regions. Plasmid pJRS943 was restricted with BsgI and XcmI to delete covR and blunt ended (17). A spectinomycin resistance-encoding cassette was isolated by SmaI restriction from pSL60-I (37) and ligated to BsgI-XcmI-restricted and blunted pJRS943 to create pEU7521. The ampicillin resistance gene from pEU7521 was removed by PvuI digestion, followed by self-ligation, to generate pEU7529. Plasmid pEU7529 was linearized by SacI and transformed into JRS2227 and JRS2281. Spectinomycin-resistant transformants containing the covR deletion were selected and were confirmed for the replacement of the covR gene by PCR as previously described (17).
RNA blot analysis.
GAS strains were cultured in THY, and growth was monitored by using a Klett-Summerson colorimeter with a red filter. Total RNA was isolated from different phases of growth as described previously (4), except that glycine was omitted from the medium. RNA was pelleted by sedimentation through 5.7 M CsCl (40) or by FastPrep (Bio 101) (4) as described previously. DNase I-treated RNA was assayed on Zeta-Probe membranes (Bio-Rad) as previously described (4). DNA probes were prepared by PCR amplification with the JRS4 chromosomal DNA as the template. The primer pairs used in this study are the following: covR, CAGCTATTCTTTGATATACTCTTTAGAG (5Cov) and Cov-GusAS; gyrA, GATCTGCAGGAATACGACTCATTTCTCTTTATCCC and GTCATCCTGACCGCTTGTCAAAAGG; hasA, GAGAGAGAATTCACCTAGGAGTGTTTGATTTT (intHasA1) and GAGGAGGAATTCAGATGCCGAGTCATTA (intHasS1); sagA, GGAGGTAAACCTTATGTTAA (SagAL) and AGATTATTTACCTGGCGTAT (SagAR).
GusA assays.
For plate assays, 10-μl volumes of overnight GAS cultures were spotted onto THY agar plates containing 200 μg of X-Glu (Gold Biotechnology Inc.) per ml. GAS strains that produced low GusA activity were white, while strains that produced high GusA activity were blue. Specific activity of GusA was assayed from GAS cultures grown to late exponential phase as previously described (14). For some experiments, GAS cultures were grown in THY broth buffered with 100 mM Tris-HCl at pH 7.2. The rate of hydrolysis was standardized by comparison to known concentrations of glucuronidase (Sigma catalog no. 500-0006). One unit of GusA activity was defined as that which liberates 1 μg of phenolphthalein (from phenolphthalein glucuronide)/h/mg of protein of GAS lysate at 37°C, pH 6.8. The protein concentration was determined by the Bio-Rad protein assay standardized against bovine serum albumin.
RESULTS
Construction of a Pcov-gusA reporter strain and isolation of rocA mutants.
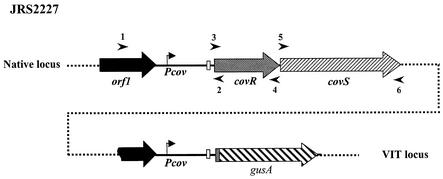
To facilitate studies of the regulation of the CovRS operon, we constructed a Pcov-gusA reporter strain. The entire promoter region of covR, including the untranslated leader sequence and the first two codons of the covR ORF, was fused to a gusA reporter gene to create a translational fusion. By using the pVIT system, this Pcov-gusA reporter fusion was then integrated at an ectopic location on the chromosome of strain RTG229 (20), an M6 GAS strain derived from JRS4, to generate JRS2227 (Fig. 1). In this strain, the native covRS region remains unaltered.
FIG. 1.
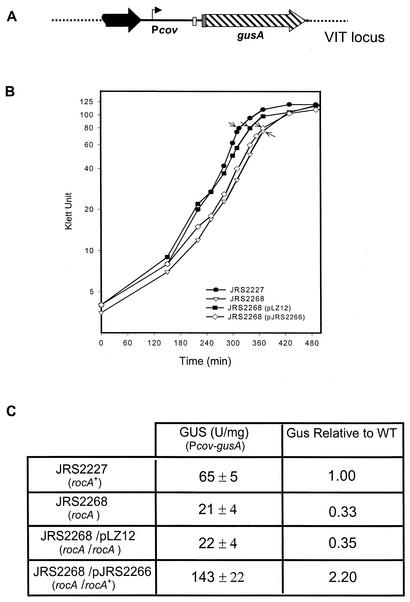
Construction of Pcov-gusA reporter strain. The promoter region of covR, along with the sequence encoding the first two amino acids of CovR (amplified by primers 1 and 2 [arrowheads]), was fused to the gusA reporter gene isolated from plasmid pNZ8008. The Pcov-gusA reporter construct was inserted into the GAS chromosome at the VIT locus, which is not linked to the native covRS locus. Native covRS and the region of covR that is present at the VIT locus are shown. Symbols: bent arrow, covR promoter; white box, ribosome binding site of CovR; checkered box in front of gusA, first two amino acids of CovR; broken black arrow, part of the upstream ORF present in the Pcov-gusA fusion. Arrowheads indicate primers used for PCR analysis. The orientation of Pcov-gusA relative to the native covRS locus is unknown.
