Abstract
We characterized a Staphylococcus aureus norA gene expression regulator, NorR, initially identified from its binding to the norA promoter. The norR gene was 444 bp in length, located ∼7 kb upstream from the norA gene, and encoded a predicted 17.6-kDa protein. Overexpression of norR in wild-type S. aureus strain ISP794 led to a fourfold decrease in sensitivity to quinolones and ethidium bromide and an increase in the level of norA transcripts, suggesting that NorR acts as a positive regulator of norA expression. Overexpression of norR in sarA and agr mutants did not alter quinolone sensitivity or levels of norA transcription, indicating that the presence of these two global regulatory systems is necessary for NorR to affect the expression of norA. Insertion and disruption of norR in ISP794 increased resistance to quinolones by 4- to 16-fold but had no effect on norA transcription, suggesting that NorR acts as a repressor for another unidentified efflux pump or pumps. These mutants also exhibited an exaggerated clumping phenotype in liquid media, which was complemented fully by a plasmid-encoded norR gene. Collectively, these results indicate that NorR is a multifunctional regulator, affecting cell surface properties as well as the expression of NorA and likely other multidrug resistance efflux pumps.
Staphylococcus aureus is a leading cause of human infectious diseases worldwide, ranging from superficial skin lesions to systemic and life-threatening infections, such as osteomyelitis, endocarditis, pneumonia, and septicemia. The virulence of S. aureus has been associated with the production of a large number of extracellular toxins, enzymes, and cell-surface-associated proteins, encoded by diverse genes, the expression of which is controlled by the accessory gene regulator locus (agr) (35, 50). The agr regulatory effector is a 510-base RNA molecule (RNAIII) (42). The agr locus regulates most target genes at the level of transcription, but was also shown to affect translation of some genes (35, 42). sarA is the second known global regulatory locus that was also involved in the regulation of virulence factors in S. aureus (4, 34). SarA activates α-toxin gene transcription but represses transcription of genes for serine protease and protein A (7, 9, 10). SarA acts partly through the agr regulatory pathway by binding to agr promoters and stimulating the transcription of agr (8, 11, 45). Since agr and sarA loci play such important roles in diverse gene regulation, they may also participate in regulation of efflux pump expression, which causes resistance to multiple antibiotics, including fluoroquinolones in S. aureus.
Fluoroquinolones are synthetic antimicrobial agents and have been used for treatment of a broad range of bacterial infections (22). Increases in resistance to diverse antibiotics, including fluoroquinolones, have limited the choice of antimicrobial agents in some clinical settings. The genetics and mechanisms of bacterial resistance to fluoroquinolones have been studied extensively. Fluoroquinolones act on DNA gyrase and topoisomerase IV to inhibit bacterial DNA replication (12). Mutations in gyrA and gyrB encoding the subunits of DNA gyrase (21, 23, 26) and grlA and grlB encoding the subunits of DNA topoisomerase IV of S. aureus (13, 38) lead to quinolone resistance in gram-positive and gram-negative bacteria. One of the most intriguing mechanisms underlying resistance to fluoroquinolones as well as a range of other antimicrobial agents involves the extrusion of a variety of structurally unrelated compounds due to active efflux by membrane pumps (19, 51). On the basis of bioenergetic and structural criteria, the multidrug transporters have been divided into five major families (5): the ATP-binding cassette family (ABC), the major facilitator superfamily (MFS), the multidrug and toxic compound extrusion transporters (MATE), the drug/metabolite transporters (DMT), and the resistance/nodulation/division transporters (RND) (31, 44). The S. aureus NorA protein belongs to the MFS group frequently found in bacteria.
Overexpression of the NorA multidrug resistance (MDR) efflux pump causes resistance to some quinolones (25, 27, 37, 39, 53). It is characterized by the presence of 12 transmembrane segments (37, 39) and is related to Bmr, an efflux pump of Bacillus subtilis (1, 36, 55). NorA protects the cell from a number of lipophilic and monocationic compounds, such as ethidium bromide, cetrimide, benzalkonium chloride, tetraphenylphosphonium bromide, and acriflavine, as well as hydrophilic quinolones (25, 27). While the physiological function of NorA as a self-sufficient multidrug transporter was demonstrated with cytoplasmic membrane vesicles and reconstituted proteoliposomes (54), the regulation of the expression of the NorA efflux pump is still not well understood.
Previous studies demonstrated that the expression of norA is increased by mutations in the promoter region that increase mRNA stability, reported in the case of the chromosomal quinolone resistance locus, flqB, which is linked to fus and the transposon insertion Ω1108 on the SmaI D fragment (17), and by mutation in the arlS gene that alters the two-component regulatory system arlRS, reported in the case of the insertion-disruption of arlS by Tn917LTV1 in the S. aureus chromosome, but it is not known if the effects of arlRS on norA expression are direct or indirect (15, 16). In order to identify direct regulatory elements involved in norA expression, we have purified the 17.6-kDa protein that binds to the norA promoter, identified its gene, and characterized the effects of its disruption and overexpression. Our data show that this putative regulatory protein, which we have named “NorR,” functions as an activator of norA expression but is also multifunctional, affecting cell surface properties and acting as a negative regulator for expression of other effectors of the MDR phenotype that likely represent other as-yet-uncharacterized MDR efflux pumps.
(This work was presented in part at the 101st General Meeting of the American Society of Microbiology, Orlando, Fla., 20 to 24 May 2001.)
MATERIALS AND METHODS
Bacterial strains, plasmids, growth media, and other materials.
The bacterial strains and plasmids used in this study are listed in Table 1. Staphylococci were cultivated in brain heart infusion broth (BHI) (Difco, Sparks, Md.) at 37°C unless otherwise stated. Escherichia coli cells were grown in Luria-Bertani (LB) medium. Lysostaphin was obtained from AMBI Products, Inc., New York, N.Y. Ciprofloxacin and moxifloxacin were obtained from Bayer Corp., Westhaven, Conn. Sparfloxacin was obtained from Parke-Davis Pharmaceutical Research Division, Ann Arbor, Mich. 2′-(4-Ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5′-bi-1H-benzimidazole (Hoechst 33342), nalidixic acid, norfloxacin, ethidium bromide, cetrimide, tetracycline, erythromycin, reserpine, and chloramphenicol were obtained from Sigma Chemical Co., St. Louis, Mo. All primers used in this study were synthesized at the Tufts University Core Facility, Boston, Mass., and are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotypes or relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | 8325-4 r− | 30 |
| ISP794 | 8325 pig-131 | 49 |
| MT23142 | 8325 pig-131 flqB | 39 |
| MT1222 | 8325 pig-131 grlA flqB gyrA | 39 |
| BF16 | 8325 pig-131 arlS::Tn917LTV1 | 15 |
| QT1 | ISP794 norR::cat | This study |
| QT2 | ISP794 norR::cat | This study |
| KL820 | RN4220 norA::cat | 25 |
| RN6390 | 8325-4 Hla+ Prt+ | 41 |
| RN6911 | RN6390 agr::tetM | 42 |
| ALC136 | RN6390 sar::Tn917LTV1 | 10 |
| ALC135 | RN6390 agr::tetM sar::Tn917LTV1 | 6 |
| SH1000 | Functional rsbU derivative of 8325-4 rsbU+ | 24 |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 phoA hsdR17(rk− mk−) supE44 λ−thi-1 gyrA96 relA1 | Gibco-BRL |
| BL21(DE3) | E. coli B F−dcm ompT hsdS(rB− mB−) galλ(DE3) | Stratagene |
| Plasmids | ||
| pGEM3-zf(+) | 2.9-kb E. coli cloning vector, Apr | Promega |
| pCL52.2 | Temperature-sensitive E. coli- S. aureus shuttle vector | 47 |
| pL150 | Shuttle cloning vector (Apr Cmr) | 47 |
| pSK950 | 10.5-kb plasmid carrying the attP site of phage L54a, replicon of pE194; Tcr Emr (S. aureus) | 40 |
| pTrcHisA | Cloning and His-tag-expressing vector in E. coli | Invitrogen |
| pQT1 | pGEM3-zf(+)-norR | This study |
| pQT2 | pGEM3-zf(+)-norR::cat | This study |
| pQT3 | pCL52.2-norR::cat | This study |
| pQT4 | pSK950-norR | This study |
| pQT5 | pTrcHisA-norR | This study |
TABLE 2.
