Abstract
The cAMP-catabolite activator protein (CAP) complex is a pleiotropic regulator that regulates a vast number of Escherichia coli genes, including those involved in carbon metabolism. We identified two new targets of this complex: argG, which encodes the arginosuccinate synthase involved in the arginine biosynthetic pathway, and metY, which encodes one of the two methionine tRNA initiators, tRNAf2Met. The cAMP-CAP complex activates argG transcription and inhibits metY transcription from the same DNA position. We also show that ArgR, the specific repressor of the arginine biosynthetic pathway, together with its arginine cofactor, acts on the regulation of metY mediated by CAP. The regulation of the two divergent promoters is thus simultaneously controlled not only by the cAMP-CAP complex, a global regulator, but also by a specific regulator of arginine metabolism, suggesting a previously unsuspected link between carbon metabolism and translation initiation.
All forms of life degrade carbon-containing molecules. The cAMP-catabolite activator protein (CAP) complex is a global regulator involved in the regulation, repression as well as activation, of a vast number of Escherichia coli genes. Initially, its role was solely thought to control of the use of alternative carbon sources when glucose was lacking. Indeed, the largest group of targets controls the catabolism of carbohydrates, amino acids, and nucleosides. However, it is now clear that CAP also controls the expression of genes involved in many noncatabolic functions, including genes encoding membrane proteins, involved in metabolic transport (e.g., proP [39]), in carbon starvation, and in resistance to stress (e.g., gadA [8]). The absence of an obvious link between these various targets suggests that CAP controls the expression of genes involved in adaptation to growth conditions under limited nutrient supply. The promoters of these genes are usually regulated by multiple factors, and CAP tends either to be involved in coactivation together with a second activator or to act in tandem with a repressor (for example, at the lac promoter [33]). CAP seems therefore to sense a global signal (e.g., glucose starvation as reflected by the intracellular cAMP concentration), whereas the specific regulator monitors the level of a specific metabolite that may or may not be present.
In the present work, the two-dimensional protein pattern of a CAP-deficient strain revealed an alteration in the level of ArgG, the arginosuccinate synthase involved in the arginine biosynthetic pathway. The study of the DNA promoter region of this gene showed the presence of the divergent promoter of metY, which encodes one of the two methionine tRNAs required for the initiation of protein synthesis. Since argG is known to be regulated by a specific repressor, ArgR, we studied the direct effect of CAP and ArgR, in vivo and in vitro, on the transcription of argG and metY. Finally, we demonstrated the coupling between the arginine biosynthetic pathway and a gene responsible for the initiation of protein synthesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli K-12 derivatives and plasmids used in this work are listed in Table 1. The crp gene was mutated by inserting an Smr/Spcr cassette into the BclI site of the crp gene, yielding crp::Sm. This mutation was introduced into strain MG1655 by allelic replacement using the M13mp8 phage, giving rise to strain to BE1815 (5). The metZΔ2::cat mutation carried by strain IBPC 6411 (transduced from strain TK2 [21] and kindly provided by M. Springer) was introduced into MG1655 by transduction, giving rise to strain BE1623. Plasmid pDIA530 was constructed as follows: the fragment containing positions −417 to +90 of the argG promoter according to reference 9 was PCR amplified from genomic DNA using the Expand high-fidelity PCR system (Roche) and synthetic oligonucleotides 5′-CAGAAGGATCCTTTCAAATCCC-3′, containing a BamHI site, and 5′-GGAGAAGCTTGAGAATCGTCGTC-3′, containing a HindIII site. After purification, the fragment was cloned into the BamHI and HindIII restriction sites of PKK232-8 (Pharmacia). Plasmid pDIA560 was similar to pDIA530 except that the CAP binding site was mutated by PCR amplification with the synthetic oligonucleotide 5′AATCTGCAGGCATTATAGTAATCCACGCTCGATTTTGTCAACGTTTATTGC-3′. Plasmid pDIA539 contained the 268-bp argG promoter (see below), cloned into the HincII blunted site of pJCD01 (24). Strains were grown at 37°C in M9 minimal medium (26) supplemented with mannose (0.4%), thiamine (5 mg/ml), a mixture of all amino acids with or without arginine (0.005% of each), and when necessary 100 μg of ampicillin/ml. All experiments were performed in accordance with the European regulation requirements concerning the contained use of genetically modified organisms of group I (agreement no. 2735).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Prototrophic E. coli K-12 | CGSC 6300a |
| BE1815 | MG1655 crp::Sm | This study |
| IBPC 6411 | thi-1 argE3 his-4 proA2 lacY1 galK2 mtl-1 xyl-5 tsx-29 supE44 rpsL pps metZΔ2::cat | M. Springer |
| BE1723 | MG1655 metZΔ2::catZ | This study |
| Plasmids | ||
| pJCD01 | PUC19 derivative containing a polylinker flanked by the divergent terminators rpoCt and rrnBT1T2 | 24 |
| pKK232-8 | Cloning vector for promoter analysis | Pharmacia |
| pDIA530 | pKK232-8 derivative carrying the −417 to +90 argG promoter region | This study |
| pDIA539 | pJCD01 derivative carrying the −212 to +56 argG promoter region | This study |
| pDIA560 | As pDIA530 but with a mutated CAP site: AGTGAtccacgCCACA→ AGTAAtccacgCTCGAb | This study |
CGSC, E. coli Genetic Stock Center, Yale University.
Underlining indicates mutated residues.
Two-dimensional gel electrophoresis.
Ten micrograms of total proteins, extracted from a culture at an optical density at 600 nm (OD600) of 0.4 to 0.5, was resolved on a two-dimensional (2D) gel (23). Polypeptides were detected by silver staining (29). Proteins were quantified as previously described (4). Spots of interest were excised from multiple gels and subjected to internal amino acid (37). The amino acid sequence homology search was carried out using the BLASTP software (2).
Quantitative analysis of mRNA.
RNA was prepared, subjected to a slot blot, and quantified as previously described (36). The experiment was performed using two independent cultures with a probe corresponding to a 700-bp fragment of argG, generated by PCR amplification using the PCR DIG probe synthesis kit (Roche). Quantitation was made with the Bio-Rad Multi-Analyst system.
