Abstract
A chlorate reductase has been purified from the chlorate-reducing strain Pseudomonas chloritidismutans. Comparison with the periplasmic (per)chlorate reductase of strain GR-1 showed that the cytoplasmic chlorate reductase of P. chloritidismutans reduced only chlorate and bromate. Differences were also found in N-terminal sequences, molecular weight, and subunit composition. Metal analysis and electron paramagnetic resonance measurements showed the presence of iron and molybdenum, which are also found in other dissimilatory oxyanion reductases.
Pseudomonas chloritidismutans (strain AW-1) is a recently isolated facultative anaerobic chlorate-reducing bacterium (22). In chlorate-reducing bacteria, chlorate is reduced to chlorite by a chlorate reductase, and in a second reaction, chlorite is disproportionated to chloride and oxygen by a chlorite dismutase. Two closely related strains of P. chloritidismutans, Pseudomonas stutzeri DSM 50227 and DSM 5190T (with 16S ribosomal DNA similarities of 100% and 98.6%, respectively), were not able to grow by dissimilatory chlorate reduction. Accordingly, both P. stutzeri strains lack chlorite dismutase activity. However, in addition to nitrate reductase activity, these strains also showed chlorate reductase activity, which has been observed before for other denitrifying bacteria (6, 15). P. chloritidismutans differed from other (per)chlorate-reducing bacteria in that it was only able to use chlorate as a terminal electron acceptor. Other chlorate-reducing bacteria can also couple the reduction of perchlorate or nitrate to growth (1, 13). Although cell extracts also showed bromate reductase activity, P. chloritidismutans could not grow on bromate (22).
Up to now, two enzymes that can reduce chlorate and/or perchlorate have been purified and characterized. A chlorate reductase C has been purified from the denitrifying strain Proteus mirabilis, as well as two nitrate reductases (15). The only known substrate of chlorate reductase C is chlorate, which was reduced to chlorite. It was not demonstrated that Proteus mirabilis was able to couple the reduction of chlorate to growth. A second (per)chlorate reductase has been purified from strain GR-1 (10). Experiments showed that one enzyme is responsible for both chlorate and perchlorate reduction activity. Besides (per)chlorate, nitrate, iodate, and bromate were also reduced by the (per)chlorate reductase of strain GR-1. Perchlorate-grown cells were unable to oxidize nitrate or nitrite, indicating that another nitrate reductase may be involved in nitrate-grown cells (17). The purified chlorate reductase of P. chloritidismutans reported here is the first chlorate reductase derived from a chlorate-reducing bacterium that is capable of only dissimilatory chlorate reduction.
P. chloritidismutans (DSM 13592) was grown under strictly anaerobic conditions at 30°C, as described before (1). Strain GR-1 (DSM 11199) was grown as described previously (10, 17). Chlorate (10 mM) was used as the electron acceptor, while acetate (10 mM) was used as the electron donor and carbon source. Cell extracts of P. chloritidismutans lost chlorate reduction activity when stored under air. Therefore, the preparation of cell extracts and all purification steps were done in an anaerobic glove box containing an atmosphere of 96% N2 and 4% H2, and purified enzyme fractions were kept on ice (4°C). Cells were collected by centrifugation at 9,000 rpm for 10 min at 4°C. The cell pellet was suspended (1:2 [wt/vol]) in 15 mM potassium-sodium phosphate buffer, pH 7.2 (buffer A), and cells were disrupted by ultrasonic disintegration (Sonics & Materials Inc., Danbury, Conn.). Cell debris and whole cells were removed by centrifugation at 13,000 rpm for 10 min at 4°C. The supernatant fraction was subjected to an ultracentrifugation step, 110,000 × g for 1 h at 4°C. This resulted in a red supernatant containing the cytoplasmic and periplasmic fraction. The pellet (membrane fraction) was suspended in buffer A.
