Abstract
Tuberculosis is one of the leading preventable causes of death. Emergence of drug-resistant tuberculosis makes the discovery of new targets for antimycobacterial drugs critical. The unique mycobacterial cell wall lipids are known to play an important role in pathogenesis, and therefore the genes responsible for their biosynthesis offer potential new targets. To assess the possible role of some of the genes potentially involved in cell wall lipid synthesis, we disrupted a mas-like gene, msl7, and a chalcone synthase-like gene, pks10, with phage-mediated delivery of the disruption construct, in which the target gene was disrupted by replacement of an internal segment with the hygromycin resistance gene (hyg). Gene disruption by allelic exchange in the case of each disruptant was confirmed by PCR and Southern blot analyses. Neither msl7 nor pks10 mutants could produce dimycocerosyl phthiocerol, although both could produce mycocerosic acids. Thus, it is concluded that these gene products are involved in the biosynthesis of phthiocerol. Both mutants were found to be attenuated in a murine model, supporting the hypothesis that dimycocerosyl phthiocerol is a virulence factor and thus the many steps involved in its biosynthesis offer potential novel targets for antimycobacterial therapy.
Two billion people are latently infected with Mycobacterium tuberculosis, and 5 to 10% of them will develop active tuberculosis at some time during their life (World Health Organization, http://www.who.int/gtb/publications/globrep01/, 2001). Currently 8 million new cases of active tuberculosis are diagnosed annually, with 2 million deaths per year. With the advent of multidrug-resistant tuberculosis, this disease has become a major public health problem. The Centers for Disease Control and Prevention have designated drug-resistant M. tuberculosis a class C organism, with the potential to create major public health problems in population centers. Effective defense against this pathogen requires identification of new targets for development of antimycobacterial drugs. M. tuberculosis is a highly successful pathogen because it can evade host defenses and mycobacterial drugs. The unusually lipid-rich (50 to 60%) cell walls of this pathogen constitute an impermeable barrier that helps the pathogen enter monocytes/macrophages and grow in them (4, 9, 16, 18). Thus, it is not surprising that this pathogen contains a large number of genes involved in lipid synthesis.
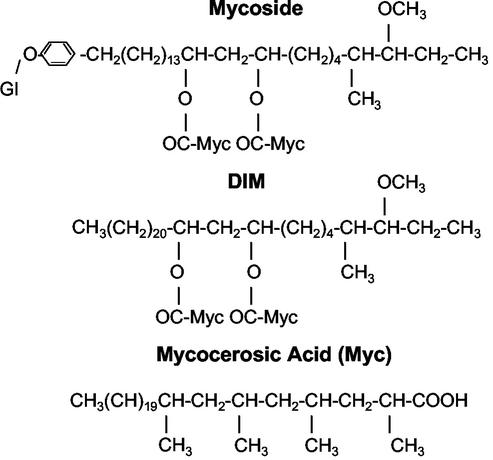
Identification of the genes involved in biosynthesis of the unique cell wall lipids of mycobacteria started with the cloning of the mycocerosic acid synthase (mas) gene (17). In this case, the cloning was done based on the amino acid sequence of the purified multifunctional enzyme that catalyzes the elongation of n-C20 fatty acid (and homologues) to generate a major class of fatty acids called mycocerosic acids (20). These acids are esterified to a long-chain diol, phthiocerol, to generate dimycocerosyl phthiocerol (DIM), which has been found to be a virulence factor (5, 8, 22). In some mycobacterial species, including some clinical isolates of M. tuberculosis, they are also esterified to phenolphthiocerol, which is glycosylated to produce mycosides (9, 15) (Fig. 1).
FIG. 1.
Structure of dimycocerosyl esters of glycosylated (Gl) phenolphthiocerol (mycoside) and phthiocerol (DIM).
A plausible pathway for the biosynthesis of phthiocerol has been postulated on the basis of biochemical analogies. Based on the nature of the reactions that must be involved in this synthesis, a set of multifunctional proteins have been postulated to be involved in this process (15). On the basis of the catalytic domains required to catalyze such a sequence of reactions, a segment of mycobacterial genome that could encode such proteins was found. Strong support for the involvement of this segment of the genome in DIM synthesis was provided when it was demonstrated that disruption of this gene segment in Mycobacterium bovis BCG in fact prevented the synthesis of DIM (2). Transposon mutagenesis confirmed this conclusion also for M. tuberculosis (5, 8). However, the enzymology of biosynthesis of DIM has not been elucidated, and not all of the genes involved in the synthesis of DIM have been identified.
The mycobacterial genome contains a large number of pks genes, including mas-like genes (msl). It also contains several chalcone synthase-like genes that have been found only in plants until the recent discovery of bacterial homologues (7, 13, 19). pks10 is one such gene found in M. tuberculosis. To examine the possible biological functions of pks1/pks15 and pks10, we used a gene disruption approach. In this paper, we report that disruptions of msl7 (pks1 and pks15) and a chalcone synthase-like gene (pks10) in M. tuberculosis results in DIM deficiency and attenuation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. tuberculosis H37Rv (ATCC 25618) was grown either in Middlebrook 7H9 broth supplemented with 10% OADC (oleic acid, albumin, and dextrose; 7H9-OADC; BBL) enrichment plus 0.2% glucose and 0.05% Tween 80 at 37°C in roller bottles or on Middlebrook 7H10-OADC agar plates incubated at 37°C in sealed plastic bags. Mycobacterium smegmatis mc2155 was grown in liquid Luria-Bertani (LB) medium with 0.05% Tween 80 for competent cell preparation and in Middlebrook 7H9 broth (Difco) with 0.05% Tween 80 for transduction. Escherichia coli DH5α (Life Technology) and HB101 were used as host strains for cloning experiments and were grown on Luria-Bertani broth or agar. When required, antibiotics were added to the culture media at the following concentrations: ampicillin, 100 μg/ml for E. coli; hygromycin B, 150 μg/ml for E. coli and 50 μg/ml for M. tuberculosis.
General DNA techniques.
All recombinant DNA techniques were performed according to standard procedures (21). DNA restriction and modifying enzymes were obtained from New England Biolabs (Beverly, Mass.) and used according to the manufacturer's suggestions.