Gel mobility shift DNA binding assay.
A 268-bp fragment corresponding to the promoter region of argG (−212 to +56) according to reference 9 was amplified from genomic DNA by PCR using Pfu DNA polymerase (Stratagene) and synthetic oligonucleotides 5′-GTGTACCGAGACGGGACG-3′ and 5′-TTAACTGATGATGAGCCTGG-3′ (one of which was labeled with [γ-32P]ATP by use of T4 polynucleotide kinase). The PCR product was purified with the High pure PCR product purification kit (Roche). Wild-type and mutated H159L CAP proteins were purified according to the method of Ghosaini et al. (13). Gel retardation experiments were performed as previously described (10), with some modifications: cAMP-CAP was bound to the labeled DNA fragment (0.2 nM) in a HEPES-Mg-K glutamate reaction buffer (40 mM HEPES [pH 8], 10 mM MgCl2, 100 mM K-glutamate) in the presence of 160 μM cAMP, at room temperature for 20 min.
DNase I footprinting experiments.
The binding of cAMP-CAP and/or ArgR (kindly provided by D. Charlier) to the 268-bp DNA fragment encompassing the argG promoter was performed in HEPES-Mg-K glutamate buffer in the presence of 160 μM cAMP as previously described (10). After 20 min at room temperature, DNase I was added at a final concentration of 0.1 μg/ml. Reaction mixtures without any regulator were incubated at 37°C for 15 s and those containing CAP or/and ArgR were incubated at 37°C for 25 s. Reactions were stopped and were subjected to electrophoresis after heating at 90°C. Protected bands were identified by comparison with the same fragment treated for A+G sequencing reactions (25).
Chloramphenicol acetyltransferase assay.
Bacteria carrying pDIA530 or pDIA560 were grown to log phase. Three 1.5-ml samples from two independent cultures were centrifuged for 5 min at 13,000 × g. Pellets were resuspended in 500 μl of 100 mM Tris-HCl (pH 7.8) and treated as described previously (35, 36).
In vitro transcription assays.
In vitro transcription experiments were performed at least two times with pDIA539 (1.2 nM) in buffer containing 40 mM Tris-HCl (pH 8), 10 mM MgCl2, 100 mM KCl, 0.5-mg/ml bovine serum albumin, 160 μM cAMP, and 1.4 mM dithiothreitol. After 20 min at room temperature, 14 μl of the mixture containing CAP or H159L CAP and/or ArgR at different concentrations was incubated at 30°C for 3 min. Then, 3.5 μl of a mixture containing nucleoside triphosphates, [α-32P]UTP, and RNA polymerase (15 nM) was added to perform polymerization at 30°C for 10 min. The reaction was stopped by adding 1% sodium dodecyl sulfate and xylene blue formamide. After heating at 70°C and electrophoresis gel, the data were quantified with a PhosphorImager (Molecular Dynamics).
β-galactosidase assay.
Overnight cultures were freshly diluted to an OD600 of 0.1 and were incubated 30 and 60 min at 30°C in M63 medium (26) supplemented with isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) and glycerol (0.4%) ± arginine (10 mM). β-galactosidase activity was then determined by the method of Miller (27). Each assay was performed with two independent cultures.
RESULTS
The expression of argG is reduced in a crp mutant.
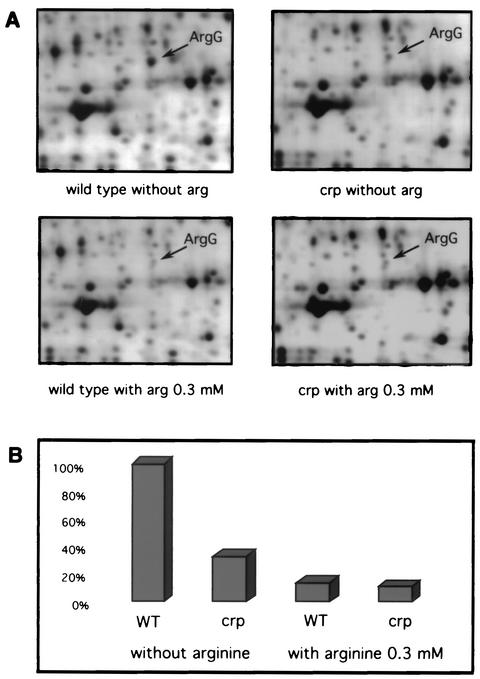
Preliminary two-dimensional experiments with argH mutants (which are unable to synthesize arginine) showed that the level of several polypeptides was altered in a crp background (data not shown). Among them, arginosuccinate synthase was identified by microsequencing and a BLASTP search: the internal sequence of the protein was TFSDDVEMMLEANRI, which is 100% identical to an ArgG peptide (Swissprot accession number p22767). To rule out any effect of the argH mutation, a second set of experiments was performed in a wild-type background. Figure 1A shows representative 2D patterns of silver-stained proteins isolated from wild-type MG1655 and its crp derivative, BE1815, grown in M9 minimum medium supplemented with mannose and with or without arginine. The ArgG accumulation level was quantified (Fig. 1B): in the crp strain, BE1815, it corresponded to 33% of the wild-type level. In the presence of 0.3 mM arginine, both strains contained around 10% of the level found in the wild type in the absence of arginine. This was expected since arginine is a repressor of arginine biosynthesis genes (9).
FIG. 1.
(A) 2D gels of the wild-type strain (MG1655) and the crp mutant strain (BE1815) grown in minimal medium with or without 0.3 mM arginine. (B) Relative amounts of the ArgG protein in the four conditions.
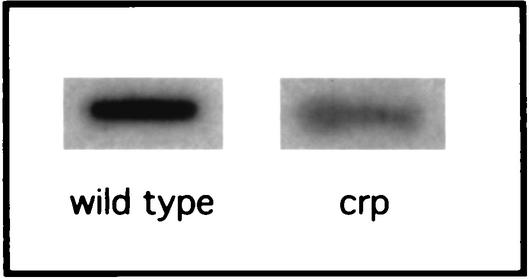
To determine whether the variations in protein levels observed correlated with a difference in the level of mRNA, a slot blot experiment with a chemiluminescent argG probe was performed with total RNA extracted from an exponential-phase culture grown in minimal medium without arginine (Fig. 2). A quantitative analysis showed that the mutant contained just 27% of the amount of argG mRNA contained by the wild type. This demonstrated that the regulation of ArgG synthesis by the cAMP-CAP complex is directly correlated with the amount of its mRNA.
