Abstract
Genome projects are generating large numbers of potential new targets for drug discovery. One challenge is target validation, proving the usefulness of a specific target in an animal model. In this paper, we demonstrate a new approach to validation and assay development. We selected in vitro specific peptide binders to a potential pathogen target. By inducing the expression of a selected peptide in pathogen cells causing a lethal infection in mice, the animals were rescued. Thus, by combining in vitro selection methods for peptide binders with inducible expression in animals, the target's validity was rigorously tested and demonstrated. This approach to validation can be generalized and has the potential to become a valuable tool in the drug discovery process.
Complete or nearly complete genome sequences of human pathogens such as Escherichia coli (1), Haemophilus influenzae (2), Staphylococcus aureus (http://www.tigr.org), and Enterococcus faecalis (http://www.tigr.org) are now known. These genomes have been annotated and potential targets for drug discovery have been identified. Selecting the most useful targets is limited by the need to define those targets essential to the disease process in an animal model and by the lack of biochemical characterization of many of them.
Technologies currently available to validate targets, such as gene knockouts (3, 4), signature-tagged mutagenesis (5, 6), genomic analysis and mapping by in vitro transposition (7), anti-sense approaches (8), or temperature-sensitive mutants (9), are not designed to test whether the wild-type protein target is essential for pathogen growth in the disease state. In particular, it is desirable to demonstrate the reversal of the disease state by modulating the function of the target in vivo.
We imagined that a target-specific peptide would be useful in target validation. Hypothetically, small peptides of 10 to 20 amino acids can be selected to interact with virtually any binding cleft on a protein. In some cases, these clefts may be located at the active site and peptide binding may disrupt protein function. One advantage of peptides is that they can be expressed intracellularly, by using gene-expression systems. This function facilitates testing the peptide in vivo to determine whether it has a phenotype that relates to a specific disease model.
To pursue this possibility, we chose prolyl-tRNA synthetase (ProRS) as a target (10). This enzyme catalyzes the attachment of proline to tRNAPro. Although drugs directed at ProRS have never been developed, it is an essential protein, and for that reason it is an obvious candidate as a target for a new antiinfective. Peptides that specifically inhibit ProRS were obtained by selection, and one of these selected peptides was pursued in depth in an animal model to determine whether ProRS was indeed suitable as a target for new antiinfectives.
Materials and Methods
Selection of Peptide Binders.
Affinity selection of peptides encoded as a fusion to the N terminus of the gene III protein of the filamentous fd phage was conducted as described (11). Phage libraries were either PHD7 and PHD12 (displaying either 7 or 12 residue peptides; New England Biolabs), a random 12-mer library supplied by B. Kay (University of Wisconsin, Madison), or libraries of random 11-residue peptides in which the central residue of each library was fixed with a different residue. Library complexity was typically >10−8. Biotinylated target protein and phage were incubated together in solution before the capture of the target phage complex on streptavidin-coated paramagnetic particles (12).
Construction of Expression Plasmids.
The fusion gene of a peptide with the sequence of SREWHFWRDYNPTSR and glutathione S-transferase (Pro-3/GST) was PCR-amplified using pGEX-4T2 (Amersham Pharmacia) and a combination of two forward primers, Pro3 (5′-CCAACAACATATGTCCCGTGAATGGCACTTCTGGCGTGACTAC) and Pro3/GST (5′-TTCTGGCGTGACTACAACCCGACCTCCCGTGGGGGTGGAGGCATGTCCCCTATACTA), and a reverse primer, GST/R (5′-AGTTGAATTCTTAATCCGATTTTGGAGGATGG). The resulting PCR product was then amplified with primers Pro3 (KpnI) (5′-CAAGGTACCCATGTCCCGTGAATGGCAC) and GST/R (BamHI) (5′-CGCGGATCCTTAATCCGATTTTGGAGGATGG). The amplified DNA was digested with KpnI and BamHI restriction endonucleases and cloned into the KpnI/BamHI sites of the expression vector pPROtet (CLONTECH). The vector was transformed into E. coli DH5αPRO cells that constitutively express the Tet repressor. pPROtet uses the PL promoter of phage lambda combined with the Tet operator of the Tn10 tetracycline resistance operon to direct the regulated expression of genes. A construct containing Pro-3/GST in pPROtet, termed pC3844, was identified by PCR expression screening and confirmed by DNA sequencing. The plasmid contains a Glu-Gly-Gly-Gly linker between the Pro-3 peptide and GST. As a control, the GST gene was also cloned into pPROtet, resulting in plasmid pC3868.