We used insertional mutagenesis to identify potential activators of covR expression. For this purpose, we used the insertion sequence ISS1 because it appears to insert itself randomly into the genome of gram-positive bacteria, including various streptococci (39, 59), and because it rarely inserts itself more than once into the same cell (39, 51, 59). We introduced this transposon into JRS2227 on pGhost9::ISS1, a plasmid whose replication is temperature sensitive (38). An erythromycin-resistant (Ermr) transformant containing pGhost9::ISS1 was grown overnight at 30°C, and Ermr colonies containing the transposon were isolated at 37°C. In three different experiments, the transposition frequency (as measured by the number of Ermr colonies divided by the total number of colonies at 37°C) was between 0.3 and 0.5%, which is comparable to frequencies obtained for ISS1 transposition in other streptococci (51, 59, 60).
Among the operons that are negatively regulated by CovR is the has operon, required for production of the hyaluronic acid capsule (17). We reasoned that inactivation of an activator of Pcov would cause increased transcription of Phas because there would be less CovR expression. This would result in a mucoid colony phenotype in the JRS4-derived GAS strain JRS2227, which produces very little capsule. Therefore, we screened the JRS2227 colonies containing inserted ISS1 for a mucoid phenotype. Mucoid colonies were then tested by a plate assay by using X-Glu to monitor expression of GusA from the Pcov-gusA reporter fusion in the strain. A defect in expression of Pcov results in production of white colonies. Among the mucoid colonies, approximately 30% were white.
To eliminate any possible insertion in the covRS operon, the size of the covRS region was verified by PCR amplification with primers 1 plus 2, 3 plus 4, and 5 plus 6 (Fig. 1), which include the region upstream of the covRS operon, as well as the entire covRS operon. The clones that showed wild-type-size fragments, suggesting an intact covRS region, were chosen for further analysis.
The site of the ISS1 insertion was identified as described in Materials and Methods for 31 of the mucoid white colonies. In all 31 cases, Southern hybridization analysis showed that ISS1 insertion had occurred at only one location. Among these mutants, more than one independent insertion had occurred in eight different genes. Four of these genes (SPy1032 [hylA], SPy481, SPy1505, and a conserved ORF [SPyM18_0587] absent from the M1 and M3 genomes but present in the M5 and M18 genomes) had multiple insertions in the same location and thus could be sisters. Multiple independent insertions at different locations were found in each of the four remaining genes, SPy 1981 (relA), SPy1605, SPy159, and SPy0534 (aroE). Although none of the genes into which ISS1 had inserted itself showed homology to any known transcriptional regulator, SPy1605 showed some homology to sensor kinases (see below), which are involved in gene regulation. Therefore, we focused our studies on SPy1605, which we call rocA (for “regulation of covR”; see below). The rocA gene encodes a polypeptide of 451 residues. Two independent ISS1 insertions had occurred in this gene and were located within codons 25 and 91, respectively. The rocA gene is preceded by a putative promoter region upstream of the translational start codon and followed by a potential rho-independent terminator just downstream of the stop codon (Fig. 2A). The gene downstream of rocA is transcribed in the opposite direction, which makes it unlikely that the ISS1 insertions would have polar effects.
RocA regulates the expression of covR.
To be sure that the phenotype of the rocA insertion mutation did not result from additional spontaneous mutations elsewhere in the genome, we inactivated the rocA gene in the nonmutagenized Pcov-gusA reporter strain JRS2227. A PCR-amplified fragment internal to the coding region of rocA was cloned into the suicide vector pUC-Spec (28) to produce pEU7457. Following introduction of this plasmid into JRS2227, recombinants in which pEU7457 had been integrated into rocA were selected as Specr colonies. This integration results in inactivation of the rocA gene (Fig. 2B). In these integrants, the junctions between the integrated plasmid and the chromosome were identified by PCR analysis (by using primers 1 plus 2 and 3 plus 4 [Fig. 2B]). In addition PCR with primers 1 plus 4 failed to amplify a full-length copy of rocA.
Transcription of Pcov was quantitated in these strains by measuring the activity of GusA produced from the Pcov-gusA reporter fusion (Fig. 3A). For this assay, cells were grown in THY broth and harvested late in the exponential phase (Fig. 3B). We found that the rocA mutant strain (JRS2268) showed threefold less Pcov-gusA expression than its rocA+ parent (JRS2227, Fig. 3C). To be sure that this effect was caused by the rocA mutation, we used complementation. We cloned a DNA fragment including rocA with its potential promoter region into pLZ12, which replicates in GAS, to generate pJRS2266 (Fig. 2A). As a control, we used JRS2268 (rocA) containing the vector pLZ12. This control strain had the same GusA activity as strain JRS2268, while the complemented strain, JRS2268/pJRS2266, expressed about twofold more GusA than the parental rocA+ strain, JRS2227. Thus, when RocA was overexpressed from the multicopy plasmid, Pcov expression was not only restored but was increased above the wild-type level (Fig. 3C). Therefore, it appears that RocA activates expression from the Pcov promoter.