Enzyme purification was started by loading the red supernatant fraction onto a Q-Sepharose column (2 by 30 cm) equilibrated with 50 mM Tris-HCl buffer, pH 7.5. The chlorate reductase eluted at the start of a linear gradient of 0 to 1 M potassium chloride in 50 mM Tris-HCl buffer, pH 7.5. Active fractions were pooled and desalted by ultrafiltration (150-ml stirred cell; filter pore size, 10 kDa; Filtron Technology). The concentrated enzyme fraction was diluted with 10 mM Tris-HCl, pH 7.2. This fraction was loaded on a hydroxyapatite column equilibrated with 10 mM Tris-HCl, pH 7.2. A linear gradient was established from 10 mM Tris-HCl, pH 7.2, to 450 mM potassium phosphate, pH 7.2. Active fractions were combined and loaded onto a Mono-Q column which was equilibrated with 50 mM Tris-HCl buffer, pH 7.5. A linear gradient was established from 0 to 1 M potassium chloride in 50 mM Tris-HCl buffer, pH 7.5.
A 400-μl aliquot of the active fraction was subsequently loaded onto a Superdex 200 column (1.6 by 70.5 cm) equilibrated with 50 mM potassium phosphate buffer, pH 7.0, containing 100 mM NaCl. Chlorate reductase activity was measured anaerobically in stoppered glass cuvettes by monitoring the oxidation of reduced methyl viologen at 578 nm and 30°C in a Hitachi spectrophotometer (U-2010) as described earlier (6). The following electron acceptors (all sodium salts) were tested: ClO4−, ClO3−, ClO2−, NO3−, NO2−, BrO3−, SO42−, SeO42−, IO3−, and IO4−. The enzyme activity was measured at different pHs (5.5 to 10) and temperatures (30 to 90°C). Kinetic parameters were obtained by a computer-aided direct fit of the Michaelis-Menten curve. The chlorate concentration was varied between 10 μm and 4 mM.
Chlorite dismutase and catalase activities were determined by measuring oxygen production with a Clark-type electrode (Yellow Springs Instruments, Yellow Springs, Ohio) as described earlier (22). One unit of activity is defined as the amount of enzyme required to convert 1 μmol of substrate per minute. The protein content of the enzyme fractions was determined by the method of Bradford, with bovine serum albumin as the standard (4).
For localization of the chlorate reductase, a culture of P. chloritidismutans or strain GR-1 was centrifuged for 10 min at 9,000 rpm and 4°C. The cell pellet was suspended (1:2 [wt/vol]) in EDTA buffer (50 mM Tris, 50 mM EDTA, 170 mM Na2CO3, pH 9) (20). This cell suspension was incubated for 30 min at room temperature, followed by a centrifugation step (13,000 rpm, 10 min at 4°C). The red supernatant (periplasmic fraction) was separated from the pellet. The pellet was suspended in buffer A (1:2 [wt/vol]), disrupted by ultrasonic disintegration, and further processed as described above.
A gentler method to disrupt the cells is a freeze-thaw procedure (23). A cell pellet was suspended in buffer A. After the suspension was frozen in liquid N2, it was thawed by immersing it in running water (40°). This procedure was repeated four times. A small amount of DNase I (Sigma) was added to reduce the viscosity due to the released DNA. The solution was centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant (cell extract) was centrifuged at 110,000 × g for 1 h at 4°C. The supernatant (cytoplasmic and periplasmic fraction) was separated from the pellet. The pellet (membrane fraction) was suspended in buffer A.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (12) with 12% acrylamide gels. To calculate the molecular mass of the subunits and the native enzyme, the SDS-PAGE and Superdex 200 column were calibrated with standard proteins. For determination of the N terminus of the subunits, the subunits were blotted onto a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad) following the manufacturer's instructions. The N-terminal sequences were determined as described earlier (18).
Electron paramagnetic resonance spectra were recorded on a Bruker ER-200D spectrometer with peripheral equipment and data handling as described previously (16). The modulation frequency was 100 kHz. The presence of metals was measured by inductively coupled plasma mass spectrometry (Elan 6000; Perkin-Elmer).