Primers used for this study
| Primers | Primer sequencea | Comments |
|---|---|---|
| NorRBa | 5′-AAACGGATCCTATGTCTGGAGG-3′ | Upstream of norR, BamHI site added |
| NorRPs | 5′-TATGCTGCAGCGAACATTTATGA-3′ | Downstream of norR, PstI site added |
| NorREc | 5′-AAACTATCAAGAATTCCTCGCTA-3′ | Downstream of norR, EcoRI site added |
| catpvu1 | 5′-GATCCAGCTGAAGCACCCATTAGT-3′ | Upstream of cat gene in plasmid pL150, PvuII site added |
| catpvu2 | 5′-GATCCAGCTGAGTGACATTAGAAA-3′ | Downstream of cat gene, PvuII site added |
| norA1 | 5′-TGCAATTTCATATGATCAATCCC-3′ | Upstream of norA (−35), amplifies a 150-bp norA promoter |
| norA2 | 5′-AGATTGCAATTCATGCTAAATATT-3′ | Downstream of norA (−10), amplifies a 150-bp norA promoter |
| norR1 | 5′-AAAATGATAACATATATATATTAA-3′ | Upstream of norR putative promoter |
| norR2 | 5′-CGTTTTTTTCTCTTTTCGGATTGGT-3′ | Downstream of norR putative promoter, before the putative Shine-Dalgarno region |
Endonuclease restriction sites added to the primer sequences are underlined.
MIC determinations.
MICs of quinolones, ethidium bromide, cetrimide, and Hoechst 33342 were determined out by serial agar dilution on Trypticase soy agar (TSA). All plates were incubated at 30 or 37°C for 24 h before being read. Determinations of MICs of quinolones and other chemical compounds for transformants containing pSK950 and pQT4 were done on TSA containing 5 μg of erythromycin per ml to ensure maintenance of the plasmid, with incubation at 30°C. The effect of reserpine on quinolone susceptibility was determined by broth (BHI) dilution in the presence and absence of 10 and 20 μg of reserpine per ml.
DNA isolation.
Plasmid DNA isolation was performed with the Qiagen midiprep kit (Qiagen, Inc., Valencia, Calif.) as recommended by the manufacturer. S. aureus was transformed with plasmid DNA by electroporation, as previously described (39). Chromosomal DNA from S. aureus was prepared with the Easy DNA kit (Invitrogen Life Technologies, Carlsbad, Calif.) as recommended by the manufacturer.
Southern hybridization.
Restriction endonuclease-digested staphylococcal chromosomal DNA was resolved by electrophoresis at 100 V in 0.9% agarose for 8 h. The DNA was transferred to Hybond-N+ nylon membrane by alkaline blotting (Amersham, Pharmacia Biotech, Little Chalfont, United Kingdom). Target genes were detected by hybridization with a gel-purified DNA probe that was nonradioactively labeled with the ECL (enhanced chemiluminescence) direct nucleic acid labeling kit (Amersham, Pharmacia Biotech, United Kingdom).
Cloning and overexpression of norR.
To clone the norR gene, primers based on flanking sequences (NCTC8325; Oklahoma University) (B. A. Roe, Y. R. Tian, H. Jia, S. Li, S. Lin, S. Kenton, H. Lai, J. D. White, A. Dorman, F. Z. Najar, S. Clifton, V. Worrell, and J. Iandolo, Staphylococcus aureus Genome Sequencing Project, 2002) were synthesized by the Tufts University Core Facility (Boston, Mass.). A 1.3-kb fragment was amplified by PCR from S. aureus ISP794 chromosomal DNA with primers norRBa and norRPs, which generated flanking BamHI and PstI sites, respectively (Table 2). The amplified norR gene was digested with PstI and BamHI and then ligated into the PstI and BamHI site of the plasmid pGEM3-zf(+) to yield pQT1 and introduced into E. coli DH5α. Plasmids extracted from ampicillin-resistant colonies were screened for the norR fragment insertion by restriction endonuclease digest patterns and confirmed by DNA sequencing.
To generate a plasmid for expression of norR in S. aureus, the norR gene was subcloned in E. coli into the temperature-sensitive shuttle plasmid pSK950 to yield pQT4. This plasmid was then electroporated into S. aureus RN4220 (8325 r−) to generate transformants, and the structure of pQT4 in S. aureus was confirmed by restriction mapping. Electrocompetent strain ISP794 and other strains were transformed with this plasmid isolated from RN4220. Tetracycline- and erythromycin-resistant colonies isolated at 30°C were confirmed to have intact pQT4 by restriction mapping.
Construction of a norR mutant by allelic exchange.
To generate a norR mutant, the 800-bp DNA fragment containing the cat gene was amplified from plasmid pLI50 with primers catpvu1 and catpvu2. The PCR product was digested with PvuII and then ligated into an AccI site, previously blunted with Klenow fragment enzyme and deoxynucleoside triphosphates (dNTPs), within the putative norR coding region of plasmid pQT1. The resultant plasmid, containing the 2.1-kb norR::cat fragment, termed “pQT2,” was confirmed by restriction mapping and sequencing. The 2.1-kb norR::cat fragment was subcloned into the temperature-sensitive shuttle plasmid pCL52.2 to yield pQT3. This plasmid was then introduced into RN4220 by electroporation to generate chloramphenicol- and tetracycline-resistant transformants. Putative transformants were confirmed by restriction mapping and DNA sequencing. Electrocompetent ISP794 was subsequently transformed with pQT3 isolated from RN4220. Colonies grown at 30°C that were resistant to chloramphenicol and tetracycline were selected for the allelic exchange after screening. ISP794 harboring pQT3 was grown in BHI broth with tetracycline (3 μg/ml) at 30°C, diluted 1:1,000 in fresh medium, and propagated at 42°C for 24 h. The culture was grown again at 30°C without selection for 48 h. Chloramphenicol-resistant, tetracycline-sensitive colonies, representing possible double-crossover events, were screened for and tested for cat insertion into norR by Southern hybridization, PCR, and sequencing of the PCR fragment containing the junctional fragment.