FIG. 3.
Expression of the covR gene in the wild-type and rocA mutant strains. (A) A gusA reporter system was used to measure the expression from the covR promoter at the VIT locus (symbols are as in Fig. 1). (B) Arrows indicate the times at which the samples were assayed for GusA activity. (C) The values shown are units of glucuronidase activity (with standard errors of the mean of experiments repeated at least four times). The relative ratio of GusA activity with respect to that of the wild type (WT) is shown.
CovR is not required for the activation of Pcov by RocA.
Because CovR represses its own transcription by binding to DNA in the Pcov region (Gusa and Scott, personal communication), it seemed possible that RocA could activate Pcov by affecting the interaction of CovR with the Pcov promoter DNA. If RocA acts on Pcov through CovR, then in the covR rocA double mutant, Pcov expression should be the same as in a covR single mutant. If RocA and CovR act independently on Pcov, however, the decrease in transcription in the rocA mutant should be independent of the status of covR (mutant or wild type). To assay this, GusA specific activity was measured at the mid-exponential phase of growth. The results (Table 1) show a 2.4-fold decrease in Pcov transcription in the rocA mutant compared to that in the rocA+ parent. The decrease was seen in both covR+ and covR mutant strains. Similarly, inactivation of covR resulted in a 2.7-fold increase in Pcov-gusA transcription in both the rocA+ and rocA mutant strains. This strongly suggests that the two systems that regulate Pcov are independent.
TABLE 1.
Regulation of covR expression by RocA is independent of functional CovR
| Strain | Genotype | Gus activity (U/mg)a |
|---|---|---|
| JRS2227 | covR+rocA+ | 136 ± 23 |
| JRS2281 | covR+rocA | 50 ± 6 |
| JRS2289 | covR rocA+ | 332 ± 62 |
| JRS2290 | covR rocA | 123 ± 10 |
Regulation of covR expression was measured from the Pcov-gusA fusion at mid-exponential phase. The values shown represent the average GusA activity of two independent experiments.
Effect of RocA on transcription of the has operon.
Since the original rocA insertion mutants appeared mucoid, as expected of a strain deficient in an activator of Pcov, we wanted to confirm that the increased mucoidy resulted from an increase in transcription of the capsule-encoding operon hasABC. To avoid any complications that might result from an ectopic reporter fusion assay, we measured has operon transcript directly. For this purpose, we inactivated the rocA locus in strain JRS4 by single-crossover integration of pEU7460 (Fig. 2C). The plasmid-chromosome junctions of the resulting rocA mutant strain, JRS2278, were verified by PCR (by using primers 1 plus 3 and 2 plus 4) (Fig. 2C) as described in Materials and Methods.
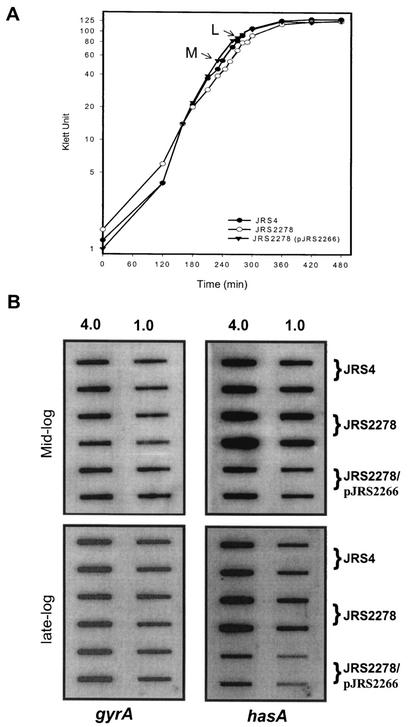
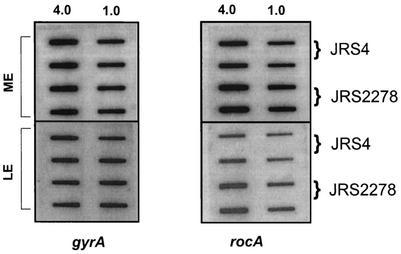
RNA was isolated from strains JRS4, JRS2278, and JRS2278/pJRS2266 in the mid- and late exponential growth phases (M and L, respectively, in Fig. 4A) and assayed by hybridization to a PCR-derived hasA probe. To ensure that equal amounts of mRNA from each strain were loaded on the filter, all samples were hybridized with a gyrA probe (Fig. 4B). We found that JRS2278 (rocA) had two- to threefold more hasA transcript than JRS4 at both stages of growth. This is as expected if RocA activates Pcov to produce more CovR repressor. The complemented strain, JRS2278/pJRS2266, produced two- to threefold less hasA transcript than the JRS4 wild-type strain. This suggests, as above (Fig. 3), that rocA is overexpressed from pJRS2266, which results in overproduction of CovR for repression of Phas.