FIG. 2.
The amount of argG mRNA in the wild-type and crp mutant strains was analyzed by slot blot hybridization with a specific 700-bp probe. Cultures were grown in minimal medium without arginine.
Identification of a CAP-binding site in the argG promoter region.
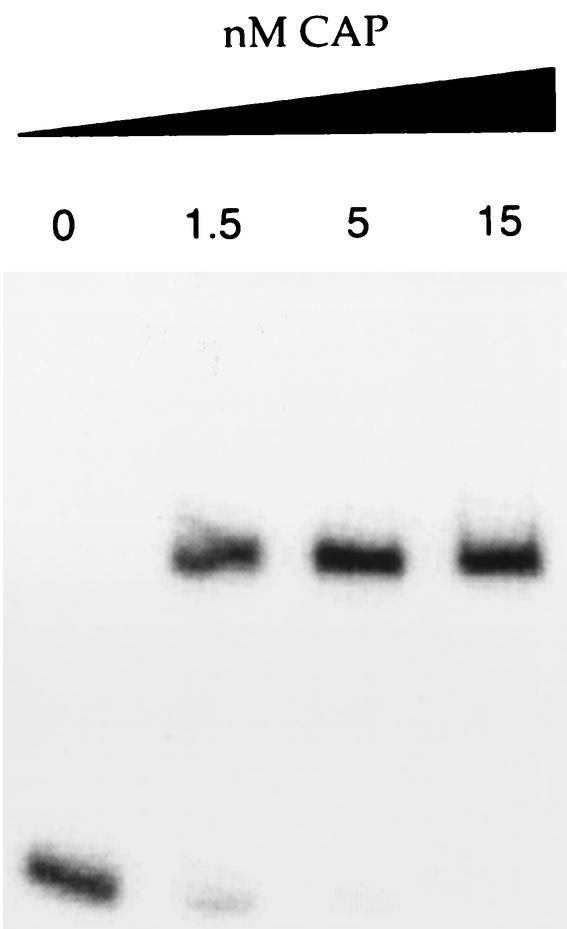
A putative cAMP-CAP-binding site close to the known consensus binding site (7) was identified, centered at −166.5 bp with respect to the transcriptional start site. To confirm this, a gel mobility shift DNA binding assay was performed with the purified cAMP-CAP complex. Figure 3 shows that the presence of cAMP-CAP, at a low CAP concentration (1.5 nM), led to a significant retardation of the DNA fragment carrying the argG promoter region between −212 and +56, suggesting that the cAMP-CAP complex binds to the argG promoter region.
FIG. 3.
Competitive gel retardation assay with CAP and the argG promoter region (positions −212 and +56 with respect to the transcriptional start site). DNA fragments were incubated with the indicated concentrations of CAP.
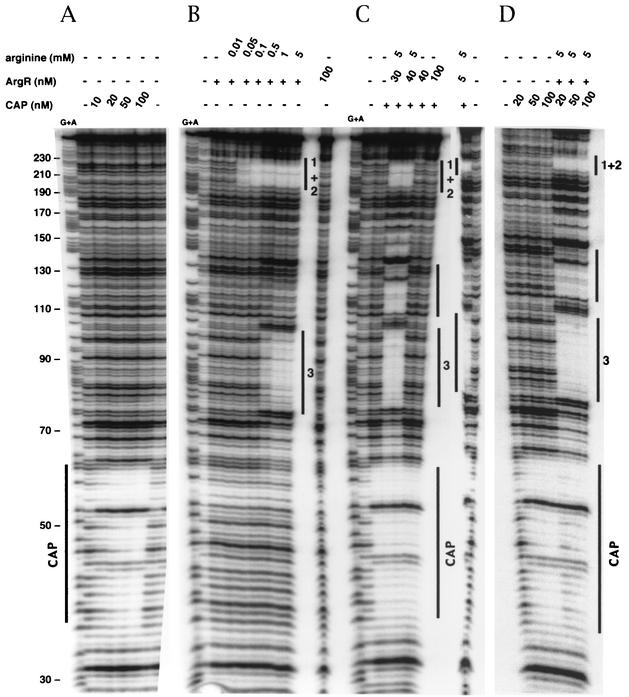
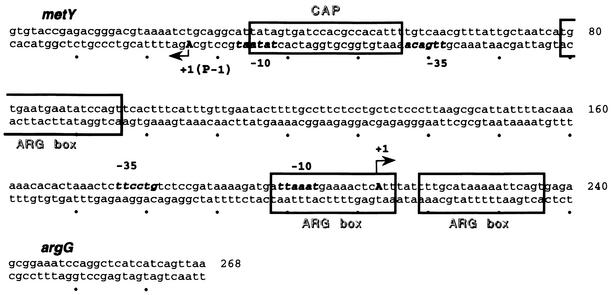
The precise location of the binding site was determined by DNase I footprinting. A footprint characteristic of the cAMP-CAP complex marked by protected bands with two hypersensitive bands 10 bp apart was detected in the presence of cAMP and 10 nM CAP (Fig. 4A). The site was fully occupied, and some hypersensitive bands were observed up- and downstream of the site in the presence of 100 nM CAP (Fig. 4A). Our data show that the cAMP-CAP complex binds specifically between positions 35 and 61, according to the numbering of the sequence in Fig. 5, which corresponds to the predicted cAMP-CAP binding site from which CAP may activate argG transcription. Moreover, the CAP-binding site overlaps both the −10 box and the upstream sequence of the metY-yhbC-nusA-infB operon P−1 promoter (15). This operon encodes successively the 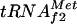 , a protein of unknown function, the transcription-translation coupling factor NusA, and the translation initiation factor IF2. This suggests that the binding of the cAMP-CAP complex at a position centered on −19.5 upstream of the metY transcription start site may repress the transcription of the metY-nusA-infB operon from the P− promoter.