DNA mobility shift analysis.
To perform the gel shift assay, a pair of primers based on the norA DNA sequence were synthesized (norA1 and norA2) (Table 2) and used to amplify a fragment from the Shine-Dalgarno sequence extending 150-bp upstream and containing the entire norA promoter region. One of the primers was biotinylated by Gibco BRL (Rockville, Md.). The gel mobility shift assay was carried out using the LightShift Chemiluminescent EMSA (elecrophoretic mobility shift assay) kit (Pierce, Rockford, Ill.), as recommended by the manufacturer. The biotin-labeled DNA was incubated with the indicated amount of purified proteins from S. aureus ISP794 in 20 μl of binding buffer (10 mM HEPES [pH 8], 60 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 0.1 mg of bovine serum albumin per ml, 0.25 mM dithiothreitol) containing 1 μg of poly(dI-dC), 200 ng of sheared herring sperm DNA, and 10% glycerol. The reaction mixture was incubated for 20 min at room temperature and analyzed by 5% nondenaturing polyacrylamide electrophoresis.
Identification of the NorR protein from cell extracts.
Cell extracts collected from 5 liters of S. aureus cells at late exponential phase (optical density at 600 nm [OD600] of 0.9) were used to purify the NorR protein as previously described (14). The 150-bp biotinylated DNA fragment described above was immobilized on magnetic beads with covalently coupled streptavidin (Dynabeads M-280; Dynal) according to the manufacturer's protocol. DNA bound to beads was incubated with protein extract in binding buffer (10 mM HEPES [pH 8], 60 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 0.1 mg of bovine serum albumin per ml, 0.25 mM dithiothreitol) containing herring sperm DNA (200 ng) for 20 min at room temperature. Beads were washed twice with binding buffer containing herring DNA and twice with binding buffer without DNA. Proteins were then eluted in binding buffer containing 0.5 M NaCl. Eluted proteins were dialyzed against water, concentrated, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The 18-kDa protein was blotted onto polyvinylidene difluoride (PVDF) membrane for N-terminal amino acid sequencing by the Edman degradation method (Dana Farber Research Institute Core Facility, Boston, Mass.).
Purification of NorR protein.
The norR gene was amplified by PCR from S. aureus ISP794 chromosomal DNA with primers norRBa and norREc, which generated flanking BamHI and EcoRI sites, respectively (Table 2). After digestion with EcoRI and BamHI, norR was ligated into the EcoRI and BamHI site of the plasmid pTrcHisA (Invitrogen, Carlsbad, Calif.) to yield pQT5 and then introduced into E. coli BL21(DE3). The purification of the histidine-tagged NorR was carried out as recommended by the manufacturer. E. coli BL21(DE3) cells harboring pQT5 were grown to mid-log phase in LB medium, at which time, isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added to the culture. After 3 h, the cells were harvested by centrifugation and then resuspended in 20 mM sodium phosphate buffer (pH 7.4). The cells were lysed with lysozyme (0.02%) and then centrifuged (100,000 × g) for 90 min. The supernatant was applied to the nickel affinity column (iminodiacetic acid [IDA]-Sepharose-Ni; Amersham Pharmacia Biotech, Uppsala, Sweden) and then washed with start buffer supplemented with concentrations of imidazole increasing from 10 to 60 mM. NorR protein was eluted with 300 mM imidazole. The homogeneity of the eluted protein was verified by SDS-PAGE.
RNA analysis.
Total S. aureus RNA was prepared by extraction from lysostaphin-treated cells grown to the postexponential phase at 37 or 30°C, by using the RNeasy mini kit (Qiagen, Valencia, Calif.). The concentration of RNA was determined spectrophotometrically as A260. For Northern blot analysis, 10 μg of total RNA was electrophoresed through a 0.9% agarose-0.66 M formaldehyde gel in morpholinepropanesulfonic acid (MOPS) and blotted onto Hybond-N+ membranes as previously described (39). DNA probes were amplified from the ISP794 chromosome and labeled with psoralen for the detection of specific transcripts (norR and norA) by using the Northern Max kit (Ambion, Inc., Austin, Tex.) as recommended by the manufacturer. Blots were hybridized with probes overnight at 42°C, washed, and autoradiographed with Kodak X-Omat film.
RESULTS
Identification of NorR from its binding to the norA promoter.
In searching for regulatory elements that directly control the expression of the norA structural gene, we had previously identified in cell extracts an ∼18-kDa protein that binds to the norA promoter, producing specific band shifts of a 150-bp DNA fragment containing the entire norA promoter region (14).
In order to identify this protein, we first isolated the protein from cell extracts of the wild-type strain ISP794 by using magnetic beads coupled to the 150-bp DNA fragment as an affinity reagent (14). The eluted proteins were separated by SDS-PAGE then transferred to PVDF membrane for N-terminal amino acid sequencing. The first 14 amino acids of the N terminus (XDQHNLXEQLCFSL) were then used to search a data bank of the S. aureus genome NCTC 8325 (Roe et al., Staphylococcus aureus Genome Sequencing Project, 2002). The database contained a putative protein of 147 amino acids and a predicted molecular mass of 17.6 kDa that contained an identical N-terminal amino acid sequence. Analysis of this amino acid sequence allowed the identification of a 444-bp open reading frame (ORF) on the S. aureus genome (Fig. 1).
FIG. 1.
Nucleotide sequence of 851 bp of S. aureus DNA containing the norR gene from ISP794 (complete sequence shown). The putative promoter regions, the Shine-Dalgarno sequence, and a putative transcription terminator are underlined. The coding region of norR is marked by the ATG start and TAA stop codons.
NorR purification and promoter binding.
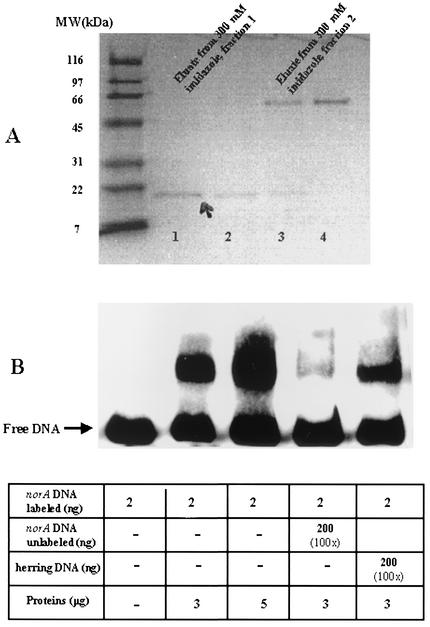
The putative gene was then expressed by cloning the 444-bp ORF coding region into pTrcHisA, a His-tag expression vector (Invitrogen). After induction with IPTG and purification on a nickel affinity column, we isolated a protein of ∼21 kDa (size includes the NorR protein [17.6 kDa] plus the His tag and the anti-Xpress antibody epitope). The SDS-PAGE gel shows one protein band at ∼21 kDa in the first eluted fraction and an additional band at ∼55 to 60 kDa in the second elution fraction (Fig. 2A). No other protein species were identified by silver staining. We speculate that the larger band is a multimeric form of NorR because it as well as the 21-kDa band is immunoreactive with antibody to the anti-Xpress antibody epitope encoded by pTrcHisA (Invitrogen) (data not shown). The purified 21-kDa NorR from the first eluted fraction (fraction 1) when incubated with the 150-bp norA promoter fragment showed a clear shift in the DNA banding pattern (Fig. 2B). With increasing concentrations of proteins, the intensity of the shifted band increased, and band shifts were reduced in the presence of 100-fold excess unlabeled 150-bp norA DNA but remained unchanged in the presence of excess herring sperm DNA, indicating specific binding to the norA promoter fragment. We named this protein “NorR” because its amino acid sequence showed homology with SarR (35% amino acid identity), a protein of the SarA family, and with the MarR (40% identity) protein of the MarR family. The aligned amino acid identity between NorR and QacR and BmrR was less than 10% for both proteins. The NorR protein and its DNA sequence are 100% conserved in the genomes of the sequenced S. aureus strains Mu50, N315, COL, and MW2.