FIG. 4.
Analysis of hasA gene transcription in the wild-type and rocA mutant strains. (A) Total RNA was isolated from JRS4 (wild type), JRS2278 (rocA), and JRS2278/pJRS2266 (rocA/rocA+) at the mid-exponential growth phase (M) and the late exponential growth phase (L). (B) On each filter, two dilutions (4.0 and 1.0 μg) of RNA were applied in vertically arranged duplicates. Membranes were probed with PCR-derived specific DNA probes internal to the coding region of gyrA and hasA (Materials and Methods). Results reported are representative of hybridization from three independent RNA isolations.
Regulation of Phas by RocA requires functional CovR.
It seemed likely that RocA decreases Phas transcription indirectly by activating Pcov to produce more CovR. To study this, we used two separate approaches, one of which relied on GusA activity produced from a Phas-gusA fusion and the other of which relied on direct measurement of the hasA mRNA level.
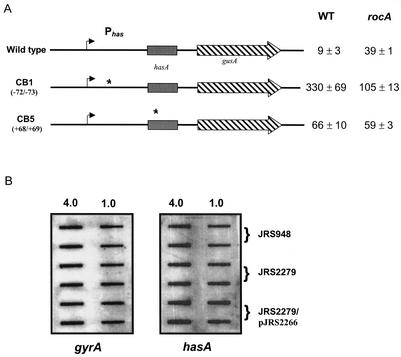
Federle and Scott (18) identified the binding sites at the Phas promoter for CovR and constructed mutants with two base substitutions at conserved T-T pairs at each of these sites. In two of these Phas mutants, Phas is still transcribed but is insensitive to repression by CovR. We reasoned that if RocA acts on Phas indirectly through CovR, a rocA mutation should not affect transcription of the Phas mutants that are not CovR regulated. In the Phas-gusA fusion with no mutations in the promoter region, the rocA strain produces approximately fourfold more GusA activity than the wild type (Fig. 5A). In contrast, in the Phas-gusA strain mutated at CovR binding site 1 (CB-1), there was no increase in GusA expression in the rocA mutant compared to that in the rocA+ strain. Instead, we observed lower GusA activity in the rocA mutant and the reason for this decrease is unknown. However, in the other Phas-gusA strain, in which CB-5 is altered, mutation of the rocA gene did not change GusA specific activity (Fig. 5A). This implies that RocA probably acts on the Phas promoter indirectly through CovR.
FIG. 5.
Regulation of the has promoter by rocA requires functional CovR. (A). Stars indicate mutations in the has promoter region that make it insensitive to CovR repression. The name of the mutant is on the left, and the location of the mutation is in parentheses. The values shown are glucuronidase units with standard errors of the mean of at least four independent experiments. WT, wild type. (B) Transcription analysis of hasA by RNA hybridization. RNA was isolated at the mid-exponential phase from JRS948 (covR), JRS2279 (covR rocA), and JRS2279/pJRS2266 (covR rocA/rocA+). Blots were prepared and hybridized as described in the legend to Fig. 4. Results are representative of hybridization from two independent RNA isolations.
To confirm this observation, we measured the amount of has transcript. As shown above (Fig. 4), JRS2278 (rocA) shows about twofold more Phas transcript than its rocA+ parent, JRS4. However, in JRS948 (covR), JRS2279 (covR rocA), and JRS2279/pJRS2266 (covR rocA/rocA+), the level of hasA transcript remained the same (Fig. 5B). This is consistent with the above result from the Phas promoter mutants and suggests that the increased has expression seen in the rocA mutant is mediated by CovR. Therefore, it appears that RocA decreases Phas transcription by activating transcription of Pcov and producing more CovR.
RocA negatively regulates its own expression.
Since it is not uncommon for transcriptional regulators to control their own synthesis, we used RNA hybridization analysis to test this for rocA. As before, we harvested RNA at the mid- and late exponential phases (Fig. 4A) and normalized to gyrA for a loading control on slot blots. At both stages of growth, there was two- to fourfold more rocA transcript in JRS2278 (rocA) than in JRS4 (rocA+) (Fig. 6). This suggests that RocA, which activates covR, negatively regulates its own expression (either directly or indirectly). Although rocA transcript appears more plentiful in the mid-exponential phase than in the late exponential phase, autoregulation persists in the later stage of growth.
FIG. 6.
RocA regulates its own expression. RNA was extracted from the JRS4 (wild-type) and JRS2278 (rocA) strains at the mid-exponential (ME) and late exponential (LE) phases as indicated in Fig. 4. Membranes were hybridized with gyrA and rocA probes. Blots were prepared and hybridized as described in Materials and Methods. The results shown are representative of hybridization from two independent RNA isolations.
Autoregulation of RocA is independent of functional CovR.