The S. aureus ProRS gene was amplified from genomic DNA (S. aureus American Type Culture Collection strain 65389) with oligonucleotides S.PRS/XhoI-5′ (5′-AATCCGCTCGAGGATTATTGCTATTGGTGCC) and S.PRS/Hind-3′ (5′-AATCGTAAGCTTTTATTTTAAGTTATCATATTT) and cloned into XhoI/HindIII sites in pACYC177 (New England Biolabs). The cloned S. aureus ProRS gene carries its own promoter and ribosome binding site and is in the same orientation as the disrupted kanamycin resistance gene in the vector. One resulting construct, pC3847, was transformed into E. coli DH5αPRO/pC3844 for growth characterization.
Protein Expression.
An overnight culture of E. coli DH5αPRO cells harboring pC3844 or pC3868 was used to inoculate fresh LB broth containing 34 μg/ml chloramphenicol and 50 μg/ml spectinomycin. Expression of Pro-3/GST or GST was induced by the addition of anhydrotetracycline (aTc) to bacterial cultures at an OD600 of 0.5. After 2.5 hr of induction, cells were pelleted from 1-ml cultures and lysed by boiling in 100 μl of SDS/PAGE sample buffer. Ten-microliter samples were loaded on a 4–20% SDS/PAGE for analysis.
Protein Purification.
E. coli DH5αPRO cells from 4-ml cultures expressing GST or Pro-3/GST were pelleted and resuspended in 200 μl of PBS containing 100 μg/ml of lysozyme and protease inhibitors mixture (Hoffmann–La Roche). The cell suspensions were incubated at 37°C for 10 min, and cells were lysed by sonication followed by centrifugation at 14,000 × g for 10 min. The cell lysate supernatant was incubated with 20 μl of glutathione/Sepharose affinity resin (Amersham Pharmacia) for 30 min and washed five times with PBS. Resin was boiled in 80 μl of SDS/PAGE sample buffer, and 20 μl of the samples was subjected to SDS/PAGE analysis.
ProRS Kinetic Characterization.
ProRS inhibition studies were performed as described (13), with saturating E. coli tRNA conditions (90 μM). For inhibition studies against proline, ATP was held at 1 mM and the proline concentration was varied between 5 and 165 μM (Km = 20 μM). For studies against ATP, proline was held at 160 μM and ATP concentrations were varied between 12.5 and 400 μM (Km = 70 μM). Initial velocity curves were generated in the presence of multiple peptide concentrations and fitted to inhibition patterns by using grafit data analysis software (Erithacus Software, Staines, U.K.).
N-Terminal Sequence Analysis.
The purified Pro-3/GST fusion protein was blotted to a poly(vinylidene difluoride) membrane (Bio-Rad) for N-terminal sequence analysis by Edman degradation.
Analysis of Charged tRNA Levels.
Cultures (400 ml) of E. coli DH5αPRO cells harboring pC3844 were grown in LB to an OD600 of 0.3 and split into duplicate cultures. One culture was induced with 50 ng of aTc per ml, and growth continued for 35 min. Cells were harvested at an OD600 of ≈0.6, and charged tRNA was quantified as described (14).
Animal Studies.
E. coli JM109 cells were cotransformed with a pSC101-based plasmid isolated from BL21PRO (CLONTECH), which encodes a constitutively expressed tetR gene and pC3844 or pC3868. The resulting JM109 strains exhibited growth behavior similar to their DH5αPRO counterparts. Mice were infected i.p. with 1 × 108 colony-forming units of E. coli JM109 cells containing pC3844 or pC3868. At 1 hr and 4 hr after pathogen inoculation, the mice were given saline, 2 mg of aTc per kg, or 50 mg of ciprofloxacin per kg (positive control) by i.p. injection. Mouse survival at 7 days postinfection is reported.
Results and Discussion
Peptide Binders to ProRS.
E. coli ProRS was panned versus a collection of linear-peptide phage display libraries. The binders were clustered into groups having related sequences. To test whether growth inhibition can be achieved via intracellular peptide expression, DNA sequences encoding several peptides from the major cluster were cloned (in the pPROtet expression system) as translational fusions to the 5′-end of the gene for GST. The fusions incorporated a flexible tetrapeptide linker. One of the peptide sequences was designated as Pro-3 and has the sequence SREWHFWRDYNPTSR. This peptide was a potent inhibitor of in vitro aminoacylation of tRNA with proline (Ki = 250 nM). The Pro-3 peptide competed with both proline and ATP.
Peptide Expression and Characterization.
Cultures of E. coli cells harboring plasmids containing the Pro-3/GST fusion or GST alone were grown to early log phase and induced with aTc. aTc is a derivative of tetracycline that is a potent inducer of the tetO/R system but has the advantage of being less toxic to E. coli than tetracycline is. SDS/PAGE analysis of cell lysates demonstrated that, upon induction, the Pro-3/GST fusion was expressed at a level comparable to that of GST alone (Fig. 1A, lanes 3 and 4 versus lanes 7 and 8, respectively). In contrast, no expression was detected in the absence of induction (Fig. 1A, lanes 1, 2, 5, and 6).