During the first purification step, the chlorate reductase was separated from a red fraction which eluted just before the chlorate reductase and contained chlorite dismutase activity. On the other columns used, the chlorate reductase eluted at the start of the linear gradient. These fractions had a brown-reddish color. The purification scheme is shown in Table 1. The active fraction, from the Superdex 200 column, was run on SDS-PAGE, and three bands were obtained, with molecular masses of 97, 38, and 34 kDa. The molecular mass for the native enzyme was about 167 kDa, as determined by gel filtration with the Superdex 200 column. These results suggest that all subunits are present in a 1:1:1 stoichiometry and that the chlorate reductase is a heterotrimer (α1β1γ1). The N-terminal sequences of this enzyme can be retrieved through the Swiss-Prot sequence database, European Bioinformatics Institute, under accession numbers P83448, P83449, and P83450. The same stoichiometry has been found for the chlorate reductase C from Proteus mirabilis (Table 2), although the sizes of the subunits were different. Chlorate reductase of strain GR-1 showed a different stoichiometry, resulting in a different molecular mass of the native enzyme (420 kDa). This enzyme was found to consist of an α and a β subunit, but a third (γ) subunit was anticipated (Table 2).
TABLE 1.
Summary of the purification of chlorate reductase from P. chloritidismutans
| Step | Vol (ml) | Protein concn (mg/ml) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Soluble fraction | 13 | 27.5 | 726 | 2 | 100 | 1 |
| Q-Sepharose | 20 | 2.8 | 335 | 6 | 46.1 | 3 |
| Hydroxyapatite | 5 | 3.5 | 155 | 8.7 | 21.4 | 4.4 |
| Mono-Q | 2 | 3.3 | 95 | 14.4 | 13.1 | 7.2 |
| Superdex 200 | 10 | 0.18 | 75 | 42 | 10.3 | 21 |
TABLE 2.
Summary of enzyme characteristics
| Enzyme | Localization | Electron acceptor(s) | Subunits (kDa) | Native enzyme composition | EPR of Mo(V) (g values) | ClO3−
|
|
|---|---|---|---|---|---|---|---|
| Vmax (U/mg) | Km (μM) | ||||||
| P. mirabilis chlorate reductase C | Membrane | Chloratea | 75, 63, 56 | Heterotrimer (α1β1γ1) | |||
| GR-1 (per)chlorate reductase | Periplasm | Perchlorate, chlorate, nitrate, iodate, bromate | 95, 40 | Trimers of heterodimers (α3β3) | 2.002, 2.017 | 13.2 | <5 |
| P. chloritidismutans chlorate reductase | Cytoplasm | Chlorate, bromateb | 97, 38, 34 | Heterotrimer (α1β1γ1) | 2.002, 2.024, 2.076 | 51.3 | 159 |
Electron acceptors other than chlorate and nitrate were not tested.
ClO4−, ClO2−, NO3−, NO2−, IO4−, IO3−, SO42−, and SeO42− were tested but did not show reductase activity.
For chlorate, Km and Vmax values of 159 μM and 51 U/mg were found, respectively. Besides chlorate, only bromate served as an electron acceptor. Under standard conditions, specific activities of 50 and 26 U/mg were found for chlorate and bromate, respectively. Because bromate could not be used as an electron acceptor for growth (1), the chlorate reductase of P. chloritidismutans is apparently not involved in another oxyanion dissimilatory reduction pathway. The enzyme from strain GR-1 showed activity with chlorate, perchlorate, bromate, iodate, and nitrate (6).
The optimum pH for the chlorate reductase was pH 7.5. At pH 5.5 and 10, the specific activity was approximately 50% of the optimal value at pH 7.5. The purified enzyme had a temperature optimum of 75°C. This is remarkable because P. chloritidismutans is a mesophilic bacterium (1), which does not grow above 42°C. The optimal temperature of the enzyme from strain GR-1 was only 45°C. However, the trimethylamine N-oxide (TMAO) reductase of Salmonella enterica serovar Typhimurium also functions at higher temperatures (below 75°C) than the optimal growth temperature, which was 37°C (11). Remarkably, besides the reduction of TMAO, this enzyme also showed chlorate and bromate but not nitrate reduction activity (11, 21).