Generation of disruption constructs for msl7 and a chalcone synthase-like gene (pks10).
The general strategy used for gene disruption with the specialized transducing phage system (10) was similar to that used previously for other msl genes (11, 22).
In order to make a disruption construct for msl7 (pks1 [Rv2946c] plus pks15 [Rv2947c]), a 2,937-bp PCR product containing part of the pks1 gene (bp 1231 to 4168 of the pks1 coding region, including the dehydratase, enoyl reductase, and ketoreductase domains) was amplified from M. tuberculosis genomic DNA, introducing BamHI sites at the 5′ and 3′ ends of the sequence (sense primer A, 5′-GGATCCGTGCAGCTGCCCACGTATGC-3′; antisense primer B, 5′-GGATCCGAGCGGCCAACCCGTCCA-3′). This product was subcloned into BamHI-digested pUC19 vector, and a 779-bp internal NcoI fragment (containing the enoyl reductase domain) was replaced with the hyg gene and used to generate recombinant phages containing the disrupted copy of msl7.
For pks10 (chs [Rv1660]) disruption, a 2,337-bp region including the total open reading frame of pks10 as well as flanking regions (bp 29760 to 32097 in MTCY06H11, accession number Z85982.1) was amplified by PCR from M. tuberculosis H37Rv genomic DNA with the following primers: sense primer A (5′-GGGAAGCTTGTCACGTTGCATGAC-3′) and antisense primer B (5′-GGGTCTAGAATACGAGCCGTGGAA-3′), introducing HindIII and XbaI restriction sites, respectively, and cloned into pBluescript SK(+) vector. A disrupted copy of pks10 was prepared by replacing a 333-bp NcoI internal fragment of the cloned gene with the hyg gene cassette. The disrupted pks10-containing fragment was used to generate specialized transducing phages.
Generation of M. tuberculosis gene-disrupted mutants.
M. tuberculosis H37Rv was grown to an optical density at 600 nm of 1.0 in Middlebrook 7H9-OADC without Tween 80. Cells from a 10-ml culture were collected by centrifugation, washed with Middlebrook 7H9-OADC with 0.3% added glycerol, and resuspended in 10 ml of Middlebrook 7H9-OADC. After overnight incubation at 37°C, cells were collected by centrifugation and resuspended in 1 ml of Middlebrook 7H9-OADC. One milliliter of lysate from msl7 and pks10 recombinant phages (2 × 1010 PFU/ml) was added to the cells to obtain a multiplicity of infection of 10. Infected cells were incubated for 4 to 6 h at 37°C, collected by centrifugation, resuspended in 1 ml of Middlebrook 7H9-OADC, and plated on Middlebrook 7H10 agar medium supplemented with 10% OADC and 50 μg of hygromycin B per ml. Hygromycin-resistant colonies were obtained after 3 to 4 weeks of incubation at 37°C.
PCR analysis.
PCR screening for disruption was performed directly on crude lysate obtained by boiling the cells by a standard protocol (21) and Platinum Taq polymerase (Life Technologies, Gaithersburg, Md.). PCR screening of msl7 transductants was carried out with a set of primers specific for the deleted segment of msl7: sense primer E, located upstream and just outside of the deleted segment, 5′-GTATGCGGCCACCCACAC-3′; and antisense primer F, located inside the deleted segment, 5′-CGCGCAGCGTGTCCCAC-3′. PCR analysis of the flanking regions was performed with sense primer C, 5′-TGAGGCGATGGTGGTGTCGATGCT-3′, and antisense primer H1, 5′-TGGACCTCGACGACCTGCAGGCAT-3′, for the 5′-flanking region and with sense primer H2, 5′-GGAACTGGCGCAGTTCCTCTGGGG-3′, and antisense primer D, 5′-ATGCGGGCCAGATCTCGGCTGCTC-3′, for the 3′-flanking region,.
pks10 mutants were screened for disruption by PCR analysis with the following primer pairs: sense primer E, 5′-GCCGTGGTCGAGCAGTATCTG-3′, and antisense primer F, 5′-CACATGCAGCACCGACGCTGA-3′, for the internal deleted segment; sense primer C, 5′-GTGTTCGCCATGATCGCCGTCGAC-3′, and antisense primer H1 for the 5′-flanking region; and sense primer H2 and antisense primer D, 5′-GTGTTCGCCATGATCGCCGTCGAC-3′, for the 3′-flanking region.
Genomic DNA isolation and Southern blotting.
M. tuberculosis genomic DNA was isolated by the GTC method with guanidine thiocyanate, Tris-HCl, and Sarcosyl solution (10). DNA samples were digested with appropriate restriction enzymes, subjected to 1% agarose gel electrophoresis, transferred to nylon membranes (Nytran Plus; Schleicher and Schuell, Keene, N.H.), and hybridized with [α-32P]dCTP-labeled probes with the random prime labeling system (Rediprime II; Amersham Pharmacia Biotech).
Test for expression of msl7 and pks10.
RNA was isolated from M. tuberculosis cells grown to the mid-exponential phase. Chilled cells isolated by centrifugation were resuspended in RNeasy lysis buffer (Qiagen), transferred to a 2-ml tube containing ceramic and silica beads (FastRNA Blue), and disrupted with a FastPrep F120 instrument (Q. Biogene). The extract collected by centrifugation was used to isolate total RNA with the RNeasy kit (Qiagen) according to the protocol provided by the manufacturer. Reverse transcription was performed with random primers and SuperScript RNase H reverse transcriptase (Life Technologies). PCR on the cDNA was done with Platinum Taq DNA polymerase (Invitrogen) and primers E and F used to test msl7 and pks10 disruptions described for PCR analysis. A control without the reverse transcriptase verified the absence of DNA contamination.
Biochemical analysis of cell wall lipids in wild type and pks gene-disrupted mutants of M. tuberculosis.