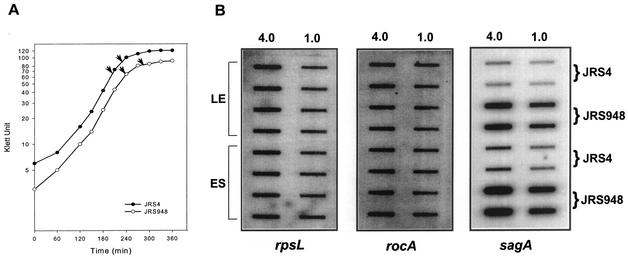
The RocA sequence contains no elements suggestive of a DNA binding protein, so its negative autoregulation is likely to be indirect. RocA activates transcription of CovR, and CovR is a negative regulator of many GAS promoters, so it seemed possible that negative autoregulation of rocA expression might also be mediated by CovR. To investigate this, we quantitated the rocA transcript levels in a covR mutant and its wild-type parent. The RNA was harvested in the late exponential and early stationary phases from cells grown in THY broth, and hybridization to an rpsL probe was used as a loading control. No difference was detected in the amount of rocA transcript in RNA from JRS4 (covR+) and JRS948 (covR) (Fig. 7). One of the genes repressed by CovR is sagA, which encodes streptolysin S (17), so an internal sagA fragment was also used to probe the same RNA to make sure the JRS948 strain had not mutated. As previously shown (17), JRS948 produces much more sagA transcript than its covR+ parent, JRS4 (Fig. 7). From these results, it appears that RocA negative autoregulation does not depend on the presence of functional CovR protein.
FIG. 7.
Regulation of rocA expression by RocA is independent of functional CovR. RNA was extracted from the JRS4 (wild-type) and JRS948 (covR) strains at the late exponential (LE) and early stationary (ES) phases (arrows in panel A). Membranes were hybridized with rpsL, rocA, and sagA probes as described in Materials and Methods (B). The results shown are representative of hybridization from two independent RNA isolations.
RocA is present in most GAS strains and not in strains of other streptococcal species.
CovR is a major global regulator in all strains of GAS examined, and it appears from the above that RocA regulates the expression of CovR. Therefore, we investigated the prevalence of rocA in different GAS strains.
To investigate the distribution of rocA among the different streptococci, we used two approaches. First, a BLAST search with the rocA nucleotide sequence against the available GAS genome sequences showed that this gene is present in the sequenced M1, M3, M5, and M18 strains. A similar search found no significant homology to genes in any sequenced strains of other streptococcal species.
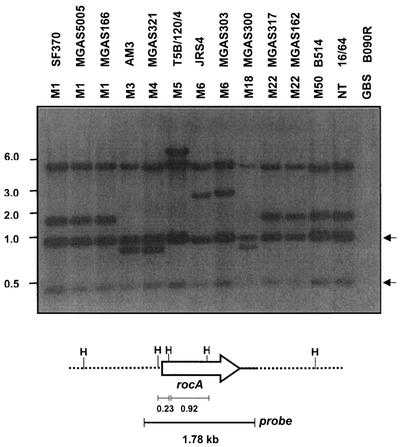
To expand these observations, we used Southern hybridization to investigate the presence of a rocA homolog in strains representing nine of the multilocus enzyme electrophoresis types of GAS (42). For this analysis, a 1.78-kb fragment including the complete rocA gene was used as a probe and the hybridization was carried out under stringent conditions (Fig. 8). This probe hybridized with two HindIII fragments that differ in size and two HindIII fragments of constant size in all 13 GAS strains tested. The HindIII fragments of variable size indicate that regions flanking rocA vary among the GAS strains and correspond to the predicted sizes for M1, M3, M5, and M18 strains. The two constant HindIII fragments were ∼0.92 and ∼0.23 kb long. The 0.92-kb fragment is internal to rocA, and the 0.23-kb fragment includes 0.14 kb of upstream DNA. These results demonstrate that rocA is present in most or all strains of GAS.
FIG. 8.
Detection of the rocA gene by Southern hybridization. Genomic DNA from the strains was restricted with HindIII and separated in a 0.7% agarose gel by electrophoresis, and the presence of the rocA gene was detected by hybridization with a 1.78-kb fragment of the rocA region as shown at the bottom. M serotypes and strain numbers are indicated at the top. GBS, group B streptococcus; NT, nontypeable. Arrows indicate the constant fragments hybridized with the probe in all GAS strains. The values on the left are molecular sizes in kilobases.
On the other hand, the rocA probe failed to hybridize with DNA derived from representative strains of streptococcal group B, C, D, E, F, G, H, L, M, N, or O or with S. pneumoniae, S. mutans, Enterococcus faecalis, or Lactococcus lactis (data not shown). These results suggest that rocA is present only in GAS and is absent from all related bacteria, including the closely related streptococci of groups C and G.
DISCUSSION
In this study, we have identified a gene, rocA, that positively regulates covR expression. The Pcov-gusA reporter gene fusion that we used to identify rocA is a translational fusion; therefore, the observed reduction of gusA expression in the mutant could be due to either transcriptional or translational regulation or both. However, when a Pcov-gusA transcriptional fusion was used, 2.2-fold less Gus activity was obtained in the rocA mutant strain than in the wild type (data not shown). This indicates that rocA regulates Pcov at the transcriptional level.