Analysis of the N-terminal sequences of the three subunits via a BLAST search in the databases did not result in any similar N-terminal sequence for the α (LNMLEPVGETLASEYPYXK; Swiss-Prot P83448) and γ (EXSEQNPNILEIKPGDTVKVXT; Swiss-Prot P83450) subunits. However, a significant similarity was found for the N-terminal sequence of the β subunit (TVKXQLSMVLDLNKEIGGQTXTAA; Swiss-Prot P83449) when this sequence was compared to the β subunit of the dimethylsulfide dehydrogenase of Rhodovulum sulfidophilum (73%) (14), the β subunit of the nitrate reductase of Haloarcula marismortui (61%) (24), the anaerobic ethylbenzene dehydrogenase subunit B of Azoarcus sp. strain EB1 (60%) (9), and the β subunit of the selenate reductase of Thauera selenatis (55%) (19). The similarity to the β subunit of the (per)chlorate reductase of strain GR-1 was 27%. A common characteristic of these enzymes is the presence of molybdenum.
Chlorate reductase activity was found mainly in the cytoplasmic fraction (Table 3). In contrast to the chlorate reductase, chlorite dismutase activity was found predominantly in the periplasmic fraction. The absence of catalase activity in the periplasmic fraction confirmed the efficiency of the localization method used. It is known that nitrate reductases are located almost exclusively in the cytoplasmic membrane (5). By disrupting cells of H. marismortui with an ultrasonic oscillating device, it was shown that nitrate reducing activity was easily released into the soluble fraction (10). With a gentler method (freeze-thaw procedure), cells of H. marismortui were disrupted, and the nitrate reductase was extracted from the resulting membrane fraction. Therefore, the enzyme was considered an extrinsic membrane protein that binds to the surface of the cytoplasmic membrane by hydrophobic interaction. This freeze-thaw procedure was also used for cells of P. chloritidismutans and resulted in the highest chlorate reductase activity in the cytoplasmic/periplasmic fraction (95.7 U) compared to the membrane fraction (8.2 U). Therefore, it is suggested that the chlorate reductase of P. chloritidismutans is cytoplasmic.
TABLE 3.
Localization of chlorate reductase, chlorite dismutase, and catalase activity in cell fractions of strain AW-1
| Fraction | ClO3− reductase (U/mg) | ClO3− reductase (U total) | ClO2− dismutase (U/mg) | ClO2− dismutase (U total) | Catalase (U/mg) | Catalase (U total) |
|---|---|---|---|---|---|---|
| Periplasmic | 6.2 | 7.5 | 236 | 284 | NAa | NA |
| Cytoplasmic | 29.6 | 47.2 | 20.6 | 32.7 | 15.7 | 24.9 |
| Membrane | 3.8 | 2 | 9.3 | 4.8 | 3.3 | 1.7 |
NA, no activity.
It is not yet clear how the respiratory pathway is arranged in chlorate-reducing bacteria. The (per)chlorate reductase of strain GR-1 is located in the periplasm, whereas the chlorate reductase described here is located in the cytoplasm. This means that the ways in which energy is conserved in these bacteria must also be different. The opposite locations of the chlorate reductase and chlorite dismutase (Table 3) imply that transport systems are required to transport chlorate from the periplasm to the cytoplasm and to transport chlorite back to the periplasm. Because chlorite is highly oxidative, it might be favorable to translocate chlorite back to the periplasm. We do not have any insight into the transport of chlorate and chlorite. However, a chlorate-chlorite antiport system might be most appropriate to avoid accumulation of toxic chlorite in the cell. A similar situation exists in Azospirillum brasilense Sp7 (5) and Paracoccus denitrificans (2, 3), both denitrifying bacteria that possess cytoplasmic nitrate reductases and periplasmic nitrite reductases. It was proposed that nitrate uptake might proceed via an energy-independent nitrate-nitrite antiport system. This process prevents intracellular accumulation of toxic levels of nitrite and allows further detoxification in the periplasm through the action of nitrite reductase.