Sodium [1-14C]propionate (50 μCi; specific activity, 55 Ci/mol) (American Radiolabeled Chemicals, St. Louis, Mo.) was added to 30 ml of 12-day-old cultures of M. tuberculosis H37Rv and its pks gene-disrupted mutants (optical density at 600 nm of 1.6 to 1.8), and incubation was continued at 37°C in roller bottles for a further 24 h. Cells were collected by centrifugation at 6,000 × g for 10 min and autoclaved. The cells were extracted with an excess of chloroform- methanol (2:1) with constant stirring at room temperature for several hours, and total lipids were extracted and assayed for total 14C with Scintiverse BD scintillation fluid (Fisher Scientific, Pittsburgh, Pa.) in a Beckman LA3801 liquid scintillation counter (22).
The lipids in the medium were also extracted and assayed for 14C. Total lipids were separated on silica gel G plates (20 by 20 cm, 0.5 mm) with hexane-ethyl ether (9:1, vol/vol), and the polar lipids that remained at the origin in this solvent system were recovered from the silica gel with chloroform-methanol (2:1, vol/vol) and subjected to thin-layer chromatography (TLC) with chloroform-methanol (9:1, vol/vol) as the developing solvent. The lipids were visualized under ultraviolet light after spraying the plates with a 0.1% ethanolic solution of 2,7-dichlorofluoresein or by spraying the plate with 5% K2Cr2O7 in 50% sulfuric acid, followed by heating at 180°C for 10 min. The 14C-labeled lipids were detected by scanning chromatograms in a Berthold Tracemaster 20 automatic TLC linear analyzer and by autoradiography.
Silica gel containing labeled material was scraped from the TLC plates, and the lipids were eluted with chloroform-methanol (2:1, vol/vol). The recovered lipids were subjected to alkaline hydrolysis followed by methylation as described previously (22). The products recovered after methylation were subjected to TLC on silica gel G plates with hexane-ethyl ether (9:1, vol/vol) as the developing solvent. The hydroxy fatty acid methyl esters were subjected to acetylation with acetic anhydride-pyridine (2:1) at room temperature overnight. The acetyl derivatives, isolated by TLC with hexane-ethyl ether (9:1, vol/vol) as the developing solvent, and the fatty acid methyl esters were analyzed by radio-gas chromatography (radio-GC).
To examine the composition of the fatty acids from the total lipids, these lipids were subjected to exhaustive alkaline hydrolysis, followed by methylation with BF3 in methanol (22). The methyl esters of fatty acids and hydroxy fatty acids were isolated by TLC and analyzed by radio-GC. The total methyl esters of fatty acids from both the wild type and the mutants were also separated by argentation-TLC (5% AgNO3 in silica gel) with hexane-ethyl ether (9:1, vol/vol) as the developing solvent (11), and each separated fatty acid fraction was analyzed by radio-GC with a Varian model 3300 gas chromatograph with a coiled stainless steel column (3.2 mm by 2 m) packed with 3% OV-1 (wt/wt) on Chrom W-HP 80/100 and a Lablogic GC-RAM radioactivity monitor with Winflow (IN/US Systems, Tampa, Fla.) software. For analysis of fatty acid methyl esters and acetylated mycolipanolic acid methyl esters, a 180 to 300°C program increasing at 15°C/min was used. For acetylated hydroxyphthioceranic acid methyl esters, isothermal analysis at 290°C was done with a helium carrier gas flow of 30 ml/min.
Bacterial growth in alveolar macrophage cell line.
Mouse alveolar macrophage cell line MH-S (ATCC CRL-2019) was obtained from the American Type Culture Collection and propagated in RPMI 1640 medium (Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum. Bacteria were grown in Middlebrook 7H9 medium to an optical density at 600 nm of 0.4 and diluted 1:400 in RPMI plus fetal bovine serum.
Two days before infection, 24-well plates were seeded with 2 × 105 MH-S cells, and cultures were infected in triplicate at 37°C with 0.1 ml of a single-cell suspension of M. tuberculosis H37Rv or its msl7 and pks10 gene-disrupted mutants (0.5 bacterial cell/macrophage; multiplicity of infection, 0.5). The bacteria were allowed to infect for 4 h, and extracellular bacteria were removed by four successive washes with warm RPMI. The infected cells were lysed in 200 ml of 0.067% sodium dodecyl sulfate in 7H9 medium for 30 min at 37°C. Serial 10-fold dilutions of the lysates were plated in triplicate on 7H10 Middlebrook agar medium supplemented with 10% OADC, 0.5% Tween 80, and hygromycin when needed. Colonies were counted after 4 weeks of incubation at 37°C. The experimental values and standard deviations were calculated from the results of three independent experiments.
Assay for virulence in murine model.
Aliquots of M. tuberculosis strain H37Rv and its mutant strains were grown in modified 7H10 liquid medium (7H10 agar formulation with agar and malachite green omitted) supplemented with 10% OADC for 1 week on a 37°C rotary shaker. Media for the mutant strains were supplemented with 50 μg of hygromycin per ml. At the end of the incubation period, culture growth was measured with a Klett-Summerson colorimeter (Klett Manufacturing, Brooklyn, N.Y.) and diluted to yield a final concentration of 1 Klett unit/ml (5 × 105 CFU/ml). The inoculum size was determined by titration in triplicate on Middlebrook 7H10 (Difco Laboratories, Detroit, Mich.) agar plates with 10% OADC enrichment.
Female C57BL/6J mice (Jackson Laboratories, Bar Harbor, Maine) were purchased at 6 weeks of age and allowed to acclimate in the facility for 1 week. Animals were housed in microisolator cages (Lab Products, Maywood, N.J.) and maintained with water and Prolab RMH 3000 rodent chow (Purina, St. Louis, Mo.) in a biosafety level 3 animal facility. Mice were randomly assigned to groups on day 1 postinfection (four mice) and day 20 postinfection (four mice). Mice anesthetized with telazol and xylazine were infected intranasally with 20 μl of a suspension containing 104 CFU. At the designated time points, mice were euthanized by CO2 asphyxiation, and their right lungs were sterilely removed. Lungs were homogenized in a 1-ml volume contained in an aerosol-resistant grinding assembly (Idea Works Laboratory Devices, Syracuse, N.Y.). Aliquots of the homogenate were serially diluted and titrated on 7H10 agar plates. Agar plates were incubated at 37°C in ambient air for 4 weeks, and viable colonies were enumerated.
RESULTS
Disruption of msl7 (pks15 and pks1) and chalcone synthase-like (pks10) genes in M. tuberculosis.