FIG. 2.
(A) SDS-PAGE analysis of NorR-His purification by Ni affinity chromatography. NorR protein in the crude cell extracts was adsorbed to the Ni column. The column was washed with buffers containing 10 and 60 mM imidazole, and the purified protein was eluted with buffer containing 300 mM imidazole. The gel was stained with Coomassie blue followed by silver staining. MW, molecular mass in kilodaltons. (B) Gel mobility shift analysis of the interaction of purified by NorR-His protein with the biotinylated 150-bp norA promoter fragment. Competing unlabeled norA promoter DNA and herring sperm DNA were used to determine the specificity of promoter binding. Protein and DNA concentrations and ratios of unlabeled to labeled DNA used in this assay are indicated in the table below the gel.
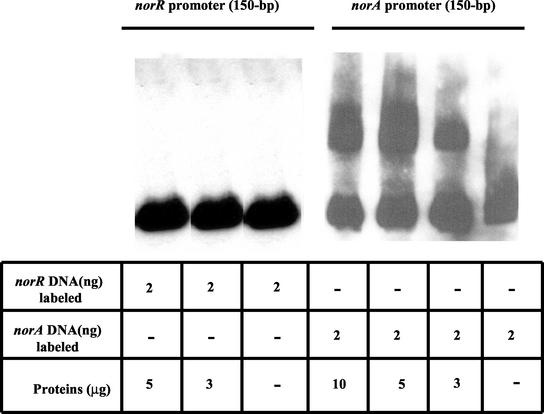
To determine whether NorR is also involved in its own regulation via direct binding, we amplified a 150-bp DNA fragment from the region upstream of the norR ATG start codon that contains two putative promoters and a putative Shine-Dalgarno region (primers are listed in Table 2). No band shift was found upon incubation of NorR with this DNA fragment (Fig. 3).
FIG. 3.
Gel mobility shift analysis of the interaction of purified NorR-His protein with the biotinylated norA and norR promoter fragments. The protein and DNA concentrations used in this assay are indicated in the table below the gel.
norR mRNA levels in wild-type and mutant strains.
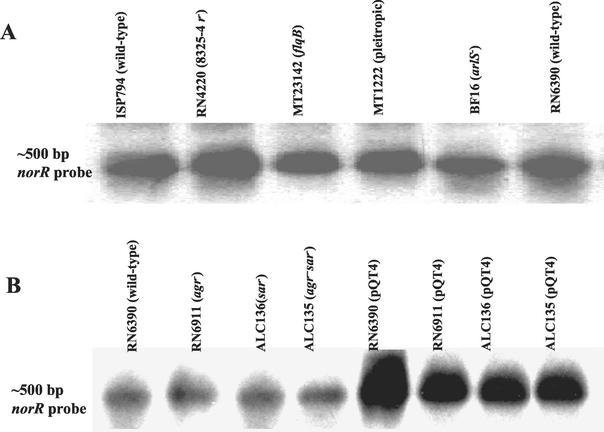
Northern blots of ISP794 (wild type), RN4220 (8325-4 r−), MT23142 (flqB), MT1222 (pleiotropic), BF16 (arlS), and RN6390 (wild type) were probed with a biotin-labeled norR fragment to ascertain norR expression in these backgrounds. A norR-hybridizing transcript was found to be ∼500 nucleotides in length by Northern analysis. Because a putative transcriptional termination signal was found 83 bp downstream from the stop codon, this norR transcript is probably monocistronic. With the same amount of total RNA, all mutant strains showed a slight decrease of twofold in the levels of norR transcription to that of ISP794, RN6390, and RN4220 at the postexponential growth phase (Fig. 4A). The decrease in norR transcripts of these mutants suggests an additional regulatory mechanism to regulate the level of MDR efflux pump possibly by NorA-mediated efflux of an effector required for norR expression.
FIG. 4.
(A) Northern hybridization of RNA extracted from S. aureus strains in the late exponential phase of growth (OD600 = 0.9) and probed with the norR gene, to observe the transcription level of norR in wild-type and mutant strains. (B) norR transcripts in agr sarA mutants with and without pQT4 (norR overexpressed).
Effects of norR overexpression. (i) Resistance to quinolones and other compounds.
To address the effect of norR overexpression in vivo, we transformed a series of wild-type and mutant strains with plasmid pSK950 into which we had cloned the norR gene (pQT4), and then we determined the MICs of quinolones and ethidium bromide for strains with the norR plasmid pQT4. The MIC determinations were carried out at 30°C in the presence of erythromycin (5 μg/ml) to ensure the stability of plasmid pQT4. In order to detect any artifact caused by erythromycin in the MIC determinations, strains with the vector plasmid pSK950 were also included in the experiment and tested in the presence of erythromycin. Strains with and without pSK950 showed no change in susceptibility to the drugs tested. Fourfold increases in the MICs of norfloxacin and ethidium bromide, which are known substrates of NorA, were seen with ISP794(pQT4). We also found a twofold increase in the MIC of ciprofloxacin, but no change in MICs of sparfloxacin, moxifloxacin, and nalidixic acid for ISP794(pQT4). ISP794(pQT4) also showed slight (twofold or less) increases in the MICs of the nonquinolone NorA substrates cetrimide and Hoechst 33342 (Table 3). To evaluate the contribution of norA overexpression to the resistance phenotype when norR was overexpressed, we used a norA knockout strain, KL820, into which we introduced plasmid pQT4. Only a slight increase in the MICs of norfloxacin, ciprofloxacin, and ethidium bromide was found in contrast to the fourfold changes in the ISP794 (norA+) background, indicating that norA overexpression accounts for most of the resistance associated with norR expression from pQT4 (Table 3). Thus, although norR could have additional positive regulatory effects on genes encoding other MDR transporters, its major effect on resistance when overexpressed appears to be mediated through norA. Mutants that overexpress norA by other mechanisms, such as BF16, MT1222, and MT23142, did not demonstrate an additional increase in the MICs of quinolones and dyes when pQT4 was introduced (Table 3).
TABLE 3.