, a protein of unknown function, the transcription-translation coupling factor NusA, and the translation initiation factor IF2. This suggests that the binding of the cAMP-CAP complex at a position centered on −19.5 upstream of the metY transcription start site may repress the transcription of the metY-nusA-infB operon from the P− promoter.
FIG. 4.
Analysis of CAP- and ArgR-binding sites in the argG and metY promoter region by DNase I footprinting assays. The labeled DNA fragment represents the coding strand of argG with cAMP-CAP complex alone (A), with 5 nM ArgR alone (+) (numbers correspond to ARG boxes) (B), with 100 nM CAP (+) and then with ArgR (C), and with 20 nM ArgR (+) and then with CAP (D). Protected regions are marked by solid lines. The coordinates were determined with G+A and correspond to the numbering used in Fig. 5. −, no protein was present.
FIG. 5.
Regulatory region of argG and metY. The nucleotide sequence corresponds to the fragment used in in vitro experiments. cAMP-CAP and ArgR-binding sites are indicated by boxes. The transcriptional start sites (+1) of argG and metY (promoter P−1) are indicated by arrows. The −10 and −35 boxes are indicated in bold.
The cAMP-CAP complex activates argG transcription in vivo.
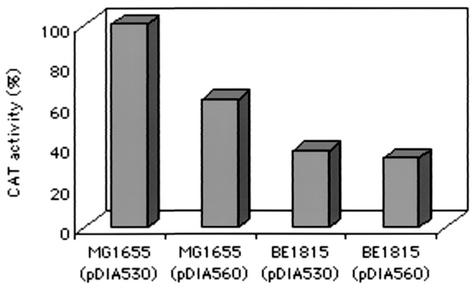
The cAMP-CAP-binding site is distant from the argG transcription start site as it is centered at position −166.5. To determine whether the cAMP-CAP complex acts directly on argG transcription, an argG-cat transcriptional fusion was constructed on plasmid PKK232.8 with an intact (pDIA530) or an altered (pDIA560) CAP binding site. The altered CAP site was designed to prevent cAMP-CAP fixation, as previously described (6), i.e., tatAGTGAtccacgCCACAttt changed to tatAGTAAtccacgCTCGAttt. A gel mobility shift DNA binding assay confirmed that the cAMP-CAP complex was unable to bind to the altered site even at a high CAP concentration (40 nM). CAT activity during log phase was measured in argG transcriptional fusions with both wild-type and mutated promoters. Three to four colonies were picked from two independent cultures (Fig. 6). For the crp+ strain, the mutated CAP-binding site on pDIA560 caused a reduction to 63% in beta-galactosidase activity of the fusion compared to the nonmutated binding site in pDIA530. However, for the crp mutant (BE1815), both the mutated and the nonmutated CAP binding sites caused the same reduction in beta-galactosidase activity, to 36% of the activity seen in the wild-type reference strain. These results are consistent with the amounts of the ArgG protein and argG mRNA (Fig. 1 and 2). The CAT activity in the wild-type strain carrying the mutated CAP binding site (pDIA560) was between that obtained with the wild-type fusion (pDIA530) in the wild-type strain and that obtained for the crp mutant strain. This suggested that the cAMP-CAP complex exerts an additional and indirect effect on argG transcription.
FIG. 6.
Effect of a crp mutation (BE1815) on argG-cat transcriptional fusion activity. pDIA530 carries the intact promoter, whereas pDIA560 carries a promoter with an altered CAP site.
It is known that argG is regulated by the hexameric ArgR repressor. In fact, ArgR, in conjunction with its corepressor arginine, represses argG transcription by binding to three ARG boxes: first to tandem ARG boxes, constituted of 18-bp boxes separated by 3 bp, located around the +1 site of argG, and then to a third single ARG box, located 101 bases upstream (Fig. 5) (9). This prompted us to study the interaction between the binding of the cAMP-CAP complex and the hexameric ArgR repressor in arg regulation.
Binding of ArgR to the argG promoter region in the presence and absence of CAP.
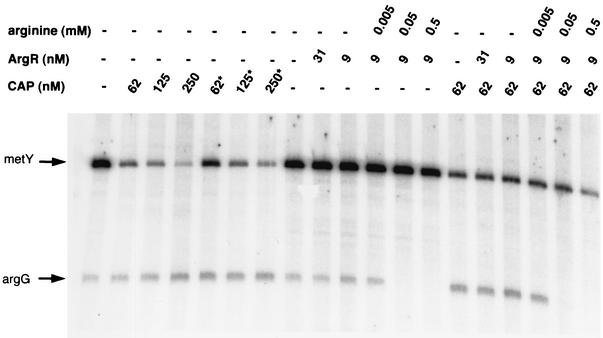
DNase I footprint experiments were first performed with ArgR (5 nM) at the argG promoter region (0.6 nM) and various arginine concentrations. It has been suggested that the degree of repression mainly depends on the concentration of arginine (14). No binding to the DNA fragment was observed in the absence of arginine, even at a high ArgR concentration (100 nM) (Fig. 4B). In the presence of 0.05 mM arginine, ArgR (7.5 nM) bound to the tandem ARG boxes around the argG +1 start site, whereas with 0.5 mM arginine, ArgR also bound to the third single ARG box (Fig. 4B). In the presence of 5 mM arginine and at a higher ArgR concentration (30 nM), a protected region was observed between the tandem ARG boxes and the single ARG box (positions 110 to 132) (data not shown; also Fig. 4C and D). This is thought to form a DNA loop, resulting from the binding to the three ARG boxes (9). DNase I footprint experiments were then performed with various concentrations of both regulators and in the presence and absence of 5 mM arginine (Fig. 4C and 4D). Again, in the absence of arginine, the ArgR regulator did not bind and had no effect on the binding of the cAMP-CAP complex to the operator. In the presence of arginine, ArgR bound to the three boxes, even at a high concentration of CAP (100 nM) (Fig. 4C and 4D). In contrast, the order with which the two regulators were added affected CAP binding. Indeed, the cAMP-CAP complex remained stably bound to DNA when ArgR was subsequently added in the presence of arginine (Fig. 4C). In contrast, when ArgR with arginine was already bound to DNA, the binding of CAP to DNA was less efficient, even at a high concentration (100 nM). However, its binding site was partially protected (Fig. 4D). Moreover, the hypersensitive bands observed between the CAP-binding site and the third ARG box with CAP alone were no longer visible in the presence of ArgR and arginine. These results suggest that the binding of each of the two regulators interferes with the binding of the other and that ArgR binding overrides CAP binding.