To investigate the function of msl7 (pks15 and pks1) and pks10 in M. tuberculosis, the corresponding open reading frames were disrupted by allelic exchange, with a phage-mediated system used to deliver the disrupted copy of each gene.
The msl7 gene is located downstream of mas and consists of two overlapping open reading frames, pks15, encoding a 496-amino-acid protein containing a β-ketoacyl synthase domain, overlapping pks1 by 3 bp, that would encode a protein of 1,616 amino acid containing acyl transferase, dehydratase, enoyl reductase, ketoreductase, and acyl carrier protein domains. The msl7 gene showed the highest homology to a putative pks from Mycobacterium leprae (accession number NP002677). Among the M. tuberculosis pks genes, msl7 shows the most similarity to pks8 (59% identity and 71% similarity to pks1 over 1,144 amino acids and 73% identity and 83% similarity to pks15 over 445 amino acids).
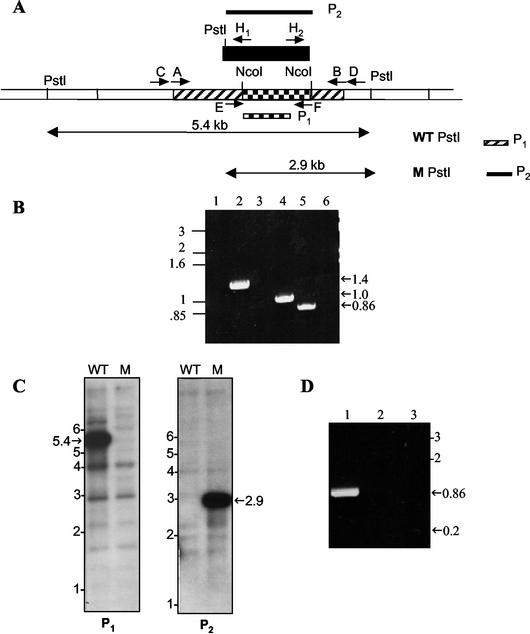
In order to make a disruption construct for msl7, a 2,937-bp PCR product containing part of the pks1 gene (bp 1231 to 4168 of pks1 coding region, including the dehydratase, enoyl reductase, and ketoreductase domains) was amplified from the M. tuberculosis genomic DNA template. A 779-bp internal NcoI fragment (containing the enoyl reductase domain) was replaced with the hyg gene and used to generate recombinant phages containing the disrupted copy of msl7 (Fig. 2A). PCR screening of the hygromycin-resistant transductants with a set of primers specific for the deleted segment of msl7 (sense primer E, located upstream and just outside of the deleted segment, and antisense primer F, located inside the deleted segment) identified two disruption mutants that failed to amplify the expected 860-bp fragment obtained with the wild-type M. tuberculosis DNA (Fig. 2B).
FIG. 2.
Disruption strategy for msl7 and evidence for gene replacement in the M. tuberculosis H37Rv mutant used for making the disruption construct. (A) Schematic representation of the construct used for disruption. Hatched, coding sequence; checkered, internal segment that was replaced with hyg gene cassette (black box); unshaded, regions of the gene outside those used to make the disruption construct. Primers A and B were used to amplify the DNA segment used to generate the disruption construct. WT PstI, PstI fragment from the wild type, expected to hybridize with probe P1, representing the fragment deleted in making the construct. M PstI, PstI fragment form the msl7 mutant, expected to hybridize with probe P2 (hyg gene). (B) PCR analysis of msl7 mutant. Lanes 1 and 2, PCR analysis of the 5′-flanking region, showing the product expected from the gene replacement mutant in lane 2 but not in the wild type (lane 1); primers C and H1. Lanes 3 and 4, PCR analysis of the 3′-flanking region, showing the expected product from the msl7 mutant (lane 4) but not from the wild type (lane 3); primers H2 and D. Lanes 5 and 6 show the PCR product representing the internal segment replaced by hyg from the wild type (lane 5) but not from the msl7 mutant (lane 6); primers E and F. (C) Southern blot analysis of M. tuberculosis H37Rv and the msl7 mutant. Genomic DNA was digested with PstI and hybridized with probe P1 (left) or P2 (right). WT, wild type; M, mutant. (D) Reverse transcription-PCR showing the presence of transcripts containing the pks1 segment of msl7 in M. tuberculosis H37Rv. For lanes 1, 2, and 3, primers E and F were used. Lane 1, wild type; lane 2, wild type without reverse transcriptase; lane 3, msl7 mutant. In all panels, sizes are shown in kilobases.
Disruption of msl7 by homologous recombination was confirmed by further PCR analysis of the flanking regions, which yielded the expected 1,382-bp 5′-flanking region and 1,062-bp 3′-flanking region (Fig. 2B). Southern blot analysis of M. tuberculosis H37Rv and the msl7 mutant along with the restriction sites used to confirm the mutant genotype are shown (Fig. 2A and C). The gene-specific probe (P1) consisting of the deleted pks1 segment that was replaced by the hyg gene hybridized to a 5.4-kb genomic fragment which was lost in the gene-disrupted mutant. The fainter hybridization signals at 4 and 2.9 kb that appeared in both the wild-type and mutant samples in Fig. 2C (left) reflect the hybridization of the gene-specific probe to homologous regions in other pks genes known to be present in the genome of M. tuberculosis. Probing with the hyg gene yielded a single shorter, 2.9-kb hybridization band in the mutant due to the PstI site present in the hyg gene cassette (Fig. 2A and C).
Recently, it was noted that pks15/1 in M. bovis BCG constitutes a single open reading frame, whereas in M. tuberculosis H37Rv, there is a single nucleotide insertion and this frameshift mutation was postulated to inactivate pks1 (6). Therefore, we tested whether the pks1 segment of msl7 is transcribed in M. tuberculosis H37Rv. Reverse transcription-PCR clearly demonstrated the presence of pks1 transcripts in the wild type, but, as expected, it was absent in the msl7 mutant (Fig. 2D). The control without reverse transcriptase did not give any product, ensuring that the PCR product was not due to DNA contamination.