Susceptibilities of strains to quinolones and other agents
| Strain (plasmid)a | Genotype | Presence/ absence of reserpineb | MIC in μg/ml in (factor change in resistance)c
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NOR | CIP | SPAR | MOXI | NAL | CTR | H33342 | EB | |||
| ISP794 | 8325 pig-131 | − | 0.5 (1) | 0.25 (1) | 0.125 (1) | 0.06 (1) | 200 (1) | 0.5 (1) | 2 (1) | 1 (1) |
| ISP794 | + | 0.5 (1) | 0.25 (1) | 0.125 (1) | 0.06 (1) | 1 (1) | ||||
| MT23142 | 8325 pig-131 flqB | − | 8 (16) | 2 (8) | 0.5 (4) | 0.06 (1) | 800 (4) | 8 (16) | 16 (8) | 8 (8) |
| MT23142 | + | 2 (4) | 0.5 (2) | 0.25 (2) | 0.06 (1) | 2 (4) | ||||
| BF16 | 8325 pig-131 arlS::Tn917LTV1 | − | 2 | 1 | 0.25 | 0.06 | 400 | 2 | 2 | 2 |
| BF16 | + | 0.5 (1) | 0.25 (1) | 0.125 (1) | 0.06 (1) | 1 (1) | ||||
| QT1 | 8325 pig-131 norR::cat | − | 4 (8) | 2 (8) | 2 (16) | 0.5 (8) | 400 (2) | 2 (4) | 2 (1) | 4 (4) |
| QT1 | + | 1 (2) | 0.5 (2) | 0.25 (2) | 0.125 (2) | 2 (2) | ||||
| QT2 | 8325 pig-131 norR::cat | − | 4 (8) | 2 (8) | 2 (16) | 0.5 (8) | 400 (2) | 2 (4) | 2 (1) | 4 (4) |
| QT2 | + | 1 (2) | 0.5 (2) | 0.25 (2) | 0.125 (2) | 2 (2) | ||||
| KL820 | RN4220 norA::cat | − | 0.33 (.6) | 0.16 (.6) | 0.06 (.4) | 0.06 (1) | 400 (2) | 2 (4) | 2 (1) | 0.33 (.3) |
| RN6390 | 8325-4 Hla+ Prt+ | − | 0.5 | 0.25 | 0.125 | 0.06 | 200 | 0.5 | 2 | 1 |
| RN6911 | RN6390 agr::tetM | − | 0.5 | 0.25 | 0.125 | 0.06 | 200 | 0.5 | 2 | 1 |
| ALC136 | RN6390 sar::Tn917LTV1 | − | 0.5 | 0.25 | 0.125 | 0.06 | 200 | 0.5 | 2 | 1 |
| ALC135 | RN6390 agr::tetM sar::Tn917LTV1 | − | 0.5 | 0.25 | 0.125 | 0.06 | 200 | 0.5 | 2 | 1 |
| MT1222 | 8325 pig-131 grlA flqB gyrA | − | 256 | 64 | 8 | 8 | 1000 | 32 | 16 | 16 |
| MT1222 | + | 64 | 32 | 4 | 4 | 4 | ||||
| ISP794(pQT4) | − | 2 (4) | 0.5 (2) | 0.125 (1) | 0.06 (1) | 200 (1) | 2 (4) | 3 (1.5) | 4 (4) | |
| MT23142(pQT4) | − | 8 | 2 | 0.5 | 0.06 | 800 | 8 | 64 | 8 | |
| BF16(pQT4) | − | 2 | 1 | 0.25 | 0.06 | 400 | 2 | 4 | 2 | |
| QT1(pQT4) | − | 2 (4) | 0.25 (1) | 0.125 (1) | 0.06 (1) | 200 (1) | 2 (4) | 2 (1) | 4 (4) | |
| QT2(pQT4) | − | 2 | 0.25 | 0.125 | 0.06 | 200 | 2 | 2 | 4 | |
| KL820(pQT4) | − | 0.5 | 0.25 | 0.06 | 0.06 | 400 | 2 | 2 | 0.5 | |
| RN6390(pQT4) | − | 1 | 0.5 | 0.125 | 0.06 | 200 | 0.5 | 2 | 2 | |
| RN6911(pQT4) | − | 0.5 | 0.25 | 0.125 | 0.06 | 200 | 0.5 | 2 | 1 | |
| ALC136(pQT4) | − | 0.5 | 0.25 | 0.125 | 0.06 | 200 | 0.5 | 2 | 1 | |
| ALC135(pQT4) | − | 0.5 | 0.25 | 0.125 | 0.06 | 200 | 0.5 | 2 | 1 | |
| MT1222(pQT4) | − | 256 | 64 | 8 | 8 | 1,000 | 32 | 16 | 16 | |
Strains harboring plasmid pQT4 were grown in the presence of erythromycin (5 μg/ml).
−, reserpine absent; +, reserpine present.
NOR, norfloxacin; CIP, ciprofloxacin; SPAR, sparfloxacin; MOXI, moxifloxacin; NAL, nalidixic acid; CTR, cetrimide; EB, ethidium bromide; H3342, Hoechst 33342.
To assess a possible interaction of norR overexpression with the agr and sarA global regulatory loci, we introduced pQT4 into sarA and agr single- and double-mutant backgrounds. In contrast to the findings with the agr+ sarA+ background, norR expression from pQT4 in the agr and sarA mutants caused no increase in the resistance phenotype (Table 3), suggesting that the resistance phenotype of norR overexpression is dependent on intact agr and sarA loci. Increased norR expression from pQT4 was maintained in these strains (Fig. 4B), and thus the dependence of the norR-overexpression resistance phenotype on intact agr and sarA cannot be explained by a dependence on these loci for norR to be expressed from pQT4.
(ii) Effect on norA transcription.
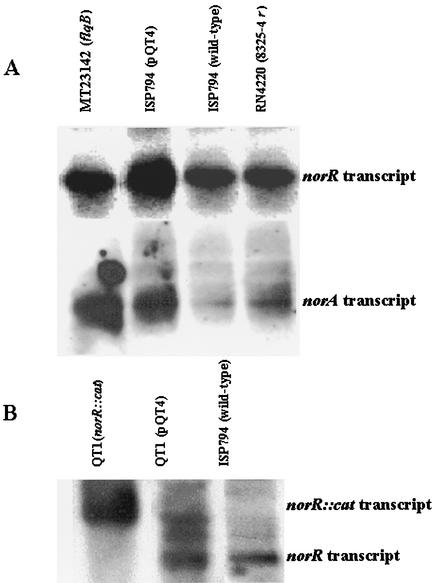
Northern blots of RNA from ISP794 with and without pQT4 and from MT23142 were probed with biotin-labeled norA and also separately with biotin-labeled norR to assess the effect of overexpression of norR on norA expression. RNA levels of norR were documented to be higher in ISP794(pQT4) than in ISP794, as expected (Fig. 5A). Notably, norA expression in ISP794(pQT4) was increased substantially to a level as high as that observed in the mutant MT23142, which carries the flqB promoter mutation responsible for the overexpression of norA in this strain (Fig. 5A). Thus, norR behaves as a positive regulator of norA transcription. Since ISP794 is a σB mutant, we also checked the effect of an overexpression of norR on norA expression by using a σB wild-type strain (SH1000) (24). The results obtained with strain SH1000 were similar to those with ISP794 (data not shown), indicating that these effects are not modulated by σB. In addition, in the agr and sarA mutants, the presence of pQT4 produced increases in norR transcripts (Fig. 4B) but no increase in norA transcripts (data not shown), further supporting the interactions of norR with the agr and sarA global regulatory systems in its effects on norA overexpression.