The cAMP-CAP complex activates argG and represses metY, whereas ArgR also represses metY but only in the presence of CAP.
In the different conditions of regulator binding, we performed in vitro transcription assays on plasmid pDIA539, which contained the metY-argG promoter region used in DNase I footprint experiments (Fig. 5). Again, a range of regulator concentrations as well as of the cofactor arginine was used. The effect of a mutated CAP protein carrying an H159L amino acid substitution in activating region I, which prevents interactions with the C-terminal domain of the α subunit of RNA polymerase (12), was also investigated. At 250 nM, CAP increased argG transcription twofold, whereas it decreased metY transcription 16-fold. The mutated protein had the same effect, although the inhibition of metY was at a slightly lower level (10-fold) (Fig. 7). This further supports the hypothesis that the binding of RNA polymerase to the metY promoter P−1 is inhibited in the presence of the cAMP-CAP complex. These results also suggest that the cAMP-CAP complex does not interact directly with RNA polymerase bound to the argG promoter: CAP could only act on DNA conformation, hence facilitating the binding of RNA polymerase to the −10 and −35 boxes of the promoter. In the absence of its corepressor arginine, the ArgR regulator had no effect on the transcription of either metY or argG, whether in the presence or in the absence of CAP (Fig. 7), in agreement with the DNase I footprinting results. With 0.05 mM arginine, when bound to the tandem ARG boxes, ArgR (9 nM) repressed argG transcription 10-fold (Fig. 7). In the presence of CAP, the inhibitory effect of ArgR plus arginine remained predominant and of the same magnitude as that on argG (Fig. 7). This result is consistent with the difference in the levels of ArgG protein measured in two-dimensional gel experiments (Fig. 1). At a higher arginine concentration (0.5 mM), when ArgR also bound to the third isolated ARG box, no further inhibition was observed at the argG promoter. In contrast, ArgR repressed the transcription of metY twofold in the presence of CAP, although ArgR had no detectable effect without CAP. This suggests that both regulators can bind to their own sites together in vitro, as shown by footprinting experiments, and that the binding of ArgR to the third ARG box, which occurs at a high concentration of arginine, reinforces the repressor effect of CAP on the binding of RNA polymerase to the metY P−1 promoter.
FIG. 7.
In vitro transcription assay by RNA polymerase from metY P−1 and argG promoters. pDIA539 was incubated with RNA polymerase in the presence or the absence (-) of regulators. Asterisks correspond to the H159L mutated CAP protein. Different transcripts are indicated by arrows.
At 37°C, metY-infB transcription is mainly terminated after metY (3, 32). Therefore, the regulation of the metY P−1 promoter in vivo was studied by monitoring the level of 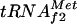 .
.
Regulation of metY by cAMP-CAP complex and ArgR in vivo.
It was previously shown that the level of β-galactosidase in the presence of IPTG reflects the efficiency of translation from the AUG start codon at the initiation step, independently of transcription, and thus the amount of 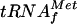 available (21). For a metZ mutant strain, in which
available (21). For a metZ mutant strain, in which 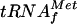 is synthesized only by metY, the level of β-galactosidase translation is similar to that measured for a wild-type strain grown in minimal medium with glucose and Casamino Acids in the presence of IPTG (21). With these carbon sources, the level of cAMP is low. In contrast, in the presence of glycerol, the cAMP level is 10-fold higher (18). Therefore, the activity of β-galactosidase was measured in cells that had been freshly diluted in minimal medium supplemented with 0.4% glycerol and with and without 10 mM arginine. After 30 and 60 min with glycerol alone, the β-galactosidase level for the metZ strain BE1723 was 70% of that obtained for the wild type (Table 2). This further supports the repressive effect of the cAMP-CAP complex observed in vitro, although the effect was less marked in vivo. This could be explained by the presence of a second metY promoter, P0, downstream of P−1 (17). Surprisingly, with glycerol and arginine, no difference was observed between wild-type and metZ strains (Table 2), suggesting that CAP had no more effect on the metY level. In this assay, a high arginine concentration (10 mM) was used to allow ArgR to bind to the third ARG box. The result obtained suggests that the binding of ArgR to the third single ARG box in the chromosome prevents the binding of the cAMP-CAP complex in vivo.
is synthesized only by metY, the level of β-galactosidase translation is similar to that measured for a wild-type strain grown in minimal medium with glucose and Casamino Acids in the presence of IPTG (21). With these carbon sources, the level of cAMP is low. In contrast, in the presence of glycerol, the cAMP level is 10-fold higher (18). Therefore, the activity of β-galactosidase was measured in cells that had been freshly diluted in minimal medium supplemented with 0.4% glycerol and with and without 10 mM arginine. After 30 and 60 min with glycerol alone, the β-galactosidase level for the metZ strain BE1723 was 70% of that obtained for the wild type (Table 2). This further supports the repressive effect of the cAMP-CAP complex observed in vitro, although the effect was less marked in vivo. This could be explained by the presence of a second metY promoter, P0, downstream of P−1 (17). Surprisingly, with glycerol and arginine, no difference was observed between wild-type and metZ strains (Table 2), suggesting that CAP had no more effect on the metY level. In this assay, a high arginine concentration (10 mM) was used to allow ArgR to bind to the third ARG box. The result obtained suggests that the binding of ArgR to the third single ARG box in the chromosome prevents the binding of the cAMP-CAP complex in vivo.
TABLE 2.
β-Galactosidase activitya
| Carbon source | Activity for strain:
|
|||
|---|---|---|---|---|
| MG1655 (Wild-type)
|
BEI723 (metZ mutant)
|
|||
| 30 min | 60 min | 30 min | 60 min | |
| Glycerol | 2,100 | 3,920 | 1,420 | 2,770 |
| Glycerol + arginine | 1,460 | 3,650 | 1,700 | 3,400 |
β-Galactosidase activity is expressed in Miller units/milligram of protein. Values are the means of two independent determinations. Activities were measured after 30 and 60 min in cultures freshly diluted to an OD600 of 0.1 in M63 medium.