Figure 1.
Inducible peptide expression. (A) Analysis of total cellular proteins from E. coli DH5αPRO cells. Mid-log phase E. coli cells harboring pC3868 (encoding GST, lanes 1–4) or pC3844 (encoding Pro-3/GST, lanes 5–8) were induced with 0 (lanes 1 and 5), 10 (lanes 2 and 6), 50 (lanes 3 and 7), or 250 (lanes 4 and 8) ng/ml aTc. Total cellular proteins were separated by 4–20% SDS/PAGE and stained with Coomassie Brilliant Blue. (B) Glutathione/Sepharose affinity resin purification of GST proteins. Lane 1, GST purified from cells carrying pC3868; lane 2, Pro-3/GST purified from cells carrying pC3844; lanes 3 and 4, pC3844 resin extract washed with 10 or 100 μM prolyl-adenylate analog during purification; lane 5, pC3844 resin extract washed with 100 μM isoleucyl-adenylate analog; lane 6, purified E. coli ProRS.
The Pro-3/GST fusion was purified with glutathione/Sepharose affinity resin. Interestingly, a protein with the same molecular weight as E. coli ProRS copurified with the Pro-3/GST fusion protein (Fig. 1B). To demonstrate that the copurified protein was E. coli ProRS, a prolyl-adenylate analog (15) was added to the wash step during purification of the fusion protein. The material purified after this wash contained significantly reduced amounts of the high-molecular weight protein; in contrast, the addition of an isoleucyl-adenylate had no effect (Fig. 1B, compare lanes 2–5). These results suggest that Pro-3/GST interacts with ProRS, at least during the purification process.
The purified fusion protein was shown by N-terminal sequence analysis (Edman degradation) to have the expected Pro-3 peptide and linker sequences (data not shown). In addition, the Pro-3/GST protein inhibited E. coli ProRS with a Ki of 180 nM. The close similarity of the Ki for the peptide and the fusion protein suggests that the interaction of the fusion protein with ProRS is through the peptide segment and, further, that GST does not interfere with this interaction.
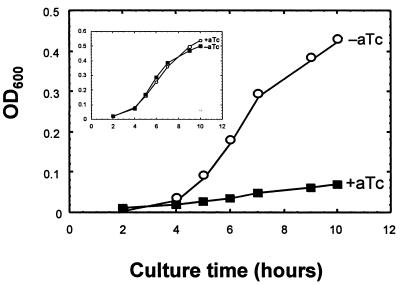
The growth of E. coli cells in culture was reduced significantly in response to expression of the Pro-3/GST fusion, whereas expression of GST alone had no effect (Fig. 2). Intracellular expression of peptides similar in sequence to Pro-3 but lacking potent ProRS inhibition (in aminoacylation assays) did not elicit any effect on cell growth (data not shown). Thus, the growth defect seen in response to Pro-3/GST expression is consistent with inhibition of ProRS.
Figure 2.
E. coli growth rates after Pro-3/GST expression. Early log phase E. coli DH5αPRO cells harboring pC3844-encoding Pro-3/GST were split into duplicate cultures, one of which was induced with 250 ng/ml aTc (+aTc) and the other of which was not (−aTc). The growth rates were determined over a 10-hr period. The inset shows an analogous experiment done with E. coli cells carrying the GST expression construct pC3868.
To define the intracellular specificity of the Pro-3/GST fusion for E. coli ProRS, tRNA was extracted from recombinant strains grown in the presence or absence of aTc. The amounts of charged tRNAPro were significantly reduced in response to Pro-3/GST expression (Fig. 3A). In contrast, levels of charged tRNAPhe and tRNAMet were unaffected. We also used a genetic complementation test to demonstrate the specificity of Pro-3/GST for ProRS. This test was based on the observation that S. aureus ProRS efficiently charged E. coli tRNAPro but was not inhibited by and did not bind Pro-3 or Pro-3/GST. (The species-specificity of the Pro-3 peptide was demonstrated with both the S. aureus and E. faecalis ProRS; in both cases, the observed Ki was >80 μM.) An E. coli strain was constructed containing two plasmids, one with S. aureus promoter elements needed for the constitutive expression of S. aureus ProRS and a second for pPROtet-inducible expression of Pro-3/GST expression (Fig. 3B). The growth of the E. coli cells expressing S. aureus ProRS was no longer inhibited by induction of Pro-3/GST expression (Fig. 3B).
Figure 3.