FIG. 5.
(A) Effects of norR overexpression on norA expression. Northern blots of RNA extracted from S. aureus strains MT23142, ISP794 (with and without pQT4), and RN4220 in the late exponential phase and probed with either the norR or norA gene. (B) norR transcripts in QT1 with and without pQT4. The norR::cat transcript is larger than norR as expected.
Properties of a norR mutant.
In order to assess further the role of NorR as a regulator, we generated a norR mutant. We first transformed the wild-type strain ISP794 with a temperature-sensitive plasmid (pCL52.2) that contained a cat gene cassette within the norR coding region (plasmid pQT3), and then transformants were first selected for resistance to chloramphenicol and tetracycline at permissive temperature (30°C). After successive growth at 42°C with tetracycline and 30°C without tetracycline to enrich for the insertion-excision event of the plasmid pQT3 in the norR gene, we screened for and identified tetracycline-sensitive, chloramphenicol-resistant colonies. All candidate norR::cat mutants were confirmed by PCR, Southern blotting, and DNA sequencing.
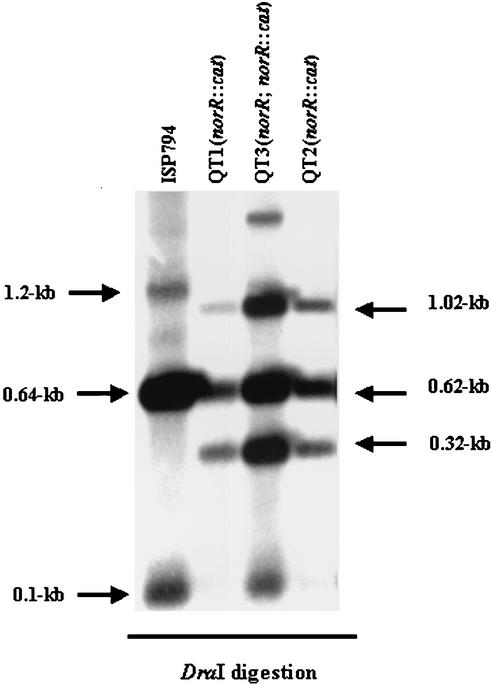
The presence of the cat gene in norR and the correct insertion-excision of the plasmid pQT3 were verified by PCR amplification with primers that flanked the norR gene. The size of the PCR fragment as well as direct sequencing of the PCR fragment confirmed that a double-crossover event had taken place between the plasmid and the chromosome. Southern hybridization of DraI chromosomal digests with fluorescently labeled norR revealed that norR mutants QT1 and QT2 carried a single norR::cat insertion in the chromosome, while QT3 resulted from a single crossover that left two copies of the norR gene (norR and norR::cat) in its chromosome (Fig. 6).
FIG. 6.
Southern hybridization of DraI restriction digests of chromosomal DNA from the S. aureus wild type and mutants. Changes occurred in the band size, and the numbers of the putative insertion mutation relative to the parental type indicate potential insertion of cat into norR. DraI cleaves both norR and cat genes (once inside each gene). The 0.7-kb norR DNA probe hybridizes with 0.64- and 0.1-kb fragments of norR gene in ISP794 DNA, while the same 0.7-kb probe hybridizes with 1.02-, 0.62-, and 0.32-kb fragments of norR::cat in DNA of mutants QT1 and QT2. The 1.2-kb fragment seen in ISP794 DNA came from incomplete enzyme digestion. QT3 resulted from a single crossover between the plasmid and the chromosome, leading to a combined hybridization pattern of wild-type and mutant norR.
The norR mutants QT1 and QT2 exhibited a resistance phenotype, with 16-fold increases in the MICs of sparfloxacin (2 μg/ml); 8-fold increases in the MICs of norfloxacin (4 μg/ml), ciprofloxacin (2 μg/ml), and moxifloxacin (0.5 μg/ml); 4-fold increases in the MICs of cetrimide (2 μg/ml) and ethidium bromide (4 μg/ml); a 2-fold increase in the MIC of nalidixic acid (400 μg/ml); and no change in MIC of Hoechst 33342 (2 μg/ml) (Table 3). The level of resistance of norR mutants to quinolones was similar to those of flqB (MT23142) and arlS (BF16) mutants that overexpress norA (Table 3), except for the MICs of sparfloxacin and moxifloxacin, which showed 16- and eightfold increases in MICs for QT1 and only a 4-fold increase and no increase in the respective MICs for MT23142 (flqB) (Table 3). In the presence of reserpine, the norR mutants QT1 and QT2 both became less resistant to quinolones and ethidium bromide. We observed a fourfold decrease in the MICs of norfloxacin, ciprofloxacin, moxifloxacin; an eightfold decrease in the MIC of sparfloxacin; and a twofold decrease in the MICs of ethidium bromide when these mutants were tested in the presence of reserpine (Table 3). The level of norA expression in Northern blots in QT1 and QT2, however, did not differ from those of wild-type strain ISP794 (data not shown), for which the level of susceptibility to quinolones and ethidium bromide was not affected by reserpine. Thus, these findings further suggest that norR also serves as a negative regulator of other effectors of multidrug resistance, likely representing as-yet-undefined MDR efflux pumps. In order to detect any effect of NorR on the transcription of arlS, we performed Northern hybridization with strains with and without norR. No change in the level of arlS transcripts was found among all strains tested (data not shown).
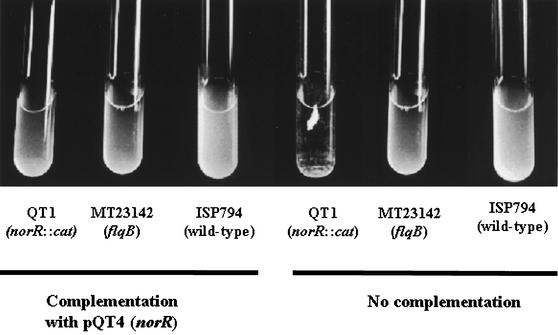
Further supporting the multiple regulatory roles of norR, the mutation in norR mutants (QT1 and QT2) also strikingly modified the growth of these strains in liquid media without a perceptible change in colony morphology on solid media. Figure 7 illustrates the clumping of the mutant in BHI broth after overnight growth.
FIG. 7.
Overnight cultures of wild-type and mutant S. aureus strains, with and without complementation by plasmid pQT4 containing the norR gene.
In order to assess whether the disruption of norR itself or a polar effect of the cat insertion was responsible for the norR::cat mutant phenotype, we transformed the strain with plasmid pQT4. Expression of norR from pQT4 in strain QT1 was documented by Northern hybridization (Fig. 5B). Complementation with plasmid-encoded norR completely reversed the clumping phenotype (Fig. 7) and resulted in a change in a resistance phenotype pattern to one similar to that of ISP794(pQT4) (Table 3). For drugs that are good NorA substrates (e.g., norfloxacin) complementation resulted in little change in resistance, likely due to increased norA expression upon restoration of norR. In contrast, for sparfloxacin and moxifloxacin, which are poor NorA substrates, norR complementation resulted in increases in drug susceptibility. Thus, disruption of norR itself is responsible for both the cell-clumping phenotype and for resistance to sparfloxacin, moxifloxacin, and likely other quinolones and other drugs with increased MICs in QT1.