In summary, the regulation of argG and metY expression is coupled by the cAMP-CAP complex but in an opposite way and in an even more complex manner with ArgR and arginine.
DISCUSSION
For E. coli K-12, arginine biosynthesis starts from glutamate and carbamoylphosphate. The enzymes responsible are coded by the arginine regulon. This regulon is composed of 12 genes, among which only a few are organized in an operon. However, all the genes in the arginine regulon are regulated by a master transcriptional regulator and repressor, ArgR, which functions as a direct sensor of arginine availability. The binding of six l-arginine molecules at the trimer-trimer interface of ArgR activates the regulator, allowing it to bind to the ARG boxes in DNA (11). The repressor binds symmetrically to four consecutive helical turns (corresponding to the two palindromic ARG boxes separated by three nucleotides, a span of nearly 40 bp) on one face of the DNA (14). The number of ArgR molecules per cell appears to be relatively high, around 600 (300 in the presence of excess arginine). Since its affinity for arginine is quite low (Kd of around 10−4 M) and the Kd of the active repressor for its operator is between 10−9 and 10−10 M, the degree of repression will mainly depend on the arginine concentration (14). argG, which is a member of the arginine regulon, encodes arginosuccinate synthase. Its promoter contains two 18-bp ARG boxes, separated by 3 bp, extending from the −10 box to the +24 residue, with respect to the +1 start site, and a third single box located 101 bp upstream of the tandem boxes (Fig. 5). The binding of the repressor to the upstream single ARG box requires much more repressor than binding to the tandem boxes. It has been suggested that binding to the three ARG boxes leads to the formation of a DNA loop and that the third site may become occupied if a loop is formed (9, 14).
In our study, we showed that argG is also regulated at a transcriptional level by the cAMP-CAP complex. This complex binds at position −166.5, with respect to the +1 start site, to the tatAGTGAtccacgCCACAttt sequence, which is very close to the consensus binding site 5′aaaTGTGAtntanaTCACAttt3′ (7). At such a distant location, CAP would have no effect on transcription. However, we demonstrated that its binding resulted in a twofold increase in argG transcription. A CAP protein that had mutated in its activating region I (H159L), such that it was unable to interact with the C-terminal domain of the α subunit of RNA polymerase (12), had the same activating effect. This suggests that in the present case, the cAMP-CAP complex does not interact directly with RNA polymerase but only facilitates its binding to −10 and −35 boxes by the stabilization of the bending of this promoter region, making it more accessible for transcription initiation. The generation of a DNA loop upon ArgR binding supports the hypothesis that the region is particularly formed (9, 14). The binding of ArgR to the tandem ARG boxes around the +1 start site, in the presence of arginine (0.05 mM), repressed the transcription of argG 10-fold, even in the presence of the cAMP-CAP complex. Thus, a gene-specific regulation, i.e., mediated by ArgR, is here clearly predominant, as observed in the case of numerous targets of CAP (e.g., lac or mal). As a consequence, the cAMP-CAP complex may increase the rate of arginine synthesis under arginine starvation conditions. The argG gene should therefore be added to the list of genes induced by CAP and involved in the biosynthesis of amino acids such as isoleucine and valine, which are derived from glutamate (7). On the other hand, CAP is known to repress gadA, coding for a glutamate decarboxylase that participates in acid resistance and synthesizes GABA from glutamate (8). This suggests that CAP may favor the consumption of glutamate for amino acid biosynthesis instead of its use in other metabolic processes.
It is interesting that from the same position, the cAMP-CAP complex is both an activator of argG and a strong repressor of the metY-yhbC-nusA-infB operon, since its binding site overlaps the sequence between the −10 and −35 boxes of the P−1 promoter of the latter (15), which encodes 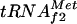 , the unknown protein YhbC, NusA, and the translation initiation factor IF2, respectively. However, an internal promoter, P2 (which directs the transcription of the protein coding part), and two intercistronic terminators, t1 and t2, are present downstream of metY. At 37°C, only a metY transcript is mainly observed, whereas under cold shock, distal genes are expressed through a transcription antitermination mechanism that is mediated by cold shock-induced Csp proteins, such as CspA (3, 32).
, the unknown protein YhbC, NusA, and the translation initiation factor IF2, respectively. However, an internal promoter, P2 (which directs the transcription of the protein coding part), and two intercistronic terminators, t1 and t2, are present downstream of metY. At 37°C, only a metY transcript is mainly observed, whereas under cold shock, distal genes are expressed through a transcription antitermination mechanism that is mediated by cold shock-induced Csp proteins, such as CspA (3, 32).