RocA appears to activate CovR expression, which acts as a repressor on several promoters. We found that the rocA mutant strain is mucoid and shows more has transcription than the wild type (Fig. 4). This is expected, since in the rocA mutant, less CovR is produced, and as a result, has transcription is derepressed. In view of the fact that has expression was increased in the rocA mutant, we measured the transcription of genes, such as sagA, ska, and speMF/sda, that are normally repressed by CovR (17). We did not find any significant differences in transcription between the wild-type and rocA mutant strains (data not shown). It is possible that the effect of the rocA mutation on these genes was very small and that the RNA slot blot assay that we used to measure transcript levels is not sensitive enough to detect subtle changes. Alternatively, the rocA mutation may not affect the transcription of the sagA, ska, and speMF genes. It is possible that the amount of CovR present in the rocA mutant is sufficient for complete repression of the sagA, ska, and speMF promoters but that more CovR is required for complete repression of the has promoter. However, it is also possible that the active form of CovR that regulates the sagA, ska, and speMF genes is different from the one that represses the has promoter and that the latter form is affected by the rocA mutation whereas the former form is not.
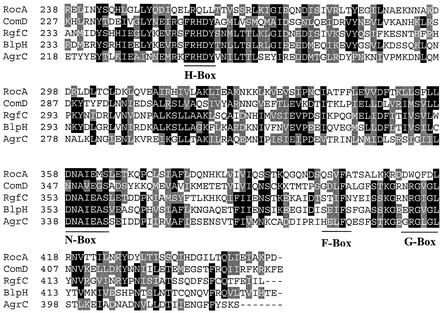
The rocA gene encodes a polypeptide of 451 amino acid residues. A BLSLTP (version 2.2.3) search of the nonredundant GenBank database showed limited homology to various sensor kinase homologs, with probability values ranging up to e−37. The homology was restricted to the C-terminal region of RocA, and the homology did not increase significantly when only the C-terminal 220 amino acids were used for a BLASTP search. Proteins that showed homologies to RocA of greater than e−10 include BlpH and ComD of S. pneumoniae (10, 24), AgrC of S. aureus (36), RgfC of S. agalactiae (52), and FasB and FasC of GAS (31), among others. Most of these sensor kinases are activated by a small peptide and are involved in quorum sensing (32).
Members of the sensor kinase superfamily exhibit clusters of highly conserved residues that are presumed to play crucial roles in signal transduction. These characteristic sequence fingerprints have been termed the H, N, F, and G boxes (56, 57). The H box contains the conserved histidine residue that is involved in autophosphorylation. The other boxes are considered to be part of the kinase subdomain and are required for kinase activity (56, 57). In the RocA sequence, we did not find a conserved H residue in the region corresponding to the H box (Fig. 9). In fact, apart from the N box, no other conserved boxes were apparent (Fig. 9). This surprising deviation from the canonical homology boxes has been reported previously for some other sensor kinases (for a review, see reference 23). For example, sensor kinases in which the conserved histidine is replaced with arginine, aspartate, or tyrosine (23, 61) and sensor kinases in which various conserved boxes are absent (12, 30, 56) have been documented previously. Thus, if RocA encodes a sensor kinase, it belongs to an unorthodox family with variant phosphorylation and kinase domains.
FIG. 9.
Comparison of RocA with homologous proteins identified by a Blast-P search. The C-terminal regions of the different sensor kinases were aligned by using ClustalW. The GenBank accession numbers of the sequences are as follows: RocA, AAK34382; ComD, CAB39530; RgfC, AAM22582; BlpH, AAK74685; AgrC, AAB63269. Putative conserved boxes are indicated below the alignment.
The N-terminal regions of sensor kinases are highly diverse and contain hydrophobic domains required for membrane insertions, known as the transmembrane (TM) helices. There are usually two to eight TM helices that, along with the surface-exposed regions, play a crucial role in signal recognition. TMPRED (http://www.ch.embnet.org/software/TMPRED-form.html) and Goldman-Engelman-Steitz hydrophobicity (15) analyses of the deduced primary amino acid residues predicted that the N-terminal 220 amino acids of RocA form seven TM helices with short (3- to 17-residue) interhelical regions. Thus, like sensor kinases, RocA is predicted to be membrane associated.
Analysis of the RocA sequence showed no obvious DNA binding motifs, such as zinc finger or helix-turn-helix motifs. In addition, a computer-based profile scan against the Pfam (http://us.expasy.org) protein families and conserved domain database (http://ncbi.nlm.nih.gov) did not show any domain related to DNA binding proteins. This strongly suggests that RocA is not a transcriptional regulator that binds directly to the promoter regions to modulate expression. Therefore, the regulation of Pcov and ProcA promoters by RocA is likely to be indirect.