DISCUSSION
NorR is a DNA-binding protein.
The regulation of expression of norA encoding a major multidrug efflux transporter in S. aureus is little understood. In our effort to identify new elements that regulate norA expression, we identifed NorR, a putative regulatory protein that has homology with SarR and MarR, two known regulators of gene expression. Purified NorR protein binds specifically to the norA promoter region, thus suggesting a direct role for NorR in expression of the norA gene. In contrast, gel mobility shift assays showed no evidence that NorR binds to its own promoter, thus suggesting that the norR locus is not directly autoregulatory.
NorR functions as an activator of norA expression.
To study further the effect of norR on norA expression, we overexpressed the norR gene from a plasmid and observed changes in the level of norA expression in S. aureus strains with different genetic backgrounds. By Northern hybridization, we found an increase in the level of norA transcripts in the wild-type strain, ISP794, harboring norR cloned on a plasmid. This increase in norA transcripts correlated with a fourfold increase in the MICs of norfloxacin and ethidium bromide, suggesting that NorR is a direct transcriptional activator of norA expression. Thus, norR is the first regulatory locus identified to act directly on the norA promoter in S. aureus.
Although there was a slight resistance phenotype in a norA knockout strain in which norR is overexpressed, the major effect of norR overexpression appears to be attributable to overexpression of NorA. This finding (i.e., the level of increased susceptibility) also implies that the additional role of NorR as a repressor of other efflux pumps (discussed below) appears to be maximal at basal levels of expression and is not further augmented by NorR overexpression.
norR mutants and other possible roles of NorR in the bacterial cell.
norR mutants also exhibited a resistance phenotype and a cell-clumping phenotype in liquid media, suggesting that norR has a complex regulatory role involving cell surface properties and likely other MDR pumps. The pleiotropic nature of the norR resistance phenotype and the reduction in resistance in the presence of reserpine argue that resistance is likely due to MDR pumps. Additional studies are ongoing to identify the specific effectors of MDR in norR mutants. The absence of increased levels of norA transcripts and some noteworthy differences in the resistance profile (resistance to sparfloxacin and moxifloxacin) of the norR mutants relative to strains overexpressing norA also argue for norR as a negative regulator of MDR pumps other than NorA that may have these more hydrophobic quinolones as substrates.
For regulatory genes in which overexpression and reduced expression both have similar phenotypes, reversal of mutant phenotypes with plasmid-encoded complementing genes may be difficult because of the precise titration of the complementing gene product needed to restore the wild-type phenotype. In this study, complementation of norR mutant QT1 with pQT4 (norR overexpressed) changed the antibiotic resistance pattern to that of ISP794(pQT4), which overexpresses norA. We propose that the resistance phenotype of norR mutants is in fact due to the disruption of NorR itself for three reasons. First, norR complementation of pQT1 reversed the mutant-clumping phenotype completely. Second, norR has a transcriptional stop site at its terminus, and its transcripts appear to be monocistronic, suggesting that insertion of the cat gene into norR does not disrupt an operon. Furthermore, the putative gene adjacent to norR is located 200 bp downstream from the norR TAA stop codon and appears to have a putative promoter region at 80-bp upstream from its ATG start codon. No change in the levels of transcripts of this ORF was seen in comparing ISP794 and QT1 (data not shown). Third, for two quinolones, sparfloxacin and moxifloxacin, which are poor NorA substrates and thus are little affected by norA overexpression, norR complementation of QT1 was complete. Work is under way to identify and characterize the efflux pump or pumps that we postulate underlie the resistance phenotype of the norR mutant and to define directly the role of norR in their regulation.
Role of NorR in regulatory networks.
NorR differs from SarR and MarR in having direct binding to a target gene promoter rather than acting indirectly on other transcriptional regulators that themselves control target gene expression. SarR and MarR are both autoregulatory, in keeping with their central role in complex regulons (2, 3, 33). Purified NorR, however, appears not to bind upstream of norR and thus is unlikely to be directly autoregulatory. It is as yet unclear what factors regulate norR expression. Other mutants that have been shown to affect norA expression do not appear to affect expression of norR. Thus, the effects of the arlRS two-component regulatory system on norA expression appear not to be mediated through changes in norR expression, suggesting that additional factors mediate increased norA expression in arlRS mutants.
Interestingly, although norR overexpression in a wild-type background causes overexpression of norA and increased resistance to NorA substrates, norR overexpression does not further increase the expression of norA or further increase resistance in mutants that exhibit cis-acting norA overexpression (flqB norA promoter mutant) (17, 39) or trans-acting overexpression (arlRS mutant) of norA (15, 16). Thus, these mutations appear to be epistatic to norR overexpression. Since sustained high levels of expression of some efflux pumps are likely harmful to the cell, it is possible that as-yet-undefined counterregulatory factors come into play when norA expression exceeds certain levels. Additional data obtained from overexpression of norR in agr and sarA mutants further indicate that the effect of norR overexpression on norA expression requires intact agr and sarA. The nature of these requirements and interactions is as yet unclear but could involve modification of NorR protein or a requirement for coordinate binding to the norA promoter by other factors under the control of the agr and sarA global regulators.
MarR in E. coli exhibits indirect effects on the expression of the AcrB MDR efflux pump (18, 32, 43) as well as affecting the expression of outer membrane proteins. Thus, NorR and MarR both mediate multiple functions within the bacterial cell that include MDR efflux pumps. Other known regulators of MDR efflux pump expression, such as BmrR of B. subtilis, QacR of S. aureus, EmrR of E. coli, and MexR of Pseudomonas aeruginosa regulate transcription of bmr, qacA, emrB, and mexAB-oprM operons encoding the MDR pumps Bmr, QacA, EmrB, and MexAB-OprM, respectively (20, 46, 52, 55). BmrR, QacR, and EmrR are also known to mediate substrate induction of transcription. EmrR is also known to regulate the expression of the plasmid-encoded mcb operon, which directs the synthesis of microcin B17. NorR is less closely related to BmrR, QacR, EmrR, and MexR than it is to MarR, and these four regulators have not been shown to have physiologic roles apart from their direct regulation of expression of their respective MDR pumps. In addition, the bmrR, qacR, emrR, and mexR genes, in contrast to norR (Fig. 2) and marR, are closely linked to the structural genes of the pumps that they regulate, consistent with their presumed targeted role of specific substrate induction of a specific pump (20, 48, 55). NorR is the first identified direct regulator of efflux pump expression in S. aureus, and it appears to function as both a positive and negative regulator of different pumps. It is noteworthy that the two best-characterized MDR pumps in Lactococcus lactis, LmrA and LmrP, are reciprocally expressed (W. Konings, personal communication), suggesting that regulators with opposing effects on expression of different pumps, like NorR, may also be present in other species. Whether or not NorR protein binds pump substrates is not known, nor is its role in mediating the quinolone induction of norA expression reported for one mutant strain (28).