Analysis of Pro-3/GST intracellular specificity. (A) Total tRNA was isolated from E. coli cells carrying pC3844 with (+) or without (−) 50 ng/ml aTc induction. The content of charged tRNAPro, tRNAMet, and tRNAPhe was quantified. (B) Growth curves for E. coli DH5αPRO carrying both pC3844 (encoding Pro-3/GST) and pC3847 (encoding the S. aureus ProRS gene) in the presence (+aTc) or absence (−aTc) of aTc. The inset shows an analogous experiment performed with E. coli DH5αPRO carrying pC3844 and pACYC177, the parental plasmid for pC3847.
Animal Studies.
The Pro-3/GST system was then studied in a lethal animal-infection model. The inducible expression system was first transferred to a virulent strain of E. coli, and the growth defect in response to plasmid-borne Pro-3/GST expression was confirmed. Mice were then infected by i.p. injection with the virulent E. coli harboring the Pro-3/GST expression system. After infection, mice were treated i.p. with aTc, and mortality was subsequently recorded. Induction of Pro-3/GST expression rescued five of five mice (Table 1). With saline in place of the inducer, none of the infected mice survived. As a further control, we showed that when bacterial cells harboring the vector alone were used, all mice succumbed, regardless of whether the inducer was added. Thus, intracellular expression of the Pro-3/GST fusion protects against a lethal pathogen infection.
Table 1.
Peptide-directed protection in a lethal animal-infection model
| Protein expression | Mouse survival
|
|
|---|---|---|
| −aTc | +aTc | |
| Pro-3/GST | 0/5 | 5/5 |
| GST | 0/5 | 0/5 |
Mice were infected i.p. with E. coli containing the inducible plasmids pC3844 (Pro3/GST) or pC3868 (GST). At 1 hr and 4 hr after pathogen inoculation, the mice were given saline (−aTc) or the inducer a Tc (+aTc) by i.p. injection. Mouse survival at 7 days postinfection is reported.
In general, peptide binders can be obtained for any target whatsoever, regardless of whether the biological function of the target is known. Some fraction of these binders will interact with a functional site and thereby inhibit activity. However, the physiological relevance, if any, of the target or target-and-peptide binder may be unclear or impossible to determine from in vitro characterization. Thus, the approach described in this paper has general value and can be applied to any potential target within a pathogen, even when the biological function of the target has not been established.
Once a target, a peptide, and the peptide binding site are validated by these methods, screening for small molecules that displace the peptide binders is a direct way to obtain new drug leads. Indeed, by using this approach with a fluorescently labeled Pro-3 peptide, we have screened small molecule libraries and identified compounds that displace the peptide and inhibit aminoacylation by ProRS.
Acknowledgments
P.R.S. is a director of Cubist Pharmaceuticals, Inc.
Abbreviations
- ProRS
prolyl-tRNA synthetase
- aTc
anhydrotetracycline
- GST
glutathione S-transferase
- Pro-3
a peptide with the sequence of SREWHFWRDYNPTSR
References
- 1.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 3.Link A J, Phillips D, Church G M. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vagner V, Dervyn E, Ehrlich S D. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 5.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 6.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 7.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inouye M. Gene. 1988;72:25–34. doi: 10.1016/0378-1119(88)90124-2. [DOI] [PubMed] [Google Scholar]
- 9.Schmid M B, Kapur N, Isaacson D R, Lindroos P, Sharpe C. Genetics. 1989;123:625–633. doi: 10.1093/genetics/123.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Söll D, Schimmel P R. In: The Enzymes. 3rd. Ed. Boyer P, editor. Vol. 10. New York: Academic; 1974. pp. 498–538. [Google Scholar]
- 11.Sparks A, Adey N, Cwirla S, Kay B. In: Phage Display of Peptides and Proteins: A Laboratory Manual. Kay B, Winter J, McCafferty J, editors. San Diego: Academic; 1996. pp. 227–253. [Google Scholar]
- 12.Hyde-Deruyscher, R., Paige, L., Hyde-Deruyscher, N., Lim, A., Fredericks, Z., Christensen, D. J., Krantz, J., Gallant, P. L., Zhang, J., Hamilton, P. T., et al. (2000) Chem. Biol., in press. [DOI] [PubMed]
- 13.Durekovic A, Flossdorf J, Kula M R. Eur J Biochem. 1973;36:528–533. doi: 10.1111/j.1432-1033.1973.tb02939.x. [DOI] [PubMed] [Google Scholar]
- 14.Hughes J, Mellows G. Biochem J. 1978;176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X Y, Hill J M, Yu G, Wang W, Kluge A F, Wendler P, Gallant P. Bioorg Med Chem Lett. 1999;9:375–380. doi: 10.1016/s0960-894x(98)00738-0. [DOI] [PubMed] [Google Scholar]