If RocA encodes a sensor kinase, it should have a cognate response regulator. In bacteria, the genes for cognate pairs of sensor kinases and response regulators are typically found together in a single operon. However, in the region containing rocA, there are no ORFs with homology to a response regulator. Although the presence of cognate response regulators at a different locus than sensor kinases is not common, it has been observed. For example, the genes for response regulators for barA of E. coli (43), expS of Erwinia carotovora (16), and gacS of Pseudomonas syringae (45) are all present far from those encoding the corresponding sensor kinases. Thus, if RocA is a sensor kinase, its cognate response regulator may be present elsewhere in the genome.
Since we observed both activation of Pcov and repression of ProcA promoters by RocA (Fig. 3 and 6), RocA may act as a sensor kinase to stimulate either two different regulators with opposite functions or a single response regulator with different activities. There are several situations in which a single sensor kinase activates two different response regulators. For example, in E. coli, sensor kinase CheA interacts with CheB and CheY to regulate chemotaxis (35). On the other hand, response regulators with two opposite functions have also been documented. For example, in the E. coli osmosensing system, response regulator OmpR functions both as an activator and as a repressor to differentially regulate the expression of the ompC and ompF genes (29, 44). Thus, whether RocA stimulates a single regulator with different activities or two different regulators remains to be explored.
RocA is present only in GAS, whereas the covRS system appears to be present in other streptococci as well, since covRS homologs have been found in group B streptococcus (21, 58), group C streptococcus (53), and S. mutans (GenBank accession no. AF393849). The streptococci that possess covRS but not rocA may modulate covR expression through a functional homolog with little or no primary sequence homology to rocA. Another possibility is that the activation of covR expression by RocA is unique to GAS. Perhaps, since strains of GAS are able to cause a very wide range of diseases, these bacteria require an additional level of covR regulation not required by other streptococci that have a more limited disease spectrum. The role of CovR in virulence gene regulation and the mechanism of regulation of covR expression have not been studied in other streptococci.
In conclusion, we have identified and characterized rocA, a novel regulatory gene involved in the activation of covR expression in GAS. Since CovR is a global regulator that controls 15% of the GAS genes, RocA may be involved in the regulation of a large network of genes. Moreover, RocA also regulates genes independently of CovR. Therefore, detailed characterization of RocA-mediated gene regulation may lead to a greater understanding of the events that occur during pathogenesis of GAS.
Acknowledgments
This work was supported by Public Health Service grant R37-AI20723 from the National Institutes of Health.
We thank Emmanuelle Maguin for providing the transposon pGhost9::ISS1, Tony Richardson and Igor Stojiljkovic for providing the plasmid pSK-Erm, Asiya Gusa for constructing the plasmid pEU7529, and Tracy Dalton for providing JRS948 RNA.
REFERENCES
- 1.Alouf, J., and H. Muller-Alouf. 1996. Cellular constituents and extracellular proteins involved in the pathogenic capacity of Streptococcus group A. Ann. Pharm. Fr. 54:49-59. [PubMed] [Google Scholar]
- 2.Bernish, B., and I. van de Rijn. 1999. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J. Biol. Chem. 274:4786-4793. [DOI] [PubMed] [Google Scholar]
- 3.Bisno, A. L. 1991. Group A streptococcal infections and acute rheumatic fever. N. Engl. J. Med. 325:783-793. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, I., P. Germon, K. McDade, and J. R. Scott. 2001. Generation and surface localization of intact M protein in Streptococcus pyogenes are dependent on sagA. Infect. Immun. 69:7029-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronze, M. S., and J. B. Dale. 1996. The reemergence of serious group A streptococcal infections and acute rheumatic fever. Am. J. Med. Sci. 311:41-54. [DOI] [PubMed] [Google Scholar]
- 6.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 7.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vos, W. M. 1986. Genetic improvement of starter streptococci by the cloning and expression of the gene coding for a non-bitter proteinase: biomolecular engineering programme—final report, p. 465-472. In E. Magnien (ed.), Biomolecular engineering in the European Community: achievements of the research programme (1982-1986)—final report. Martinus Nijhoff, Lancaster, England.