M. tuberculosis genome contains a chalcone synthase-like gene, chs (pks10), located within a cluster of pks genes, directly upstream of pks7 (Rv1661) (msl4). pks10 would encode a 353-amino-acid protein with a deduced molecular mass of 37.09 kDa which shows 74% identity and 85% similarity to M. tuberculosis pks11 (Rv1665), a second chalcone synthase-like gene located in the same region, at the end of the pks cluster. A third chalcone synthase-like gene annotated, pks18 (Rv1372), shows less homology to pks10 (28% identity and 45% similarity). Whether these chs genes are expressed in M. tuberculosis and, if they are, what roles they play remain unknown.
For pks10 disruption, a 2,337-bp region including the entire open reading frame of pks10 as well as 708 bp of the 5′-flanking and 568 bp of the 3′-flanking regions was amplified by PCR from M. tuberculosis H37Rv genomic DNA. A 333-bp NcoI internal fragment of pks10 was replaced with the hygromycin resistance gene cassette (hyg) (Fig. 3A). This pks10::hyg fragment was introduced into phAE87 to generate phAE87-pks10::hyg. The phasmid DNA was verified by restriction enzyme analysis and PCR. After infecting M. tuberculosis H37Rv with the recombinant phages, the transductants were screened by PCR with two sets of unique primers that would amplify the flanking regions (Fig. 3A). The expected 1.4-kb and a 0.8-kb 5′- and 3′-flanking product, respectively, were generated from the mutants (Fig. 3B, lanes 1 to 4). Gene disruption was further supported by the fact that the primers representing the deleted internal segment (primer pair E/F) did not yield the 0.2-kb DNA fragment that was seen with the wild type (Fig. 3B, lanes 5 and 6). Disruption of the pks10 gene was verified by Southern blot analysis with BamHI and PstI as the restriction enzymes and two probes: P1, the deleted 333-bp NcoI segment of pks10 that was replaced with the hyg gene, and P2, the hygromycin resistance gene. The hybridization bands expected from homologous recombination were found (Fig. 3D). Reverse transcription-PCR analysis showed that the pks10 transcript found in the wild type was not present in the pks10 mutant (Fig. 3C).
FIG. 3.
Disruption strategy for pks10 and evidence for pks10 gene replacement in M. tuberculosis H37Rv. (A) Schematic representation of the disruption construct. Hatched and checkered regions represent the region used to make the disruption construct (amplified with primers A and B). The checkered segment was replaced with the hyg gene cassette (black box). Primer pairs C/H1, D/H2, and E/F were used for PCR analysis of homologous recombination as described in the text. WT PstI, WT BamHI, M PstI, and M BamHI are PstI and BamHI fragments from the wild-type (WT) and pks10 mutant (M) that are expected to hybridize with probes P1, representing the fragment deleted in making the construct, and P2, the hygromycin resistance gene cassette, respectively. (B) PCR analysis of 5′-flanking (lanes 1 and 2, primers C and H1), 3′-flanking (lanes 3 and 4, primers H2 and D), and the segment replaced by hyg (lane 5 and 6, primers E and F), demonstrating homologous recombination. Lanes 1, 3, and 5, wild type; lanes 2, 4, and 6, mutant. (C) Reverse transcription-PCR analysis showing expression of pks10 in the wild type (right) but not in the pks10 mutant (left). Primers, E and F. (D) Southern blot analysis of M. tuberculosis H37Rv and pks10 mutants. Genomic DNA was digested with PstI and BamHI. Left, hybridized with pks10 segment replaced by hyg (probe P1); right, probed with hyg gene (probe P2). WT, wild type; M1 and M2, mutants.
Analysis of lipids derived from [1-14C]propionic acid.
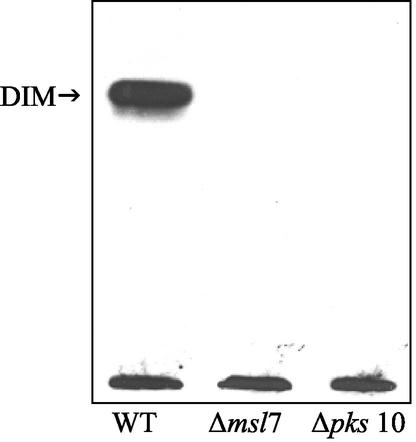
To elucidate the biosynthetic defects caused by the absence of functional msl7 and pks10 genes, we administered [1-14C]propionate as a radiotracer to label the lipids in the wild type and the mutants. The wild type and the gene-disrupted mutants incorporated similar amounts (20 to 25%) of the label into lipids. We carried out detailed analysis of the lipid classes and fatty acids in each lipid class. TLC of total lipids with 10% ethyl ether in hexane showed the absence of label in the dimycocerosyl phthiocerol (DIM) fraction in the two mutants, whereas in the wild type, 30% of the total label was contained in this fraction (Table 1, Fig. 4). The absence of DIM in these mutants was further confirmed by treatment of the plate with dichromate-sulfuric acid, followed by heating; the charred band at Rf 0.6, corresponding to DIM found in the wild type, was absent in the mutants (data not shown).
TABLE 1.
Relative distribution of 14C among the lipid classes derived from sodium [1 − 14C]propionic acid in M. tuberculosis and its msl7 and pks10 mutants
| Classa | Relative distribution of 14C (%)
|
||
|---|---|---|---|
| Wild type | msl7 mutant | pks10 mutant | |
| DIM | 30 | 0 | 0 |
| Fatty acids | 28 | 0 | 0 |
| Hydroxy fatty acids | 2 | 0 | 0 |
| PAT-I | 16 | 18 | 20 |
| Fatty acids | 14 | 14 | 16 |
| Mycolipanolic acids | 1.7 | 3 | 3 |
| Hydroxyphthioceranic acids | 0.3 | 1 | 1 |
| PAT-II | 4 | 3 | 4 |
| Fatty acids | 1 | 1 | 1 |
| Mycolipanolic acids | 1 | 0.6 | 1 |
| Hydroxyphthioceranic acids | 2 | 1.4 | 2 |
| Sulfolipids | 45 | 73 | 70 |
| Fatty acids | 10 | 10 | 10 |
| Mycolipanolic acids | 4 | 10 | 9 |
| Hydroxyphthioceranic acids | 31 | 53 | 51 |
| Very polar lipids | 5 | 6 | 6 |
| Fatty acids | 0.5 | 1 | 1 |
| Mycolipanolic acids | 0.3 | 0.5 | 0.6 |
| Hydroxyphthioceranic acids | 4.2 | 4.5 | 4.4 |
PAT, polyacyl trehaloses; DIM, dimycocerosyl phthiocerols. Very polar lipids (including diacyl trehalose) remained at and near the origin on TLC with 10% methanol in chloroform as the solvent system.