Two isoacceptor species of 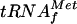 are present in E. coli K-12:
are present in E. coli K-12: 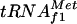 coded by a three-tandem-repeated gene metZ (20), which represents the major fraction of the initiator tRNA pool, and
coded by a three-tandem-repeated gene metZ (20), which represents the major fraction of the initiator tRNA pool, and 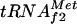 coded by metY, which represents the minor fraction (16). However, start codons AUG and GUG are recognized by both initiator tRNAs, and a metZ mutant strain is able to grow, although at a slower rate than the wild type or a metY-deficient strain (19, 21). The transcription of metZ is sensitive to ppGpp, the chemical mediator of stringent control, whereas this is not the case for metY (30). Before the present work, no regulation of the transcription of metY from its two promoters, P−1 and P0 (15, 17), had been described, although the relative fraction of
coded by metY, which represents the minor fraction (16). However, start codons AUG and GUG are recognized by both initiator tRNAs, and a metZ mutant strain is able to grow, although at a slower rate than the wild type or a metY-deficient strain (19, 21). The transcription of metZ is sensitive to ppGpp, the chemical mediator of stringent control, whereas this is not the case for metY (30). Before the present work, no regulation of the transcription of metY from its two promoters, P−1 and P0 (15, 17), had been described, although the relative fraction of 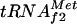 in the tRNA pool seemed to be dependent on the growth rate and on the Fis activator (31, 38). In this article, we show that a regulation mechanism for metY transcription exists, mediated by the cAMP-CAP complex and by the repressor of argG, ArgR. In fact, the cAMP-CAP complex repressed the transcription of metY from its promoter P−1 16-fold in vitro, and a significant inhibitory effect was also observed in vivo. By binding to the −10 and −35 boxes of the promoter, the cAMP-CAP complex blocked the access of RNA polymerase to promoter elements, as described for osmY (22) and cya P2 (1, 28), a mechanism that is very rarely used by CAP. ArgR only exerted its effect in the presence of CAP and a high concentration of arginine, i.e., when the third ARG box, which is centered at position −60 with respect to the +1 position of metY, was occupied by ArgR. However, the effects observed in vitro and in vivo seemed to contradict each other: ArgR repressed metY in vitro by increasing the cAMP-CAP complex repression effect, whereas in vivo it seemed to prevent the inhibitory action of CAP. It should be noted that the distance between the CAP binding site and the distal ArgR box is 42 bp, i.e., exactly four full turns of the DNA double helix, assuming a DNA pitch of 10.5 bp. This may mean that CAP and ArgR are located on the same side of the DNA helix, especially when the concentration of CAP is high, thus allowing them to act synergistically to repress the metY promoter in vitro. However, in footprinting experiments, ArgR with arginine, when bound first, partially prevented the binding of CAP, especially when CAP was added at the same concentration as ArgR. The intracellular concentration of cAMP is known to remain relatively low, even when cells are grown on glycerol, and in contrast to ArgR, the cAMP-CAP complex is a pleiotropic regulator that possesses many more available targets on the chromosome than the specific regulator ArgR. This suggests that more molecules of ArgR than of CAP are available to bind to the metY promoter. This could explain why ArgR prevents CAP binding in vivo.
in the tRNA pool seemed to be dependent on the growth rate and on the Fis activator (31, 38). In this article, we show that a regulation mechanism for metY transcription exists, mediated by the cAMP-CAP complex and by the repressor of argG, ArgR. In fact, the cAMP-CAP complex repressed the transcription of metY from its promoter P−1 16-fold in vitro, and a significant inhibitory effect was also observed in vivo. By binding to the −10 and −35 boxes of the promoter, the cAMP-CAP complex blocked the access of RNA polymerase to promoter elements, as described for osmY (22) and cya P2 (1, 28), a mechanism that is very rarely used by CAP. ArgR only exerted its effect in the presence of CAP and a high concentration of arginine, i.e., when the third ARG box, which is centered at position −60 with respect to the +1 position of metY, was occupied by ArgR. However, the effects observed in vitro and in vivo seemed to contradict each other: ArgR repressed metY in vitro by increasing the cAMP-CAP complex repression effect, whereas in vivo it seemed to prevent the inhibitory action of CAP. It should be noted that the distance between the CAP binding site and the distal ArgR box is 42 bp, i.e., exactly four full turns of the DNA double helix, assuming a DNA pitch of 10.5 bp. This may mean that CAP and ArgR are located on the same side of the DNA helix, especially when the concentration of CAP is high, thus allowing them to act synergistically to repress the metY promoter in vitro. However, in footprinting experiments, ArgR with arginine, when bound first, partially prevented the binding of CAP, especially when CAP was added at the same concentration as ArgR. The intracellular concentration of cAMP is known to remain relatively low, even when cells are grown on glycerol, and in contrast to ArgR, the cAMP-CAP complex is a pleiotropic regulator that possesses many more available targets on the chromosome than the specific regulator ArgR. This suggests that more molecules of ArgR than of CAP are available to bind to the metY promoter. This could explain why ArgR prevents CAP binding in vivo.
Finally, our results confirm the hypothesis that the different promoters of Met-tRNA initiators respond to different signals under changing physiological conditions (15). Indeed, we showed that the synthesis of one of the two tRNA initiators is coregulated with one of the arginine biosynthetic genes, argG, by both the cAMP-CAP complex and the ArgR-specific repressor of arginine biosynthesis. The function of these gene products indicates that they are coupled through their direct involvement in protein biosynthesis. However, the direct relationship between arginine and one of the two methionine tRNA initiators remains unexplained, although it was recently found that methionine seems to act on the level of RNA messenger of genes involved in arginine biosynthesis for Bacillus subtilis (34). This further supports the existence of a strong link between methionine and arginine biosynthesis, namely polyamine biosynthesis, for the synthesis of spermidine.
Acknowledgments
We are grateful to W. Maas for helpful advice and to E. Tate for critical reading. We thank M. Springer for providing us with the metZ-deficient strain and D. Charlier for providing the purified ArgR protein.
Financial support came from the Institut Pasteur and the Centre National de la Recherche Scientifique (URA 1129 and URA 2171 and FRE2364).
REFERENCES
- 1.Aiba, H. 1985. Transcription of the Escherichia coli adenylate cyclase gene is negatively regulated by cAMP-cAMP receptor protein. J. Biol. Chem. 260:3063-3070. [PubMed] [Google Scholar]
- 2.Altschul, W., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 97:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertin, P., N. Benhabiles, E. Krin, C. Laurent-Winter, C. Tendeng, E. Turlin, A. Thomas, A. Danchin, and R. Brasseur. 1999. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in Gram-negative bacteria. Mol. Microbiol. 31:319-329. [DOI] [PubMed] [Google Scholar]
- 5.Blum, P., D. Holzchu, H. S. Kwan, D. Riggs, and S. Artz. 1989. Gene replacement and retrieval with recombinant M13mp bacteriophages. J. Bacteriol. 171:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchet, A., K. Eichler, and M.-A. Mandrand-Berthelot. 1998. Regulation of the carnithine pathway in Escherichia coli: investigation of the cai-fix divergent promoter region. J. Bacteriol. 180:2599-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busby, S., and A. Kolb. 1996. The CAP modulon, p. 255-279. In E. C. C. Lyn and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. R.G. Landes Company, Georgetown, Tex.