- 12.Dutta, R., L. Qin, and M. Inouye. 1999. Histidine kinases: diversity of domain organization. Mol. Microbiol. 34:633-640. [DOI] [PubMed] [Google Scholar]
- 13.Duwat, P., A. Cochu, S. D. Ehrlich, and A. Gruss. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman, D. M., T. A. Steitz, and A. Goldman. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15:321-353. [DOI] [PubMed] [Google Scholar]
- 16.Eriksson, A. R., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11:743-752. [DOI] [PubMed] [Google Scholar]
- 17.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Federle, M. J., and J. R. Scott. 2002. Identification of binding sites for the group A streptococcal global regulator CovR. Mol. Microbiol. 43:1161-1172. [DOI] [PubMed] [Google Scholar]
- 19.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geist, R. T., N. Okada, and M. G. Caparon. 1993. Analysis of Streptococcus pyogenes promoters by using novel Tn916-based shuttle vectors for the construction of transcriptional fusions to chloramphenicol acetyltransferase. J. Bacteriol. 175:7561-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 22.Graham, M. R., L. M. Smoot, C. A. Lux Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 24.Havarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 25.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcuspyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heath, A., A. Miller, V. J. DiRita, and C. N. Engleberg. 2001. Identification of a major, CsrRS-regulated secreted protein of group A streptococcus. Microb. Pathog. 31:81-89. [DOI] [PubMed] [Google Scholar]
- 27.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 28.Husmann, L. K., D. L. Yung, S. K. Hollingshead, and J. R. Scott. 1997. Role of putative virulence factors of Streptococcus pyogenes in mouse models of long-term throat colonization and pneumonia. Infect. Immun. 65:1422-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenney, L. J. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5:135-141. [DOI] [PubMed] [Google Scholar]
- 30.Koretke, K. K., A. N. Lupas, P. V. Warren, M. Rosenberg, and J. R. Brown. 2000. Evolution of two-component signal transduction. Mol. Biol. Evol. 17:1956-1970. [DOI] [PubMed] [Google Scholar]
- 31.Kreikemeyer, B., M. D. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 32.Lazazzera, B. A., and A. D. Grossman. 1998. The ins and outs of peptide signaling. Trends Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 33.Lei, B., F. R. DeLeo, N. P. Hoe, M. R. Graham, S. M. Mackie, R. L. Cole, M. Liu, H. R. Hill, D. E. Low, M. J. Federle, J. R. Scott, and J. M. Musser. 2001. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nat. Med. 7:1298-1305. [DOI] [PubMed] [Google Scholar]
- 34.Levin, J. C., and M. R. Wessels. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A streptococcus. Mol. Microbiol. 30:209-219. [DOI] [PubMed] [Google Scholar]
- 35.Li, J., R. V. Swanson, M. I. Simon, and R. M. Weis. 1995. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 34:14626-14636. [DOI] [PubMed] [Google Scholar]
- 36.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 37.Lukomski, S., N. P. Hoe, I. Abdi, J. Rurangirwa, P. Kordari, M. Liu, S.-J. Dou, G. G. Adams, and J. M. Musser. 2000. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonald, P. M., E. Kutter, and G. Mosig. 1984. Regulation of a bacteriophage T4 late gene, SOC, which maps in an early region. Genetics 106:17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, A. A., N. C. Engleberg, and V. J. DiRita. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40:976-990. [DOI] [PubMed] [Google Scholar]
- 42.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88:2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pernestig, A. K., O. Melefors, and D. Georgellis. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276:225-231. [DOI] [PubMed] [Google Scholar]
- 44.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 45.Rich, J. J., T. G. Kinscherf, T. Kitten, and D. K. Willis. 1994. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J. Bacteriol. 176:7468-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russo, F. D., and T. J. Silhavy. 1993. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1:306-310. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Schlievert, P. M., A. P. Assimacopoulos, and P. P. Cleary. 1996. Severe invasive group A streptococcal disease: clinical description and mechanisms of pathogenesis. J. Lab Clin. Med. 127:13-22. [DOI] [PubMed] [Google Scholar]
- 49.Scott, J. R. 1972. A new gene controlling lysogeny in phage P1. Virology 48:282-283. [DOI] [PubMed] [Google Scholar]
- 50.Scott, J. R., P. C. Guenthner, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spellerberg, B., B. Pohl, G. Haase, S. Martin, J. Weber-Heynemann, and R. Lütticken. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 181:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, and R. Lütticken. 2002. rgf encodes a novel two-component signal transduction system of Streptococcus agalactiae. Infect. Immun. 70:2434-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steiner, K., and H. Malke. 2002. Dual control of streptokinase and streptolysin S production by the covRS and fasCAX two-component regulators in Streptococcus dysgalactiae subsp. equisimilis. Infect. Immun. 70:3627-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 38:1004-1016. [DOI] [PubMed] [Google Scholar]
- 55.Stevens, D. L. 1992. Invasive group A streptococcus infections. Clin. Infect. Dis. 14:2-11. [DOI] [PubMed] [Google Scholar]
- 56.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 57.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington D.C.
- 58.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thibessard, A., A. Fernandez, B. Gintz, B. Decaris, and N. Leblond-Bourget. 2002. Transposition of pGh9:ISS1 is random and efficient in Streptococcus thermophilus CNRZ368. Can. J. Microbiol. 48:473-478. [DOI] [PubMed] [Google Scholar]
- 60.Ward, P. N., T. R. Field, W. G. Ditcham, E. Maguin, and J. A. Leigh. 2001. Identification and disruption of two discrete loci encoding hyaluronic acid capsule biosynthesis genes hasA, hasB, and hasC in Streptococcus uberis. Infect. Immun. 69:392-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, J., N. Ohta, J. L. Zhao, and A. Newton. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. USA 96:13068-13073. [DOI] [PMC free article] [PubMed] [Google Scholar]