FIG. 4.
Autoradiogram of thin-layer chromatogram of lipids derived from sodium [1-14C]propionate in M. tuberculosis H37Rv (wild type [WT]), msl7 mutant, and the pks10 mutant. Total lipids were subjected to TLC on silica gel G with 10% ethyl ether in n-hexane as the solvent. A similar amount of radioactivity was used in each case. DIM, dimycocerosyl phthiocerol.
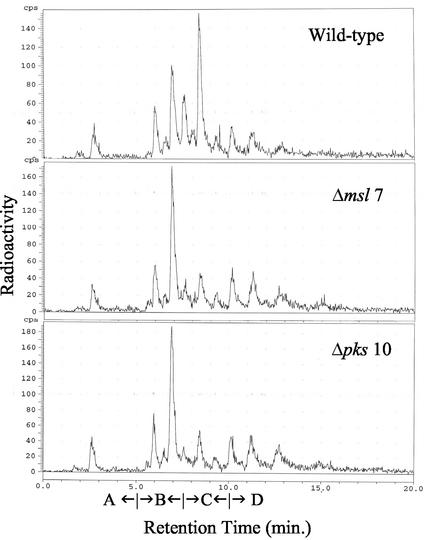
Radio-GC analysis of the methyl esters of fatty acids from the total lipids showed that the two mutants generated all the classes of fatty acids that were produced by the wild type. However, labeling of mycocerosic acids was much less in the mutants (Fig. 5). Argentation-TLC of fatty acids (as methyl esters) from the total lipids and radio-GC analysis of each fraction showed that these muta nts generated all the classes of methyl-branched fatty acids except that the labeling of the mycocerosic acid-containing fraction was much less in these mutants compared to the wild type (data not shown). That the mutants had the ability to synthesize the acyl portion of DIM was clearly indicated by radio-GC analysis of the fatty acids contained in the small amounts of labeled lipids recovered from the apolar lipids that migrated near DIM. Labeled mycocerosic acids were detected. Furthermore, radio-GC of the methyl esters from the sulfolipids in all these mutants also showed detectable levels of mycocerosic acids. Thus, these gene-disrupted mutants were found to be capable of generating mycocerosic acids. Consistent with this observation was our finding that sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) revealed the presence of mycocerosic acid synthase, and immunoblot analysis showed cross-reactivity only with this band (data not shown). However, these mutants, which did not show a deficiency in mycocerosic acid synthesis, could not produce DIM. There was also no DIM in the medium.
FIG. 5.
Radio-GC analysis of total fatty acid methyl esters derived from sodium [1-14C]propionate in M. tuberculosis H37Rv (wild type), the msl7 mutant, and the pks10 mutant. Retention time ranges for branched short-chain fatty acids (A), mycolipanoic and mycolipenic acids (B), mycocerosic acids (C), and phthioceranic acids (D) are indicated.
A detailed analysis of the more polar lipids labeled with sodium [1-14C]propionate showed that the distribution of 14C in lipids other than DIM was not affected in the msl7 and pks10 mutants except for an increase in the amount of 14C found in the sulfolipids (Table 1). The polar lipids that remained at the origin on TLC plate developed in hexane-ethyl ether (9:1, vol/vol) as the solvent system were recovered and separated by TLC with 10% methanol in chloroform as the developing solvent. The distribution pattern of 14C among the polyacyl trehaloses, sulfolipids, and diacyltrehaloses was similar among the wild type and its pks mutants except that there was a 55 to 60% increase in the labeling of sulfolipids in the mutants compared to the wild type (Table 1).
A detailed analysis of the fatty acids present in individual lipid fractions was done by a combination of argentation-TLC and radio-GC analysis of each fraction. The hydroxy fatty acids were also analyzed by radio-GC as their acetylated methyl esters. The high content of 14C found in hydroxyphthioceranic acids explains the increased labeling of sulfolipids in the mutants (Table 1). Radio-GC analysis of each fatty acid methyl ester fraction separated by argentation-TLC showed no significant differences in the fatty acid composition in individual lipids between the wild type and the two mutants. Such detailed analyses revealed identical distribution of 14C among all classes of lipids and fatty acids (data not shown).
Virulence.
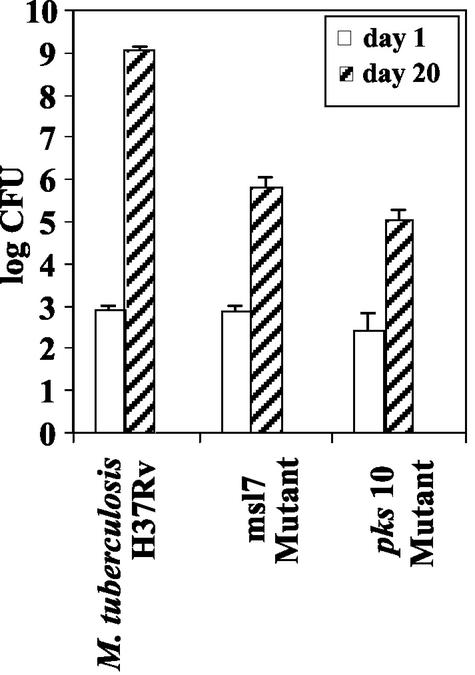
To determine whether the products of the msl7 and pks10 genes affect virulence, we compared the ability of the wild type and the mutants to grow in a murine alveolar macrophage cell line, MH-S. Starting with 2.3 × 104 ± 0.4 × 104 CFU, the wild type reached 4.3 × 105 ± 0.4 × 105 CFU in 5 days, whereas the msl7 mutant, starting with 2.3 × 104 ± 0.4 × 104 CFU, reached 2.6 × 105 ± 0.6 × 105 CFU, and the pks10 mutant, starting with 2.0 × 104 ± 0.6 × 104 CFU, reached 2.3 × 105 ± 0.3 × 105 CFU. Thus, in a 5-day growth period, both mutants showed 50% lower growth. We also tested virulence in mice in vivo after intranasal inoculation. Growth of the pathogen and its gene-disrupted mutants in murine lungs was measured over time (Fig. 6). The msl7 and pks10 mutants showed major attenuation, showing several orders of magnitude less growth compared to the wild type. The levels of growth of the wild type found in these experiments were similar to those published previously (12). At 109 CFU, the animals appeared sick, but there were no fatalities during the 20-day growth period. The growth rates of the mutants in vitro were identical to that of the wild type (data not shown).