Regulation of expression of norA is multifactorial and is known to include effects due to expression of the arlRS two-component regulatory system and mutations that affect the stability of norA mRNA (15, 17). NorR is now identified as a third component in the regulation of norA that acts at the level of transcription. Variations in band-shift patterns observed with the cell extracts obtained from the pleiotropic mutants MT1222 and BF16 (arlS) (data not shown) suggest that additional regulatory factors also likely bind to the norA promoter, and experiments are underway to identify any such additional proteins. Such proteins might also underlie the requirement for intact agr and sarA for norR to increase norA expression. As noted above, other mutants not yet genetically characterized have also been described to have a norA-mediated resistance phenotype that is inducible with norfloxacin (27-29). Thus, regulation of norA and other efflux pumps warrants further study, particularly to define the global networks that mediate regulation of efflux pump expression and to define those environmental conditions under which physiologic overexpression of various pumps reduces antibiotic action and which promote selection of resistant mutants due to efflux pump overexpression. NorR itself, functioning as both a positive and negative regulator of efflux pump expression, appears likely to have a central role in a regulatory network for MDR efflux pumps that likely also interacts in complex ways with established global regulators.
Acknowledgments
We thank Kim Lewis for providing S. aureus strain KL820; Ambrose Cheung for S. aureus strains RN6390, RN6911, ALC135, and ALC136; Chia Lee for plasmid pCL52.2; Jean Lee for plasmid pLI50; and Gordon Archer for plasmid pSK950. We thank Steve Projan for helpful comments on the manuscript. We also thank Dilek Ince for assistance with the allelic exchange experiments.
This work was supported by a grant from the United States Public Health Service, National Institutes of Health (R01 AI23988 to D.C.H.).
REFERENCES
- 1.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolhuis, H., H. W. van Veen, B. Poolman, A. J. Driessen, and W. N. Konings. 1997. Mechanisms of multidrug transporters. FEMS Microbiol. Rev. 21:55-84. [DOI] [PubMed] [Google Scholar]
- 6.Booth, M. C., A. L. Cheung, K. L. Hatter, B. D. Jett, M. C. Callegan, and M. S. Gilmore. 1997. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect. Immun. 65:1550-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, P. F., and S. J. Foster. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144:2469-2479. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien, Y. T., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991-1001. [DOI] [PubMed] [Google Scholar]
- 12.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrero, L., B. Cameron, and J. Crouzet. 1995. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 17.Fournier, B., Q. C. Truong-Bolduc, X. Zhang, and D. C. Hooper. 2001. A mutation in the 5′ untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J. Bacteriol. 183:2367-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grinius, L. L., and E. B. Goldberg. 1994. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 269:29998-30004. [PubMed] [Google Scholar]
- 20.Grkovic, S., M. H. Brown, M. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 21.Hane, M. W., and T. H. Wood. 1969. Escherichia coli K-12 mutants resistant to nalidixic acid: genetic mapping and dominance studies. J. Bacteriol. 99:238-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper, D. C. 1995. Quinolone mode of action. Drugs 49(Suppl. 2):10-15. [DOI] [PubMed] [Google Scholar]
- 23.Hooper, D. C., J. S. Wolfson, K. S. Souza, E. Y. Ng, G. L. McHugh, and M. N. Swartz. 1989. Mechanisms of quinolone resistance in Escherichia coli: characterization of nfxB and cfxB, two mutant resistance loci decreasing norfloxacin accumulation. Antimicrob. Agents Chemother. 33:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh, P. C., S. A. Siegel, B. Rogers, D. Davis, and K. Lewis. 1998. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc. Natl. Acad. Sci. USA 95:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2002. Dual targeting of DNA gyrase and topoisomerase IV: target interactions of garenoxacin (BMS-284756, T-3811ME), a new desfluoroquinolone. Antimicrob. Agents Chemother. 46:3370-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaatz, G. W., S. M. Seo, L. O'Brien, M. Wahiduzzaman, and T. J. Foster. 2000. Evidence for the existence of a multidrug efflux transporter distinct from NorA in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1404-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaatz, G. W., S. M. Seo, and C. A. Ruble. 1993. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1086-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 31.Levy, S. B. 1992. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 36:695-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maira-Litrán, T., D. G. Allison, and P. Gilbert. 2000. An evaluation of the potential of the multiple antibiotic resistance operon (mar) and the multidrug efflux pump acrAB to moderate resistance towards ciprofloxacin in Escherichia coli biofilms. J. Antimicrob. Chemother. 45:789-795. [DOI] [PubMed] [Google Scholar]
- 33.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 36.Neyfakh, A. A. 1992. The multidrug efflux transporter of Bacillus subtilis is a structural and functional homolog of the Staphylococcus NorA protein. Antimicrob. Agents Chemother. 36:484-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neyfakh, A. A., C. M. Borsch, and G. W. Kaatz. 1993. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 37:128-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship of the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, E. Y. W., M. Trucksis, and D. C. Hooper. 1994. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob. Agents Chemother. 38:1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niemeyer, D. M., M. J. Pucci, J. A. Thanassi, V. K. Sharma, and G. L. Archer. 1996. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 178:5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 42.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rechtin, T. M., A. F. Gillaspy, M. A. Schumacher, R. G. Brennan, M. S. Smeltzer, and B. K. Hurlburt. 1999. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol. Microbiol. 33:307-316. [DOI] [PubMed] [Google Scholar]
- 46.Sánchez, P., F. Rojo, and J. L. Martínez. 2002. Transcriptional regulation of mexR, the repressor of Pseudomonas aeruginosa mexAB-oprM multidrug efflux pump. FEMS Microbiol. Lett. 207:63-68. [DOI] [PubMed] [Google Scholar]
- 47.Sau, S., J. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schumacher, M. A., and R. G. Brennan. 2002. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45:885-893. [DOI] [PubMed] [Google Scholar]
- 49.Stahl, M. L., and P. A. Pattee. 1983. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J. Bacteriol. 154:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tegmark, K., E. Morfeldt, and S. Arvidson. 1998. Regulation of agr-dependent virulence genes in Staphylococcus aureus by RNAIII from coagulase-negative staphylococci. J. Bacteriol. 180:3181-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Veen, H. W., and W. N. Konings. 1997. Drug efflux proteins in multidrug resistant bacteria. Biol. Chem. Hoppe-Seyler 378:769-777. [PubMed] [Google Scholar]
- 52.Xiong, A., A. Gottman, C. Park, M. Baetens, S. Pandza, and A. Matin. 2000. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance operon by directly binding to its promoter region. Antimicrob. Agents Chemother. 44:2905-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, H., M. Bogaki, S. Nakamura, K. Ubukata, and M. Konno. 1990. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 172:6942-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, J.-L., L. Grinius, and D. C. Hooper. 2002. NorA functions as a multidrug efflux protein in both cytoplasmic membrane vesicles and reconstituted proteoliposomes. J. Bacteriol. 184:1370-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheleznova, E. E., P. N. Markham, A. A. Neyfakh, and R. G. Brennan. 1999. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell 96:353-362. [DOI] [PubMed] [Google Scholar]