- 8.Castanie-Cornet, M.-P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 9.Charlier, D., M. Roovers, F. V. Vliet, A. Boyen, R. Cunin, Y. Nakamura, N. Glansdorff, and A. Piérard. 1992. Arginine regulon of Escherichia coli K-12: a study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J. Mol. Biol. 226:367-386. [DOI] [PubMed] [Google Scholar]
- 10.De Reuse, H., A. Kolb, and A. Danchin. 1992. Positive regulation of the expression of the Escherichia coli pts operon. Identification of the regulatory regions. J. Mol. Biol. 226:623-635. [DOI] [PubMed] [Google Scholar]
- 11.Duyne, G. D. V., G. Ghosh, W. K. Maas, and P. B. Sigler. 1996. Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol. 256:377-391. [DOI] [PubMed] [Google Scholar]
- 12.Ebright, R. H. 1993. Transcription activation at class I CAP-dependent promoters. Mol. Microbiol. 8:797-802. [DOI] [PubMed] [Google Scholar]
- 13.Ghosaini, L., A. Brown, and J. Sturtevant. 1988. Scanning calorimetric study of the thermal unfolding of catabolic activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry 27:5257-5261. [DOI] [PubMed] [Google Scholar]
- 14.Glansdorff, N. 1996. Biosynthesis of arginine and polyamines, p. 408-433. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 15.Granston, A. E., D. L. Thompson, and D. I. Friedman. 1990. Identification of a second promoter for the metY-nusA-infB operon of Escherichia coli. J. Bacteriol. 172:2336-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikemura, T., and H. Ozeki. 1977. Gross map location of Escherichia coli transfer RNA genes. J. Mol. Biol. 117:419-446. [DOI] [PubMed] [Google Scholar]
- 17.Ishii, S., K. Kuroki, and F. Imamoto. 1984. tRNAf2Met gene in the leader region of the nusA operon in Escherichia coli. Proc. Natl. Acad. Sci. USA 81:409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph, E., C. Bernsley, N. Guiso, and A. Ullmann. 1982. Multiple regulation of the activity of adenylate cyclase in Escherichia coli. Mol. Gen. Genet. 185:262-268. [DOI] [PubMed] [Google Scholar]
- 19.Kenri, T., F. Imamoto, and Y. Kano. 1992. Construction and characterization of an Escherichia coli mutant deficient in the metY gene encoding tRNAf2Met: either tRNAf1Met or tRNAf2Met is required for cell growth. Gene 114:109-114. [DOI] [PubMed] [Google Scholar]
- 20.Kenri, T., F. Imamoto, and Y. Kano. 1994. Three tandemly repeated structural genes encoding tRNA(f1Met) in the metZ operon of Escherichia coli K-12. Gene 138:261-262. [DOI] [PubMed] [Google Scholar]
- 21.Kenri, T., K. Kohno, N. Goshima, F. Imamoto, and Y. Kano. 1991. Construction and characterization of an Escherichia coli mutant with a deletion of the metZ gene encoding tRNAf1Met. Gene 103:31-36. [DOI] [PubMed] [Google Scholar]
- 22.Lange, R., M. Barth, and R. Hengge-Aronis. 1993. Complex transcriptional control of the σs-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J. Bacteriol. 175:7910-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurent-Winter, C., S. Ngo, A. Danchin, and P. Bertin. 1997. Role of Escherichia coli histone-like nucleoid structuring protein in bacterial metabolism and stress response: identification of targets by two-dimensional electrophoresis. Eur. J. Biochem. 244:767-773. [DOI] [PubMed] [Google Scholar]
- 24.Marschall, C., V. Labrousse, M. Kreimer, D. Weichart, A. Kolb, and R. Hengge-Aronis. 1998. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on σs and requires activation by cAMP-CRP. J. Mol. Biol. 276:339-353. [DOI] [PubMed] [Google Scholar]
- 25.Maxam, A., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Nat. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Mori, K., and H. Aiba. 1985. Evidence for negative control of cya transcription by cAMP and cAMP receptor protein in intact Escherichia coli cells. J. Biol. Chem. 260:14838-14842. [PubMed] [Google Scholar]
- 29.Morrissey, J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117:307-310. [DOI] [PubMed] [Google Scholar]
- 30.Nagase, T., S. Ishii, and F. Imamoto. 1988. Differential transcriptional control of the two tRNAfMet genes of Escherichia coli K-12. Gene 67:49-57. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson, L., and V. Emilsson. 1994. Factor for inversion stimulation-dependent growth rate regulation of individual tRNA species in Escherichia coli. J. Biol. Chem. 269:9460-9465. [PubMed] [Google Scholar]
- 32.Regnier, P., and M. Grunberg-Manago. 1989. Cleavage by RNaseIII in the transcripts of the metY-nusA-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J. Mol. Biol. 210:293-302. [DOI] [PubMed] [Google Scholar]
- 33.Reznikoff, W. S. 1992. The lactose operon-controlling elements: a complex paradigm. Mol. Microbiol. 6:2419-2422. [DOI] [PubMed] [Google Scholar]
- 34.Sekowska, A., S. Robin, J.-J. Daudin, A. Henaut, and A. Danchin. 2001. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2:0019.1-0019.12. [DOI] [PMC free article] [PubMed]
- 35.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol resistant bacteria. Methods Enzymol. 43: 742-743. [DOI] [PubMed] [Google Scholar]
- 36.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: Role of H-NS protein and the cyclic AMP-catabolic activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Touati, E., C. Laurent-Winter, P. Quillardet, and M. Hoffnung. 1996. Global response of Escherichia coli cells to a treatment with 7-methoxy-2-nitro-naphtho(2,1-b)furan (R7000), an extremely potent mutagen. Mutat. Res. 349:193-200. [DOI] [PubMed] [Google Scholar]
- 38.Verbeek, H., L. Nilsson, G. Baliko, and L. Bosch. 1990. Potential binding sites of the trans-activator FIS are present upstream of all rRNA operons and of many but not all tRNA operons. Biochim. Biophys. Acta 1050:302-306. [DOI] [PubMed] [Google Scholar]
- 39.Xu, J., and R. C. Johnson. 1997. Cyclic AMP receptor protein functions as a repressor of the osmotically inducible promoter proP P1 in Escherichia coli. J. Bacteriol. 182:4180-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]