FIG. 6.
Growth of intranasally administered M. tuberculosis H37Rv and its msl7 and pks10 gene-disrupted mutants in the lungs of C57BL/6J mice. The experimental details are in the text.
DISCUSSION
The variety of multiple methyl-branched fatty acids found in M. tuberculosis is probably produced by unique synthases encoded by some of the many pks genes found in the mycobacterial genome (15). Since the multifunctional synthases encoded by these genes are large proteins similar in size and properties, isolation and characterization of these individual synthases is not a convenient way to determine their biosynthetic function. Therefore, a genetic approach was taken in which identification of the lipid that is missing in targeted gene disruption mutants was used to elucidate the biosynthetic function of the gene product (1, 2, 6, 11, 22).
In the present case, disruption of two different genes caused abolition of the production of DIM without causing any other qualitative change in lipid synthesis. Detailed analysis of all of the major classes of lipids and detailed analysis of fatty acids in each class derived from sodium [1-14C]propionate showed no alteration except for a substantial increase in labeling of sulfolipids. Since the major metabolic fate of methylmalonyl-coenzyme A is incorporation into mycocerosic acids (in DIM) and phthioceranic and hydroxyphthioceranic acids (in sulfolipids), blockage of DIM synthesis causes channeling of methyl-malonyl-coenzyme A into sulfolipids. The most dramatic consequence of the absence of a functional msl7 and pks10 is the abolition of DIM synthesis.
One of the most unusual lipids in the cell walls of pathogenic mycobacteria is DIM, which is composed of a long-chain diol esterified to multiple methyl-branched fatty acids called mycocerosic acids (9, 15, 18). In some species within the tuberculosis complex, such as M. bovis and some strains of M. tuberculosis such as Canetti, mycocerosic acids are esterified to a structurally analogous diol called phenolphthiocerol, in which there is a hydroxyphenyl group at the end of the alkyl chain (Fig. 1). The exact mechanism of biosynthesis of these diols is not known. Based on the structural features of this diol, a plausible biosynthetic pathway has been postulated (15).
Based on the type of the postulated reactions, the domains required for the catalysis of each step and the likely organization of these catalytic domains, and the probable organization of the genes that would encode such proteins, a set of open reading frames probably involved in this process were identified in the genome. Gene disruption provided evidence for the involvement of this cluster of open reading frames in the biosynthesis of these diols (2). However, this cluster is probably involved in synthesis of the diols from already elongated precursors (Fig. 7). The genes that encode the enzymes involved in the synthesis of these precursors remain unknown. Most probably, several pks genes are involved in producing these elongated precursors of the diols. In the present paper, we report that disruption of two pks genes, msl7 and pks10, blocks DIM synthesis, although the mutants can produce mycocerosic acids. Thus, the disruption of these pks genes probably prevents the synthesis of the diol. Since M. tuberculosis H37Rv produces only phthiocerol, not phenolphthiocerol, we cannot determine whether msl7 and pks10 are involved uniquely in phthiocerol synthesis.
FIG. 7.
Major stages involved in the biosynthesis of phenolphthiocerol and phthiocerol. CoA, coenzyme A.
Comparison of the sequence of msl7 in M. bovis BCG with its homologue in M. tuberculosis H37Rv showed the insertion of a nucleotide in the pks15 part of the latter, and this frameshift is thought to inactivate pks1 in this organism (6). However, this shift in the open reading frame creates a GTG start codon that could allow translation of the open reading frame of pks1. Our reverse transcription-PCR results clearly showed that pks1 is transcribed in M. tuberculosis H37Rv, although we do not have direct evidence for the translation of the open reading frame of the pks1 product. Our finding that pks1 disruption prevents the synthesis of DIM implies that the pks1 product is translated in M. tuberculosis H37Rv. In M. bovis BCG, disruption of pks15/1 was reported to cause selective abolition of phenolphthiocerol synthesis without affecting DIM synthesis (6). On this basis, it was postulated that pks15/1 is involved in the elongation of p-hydroxybenzoic acid to the p-hydroxyphenylalkanoic acid precursor required for synthesis of the phenolphthiocerol catalyzed by the products of the pps cluster in M. bovis BCG (Fig. 7). However, our results that disruption of pks1 blocks DIM synthesis in M. tuberculosis H37Rv, which does not produce phenolphthiocerol, are not consistent with the postulated role of pks15/1 uniquely in the synthesis of phenolphthiocerol; msl7 is probably involved in generating the precursor for phthiocerol synthesis in M. tuberculosis H37Rv.
Among the large number of pks genes present in the M. tuberculosis genome, we identified seven genes homologous to mas (22). The enzymes encoded by these mas-like genes probably catalyze the synthesis of the different classes of multiple methyl-branched fatty acids found in M. tuberculosis. The two gene-disrupted mutants presented here showed attenuation in the murine model, and they were both defective in the synthesis of DIM. Our results support previous observations that suggested that DIM is involved in virulence (5, 8).
pks10 of M. tuberculosis would encode a chalcone synthase-like protein found only in plants until recently. In recent years, more than eight bacterial homologues have been detected (7, 13, 19), and pks10 is one of them. This family of proteins catalyze the synthesis of carbon chains from coenzyme A esters without the involvement of acyl carrier protein. Although in plants such proteins use phenylpropanoyl-coenzyme A as the starting material to produce flavanoids, the recently discovered homologues, particularly the bacterial chs products, show the use of an expanding array of starter units. However, the nature of the substrates used by pks10 in M. tuberculosis H37Rv is not known. Our finding that disruption of this gene causes DIM deficiency implies that it is involved in the biosynthesis of this diol.
It is possible that p-hydroxybenzoyl-coenzyme A elongation, which is involved in the biosynthesis of phenolphthiocerol, is catalyzed by this gene product. However, DIM synthesis would require only aliphatic starter molecules, and therefore how the pks10 product may be involved in generating such starters is not clear. If the pks10 product catalyzes the elongation of hydroxybenzoyl-coenzyme A as well as elongation of the aliphatic precursor involved in the synthesis of DIM, its disruption could abolish DIM synthesis as well as phenolphthiocerol synthesis. However, pks10 has not been disrupted in an organism that produces both. If the many enzymes involved in the production of DIM and phenolphthiocerol function as a multienzyme complex that would accept precursors of phthiocerol and phenolphthiocerol, the absence of any one of the component enzymes, such as the pks10 product, may block the function of the complex and thus cause DIM deficiency. An indirect polar effect of the pks10 mutation on another pks such as pks7, which is 83 bp away, is highly unlikely, since reverse transcription-PCR analysis showed that the pks10 mutation did not affect the pks7 transcript level (data not shown).
In view of the increasing importance of finding new targets for antimycobacterial drugs, genes that are expressed during growth of the pathogen in the host would be appropriate candidates as new drug targets. By selective capture of transcribed sequences, pks10 has been found to be expressed by M. avium 48 h after infection of human macrophages (14). pks10 has also been found to be upregulated in M. tuberculosis during nutrient starvation, which mimics some of the features of persistence (3). Whether pks15/1 and pks10 are expressed in M. tuberculosis in the human host is not known. However, there is mounting evidence that DIM is a virulence factor, and our finding that disruption of pks1 and pks10 causes DIM deficiency suggests that these genes might be suitable targets for new antimycobacterial drugs. Since DIM synthesis involves many enzymes, it may be possible to find many different drugs that can intervene in the production of DIM, making it an attractive target for investigation.
Acknowledgments
This work was supported by grants AI46582-03 and AI35272-10 from the National Institutes of Health.
We thank A. K. Thirumala for valuable technical assistance.
REFERENCES
- 1.Azad, A. K., T. D. Sirakova, L. M. Rogers, and P. E. Kolattukudy. 1996. Targeted replacement of the mycocerosic acid synthase gene in Mycobacterium bovis BCG produces a mutant that lacks mycosides. Proc. Natl. Acad. Sci. USA 93:4787-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azad, A. K., T. D. Sirakova, N. D. Fernandes, and P. E. Kolattukudy. 1997. Gene knock-out reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272:16741-16745. [DOI] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, L. R., D. Enserqueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 6.Constant, P., E. Perez, W. Malaga, M.-A. Laneelle, O. Saurel, M. Daffe, and C. Guilhot. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the M. tuberculosis complex: evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbour a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 277:38148-38158. [DOI] [PubMed] [Google Scholar]
- 7.Cortes, J., J. Velasco, G. Foster, A. P. Blackaby, B. A. Rudd, and B. Wilkinson. 2002. Identification and cloning of a type III polyketide synthase required for diffusible pigment biosynthesis in Saccharopolyspora erythraea. Mol. Microbiol. 44:1213-1224. [DOI] [PubMed] [Google Scholar]
- 8.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 9.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 10.Derbyshire, K. M., and S. Bardarov. 2000. DNA transfer in mycobacteria: conjugation and transduction, p. 93-107. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 11.Dubey, V. S., T. D. Sirakova, and P. E. Kolattukudy. 2002. Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol. Microbiol. 45:1451-1459. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, K. M., M. H. Cynamon, R. K. Voladri, C. C. Hager, M. S. DeStefano, K. T. Tham, D. L. Lakey, M. R. Bochan, and D. S. Kernodle. 2001. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care 164:2213-2219. [DOI] [PubMed] [Google Scholar]
- 13.Funa, N., Y. Ohnishi, Y. Ebizuka, and S. Horinouchi. 2002. Properties and substrate specificity of RppA, a chalcone synthase-related polyketide synthase in Streptomyces griseus. J. Biol. Chem. 277:4628-4635. [DOI] [PubMed] [Google Scholar]
- 14.Hou, J. Y., J. E. Graham, and J. E. Clark-Curtiss. 2002. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences. Infect. Immun. 70:3714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolattukudy, P. E., N. D. Fernandes, A. K. Azad, A. M. Fitzmaurice, and T. D. Sirakova. 1997. Biochemistry and molecular genetics of cell wall lipid biosynthesis in mycobacteria. Mol. Microbiol. 24:263-270. [DOI] [PubMed] [Google Scholar]
- 16.Liu, J., C. E. Barry, and H. Nikaido. 1999. Cell wall: physical structure and permeability, p. 220-239. In C. Ratledge and J. Dale (ed.), Mycobacteria: molecular biology and virulence. Blackwell Science, Malden, Mass.
- 17.Mathur, M., and P. E. Kolattukudy. 1992. Molecular cloning and sequencing of the gene for mycocerosic acid synthase, a novel fatty acid elongating multifunctional enzyme, from Mycobacterium tuberculosis var. bovis bacillus Calmette-Guerin. J. Biol. Chem. 267:19388-19395. [PubMed] [Google Scholar]
- 18.Minnikin, D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, p. 95-184. In C. Ratledge and J. Stanford (ed.), The biology of the mycobacteria. Academic Press, London, UK.
- 19.Moore, B. S., and J. N. Hopke. 2001. Discovery of a new bacterial polyketide biosynthetic pathway. Chem. Biochem. 2:35-38. [DOI] [PubMed] [Google Scholar]
- 20.Rainwater, D. L., and P. E. Kolattukudy. 1985. Fatty acid biosynthesis in Mycobacterium tuberculosis var. bovis bacillus Calmette-Guerin. Purification and characterization of a novel fatty acid synthase, mycocerosic acid synthase, which elongates n-fatty acyl-coenzyme A with methylmalonyl-coenzyme A. J. Biol. Chem. 260:616-623. [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Sirakova, T. D., A. K. Thirumala, V. S. Dubey, H. Sprecher, and P. E. Kolattukudy. 2001. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276:16833-16839. [DOI] [PubMed] [Google Scholar]