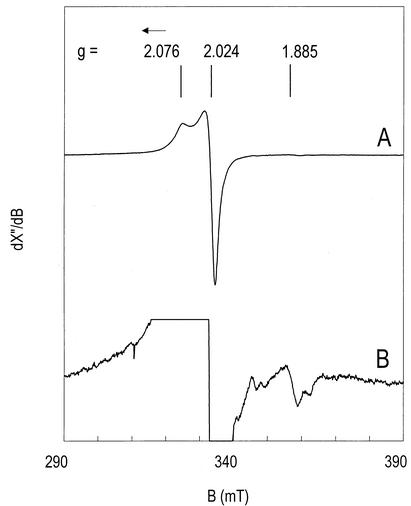
Electron paramagnetic resonance analysis of the anaerobically purified enzyme revealed an unusual signal with all g values greater than the free-electron value, ge = 2.002, namely, gz = 2.076 and gxy = 2.024 (Fig. 1, trace A). Double integration versus an external copper standard gave a spin count of 0.2 S = 1/2 per 167-kDa α1β1γ1 heterotrimer. The signal was very similar in shape and integrated intensity to that of the α3β3 heterohexameric (per)chlorate reductase from strain GR-1 (6). Previously, this signal was suggested to come from Mo(V) with Se coordination (6). In line with this suggestion, we find that the intensity of the present signal was reduced to 0.13 spins upon incubation with sodium dithionite; it diminished further to 0.05 spins upon light reduction with the deazaflavin-EDTA system.
FIG. 1.
Electron paramagnetic resonance spectra of P. chloritidismutans chlorate reductase. The enzyme concentration was 2.3 mg/ml or 14 μM. Trace A is from the enzyme isolated anaerobically; trace B is from dithionite-reduced enzyme. The amplification for trace B is 25 times that for trace A. Electron paramagnetic resonance conditions: microwave frequency, 9.43 GHz; microwave power, 126 mW; modulation frequency, 100 kHz; modulation amplitude, 0.63 mT; temperature, 16 K.
The present enzyme did contain molybdenum, but selenium was not measured. The molar ratio of [Fe] to [Mo] of ≈16 was different from the molar ratio of [Fe] to [Mo] of ≈11 found for strain GR-1 (6). Moreover, in the GR-1 enzyme, a complex 2[4Fe-4S]1+ spectrum developed upon dithionite reduction with an intensity of approximately 2 spins, and oxidation with potassium ferricyanide afforded a [3Fe-4S]1+ signal equivalent to 1 spin (6). In contrast to this, we found only very weak Fe/S signals for the P. chloritidismutans enzyme. The Fe/S spectrum integrated to approximately 1% of the protein concentration. Addition of dithionite caused an increase to some 0.02 spin (Fig. 1, trace B). The amplitude did not increase further upon light reduction. The lack of Fe/S electron paramagnetic resonance signals is puzzling; however, a few examples of [4Fe-4S] cluster-containing enzymes in which (part of) the clusters are electron paramagnetic resonance silent are known (7). Nevertheless, based on these results, we suggest that the Fe in the protein is nonheme iron.
The purification of both the (per)chlorate reductase of strain GR-1 and the chlorate reductase of P. chloritidismutans showed that molybdenum enzymes are involved in (per)chlorate reduction. In an overview by Hille (8) about mononuclear molybdenum enzymes, the dimethyl sulfoxide reductase family is described. It is remarkable that in this dimethyl sulfoxide reductase family, two other enzymes are present, the TMAO reductase and the dissimilatory nitrate reductases, both of which show chlorate reductase activity. The results shown in this paper suggest that the chlorate reductase of P. chloritidismutans is different from the nitrate reductases. Sequencing of the chlorate reductase gene coupled with crystallographic information and detailed spectroscopic characterization by electron paramagnetic resonance can give more insight into the mechanism of action of the chlorate reductase in comparison to dimethyl sulfoxide reductases.
Acknowledgments
This work was supported by the Earth and Life Sciences Foundation (ALW), which is subsidized by the Netherlands Organization for Scientific Research (NWO).
REFERENCES
- 1.Achenbach, L. A., U. Michaelidou, R. A. Bruce, J. Fryman, and J. D. Coates. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. E vol. Microbiol. 51:527-533. [DOI] [PubMed] [Google Scholar]
- 2.Boogerd, F. C., H. W. van Verseveld, and A. H. Stouthamer. 1983. Dissimilatory nitrate uptake in Paracoccus denitrificans via a ΔμH-dependent system and a nitrate-nitrite antiport system. Biochim. Biophys. Acta 723:415-427. [Google Scholar]
- 3.Boogerd, F. C., H. W. van Verseveld, D. Torenvliet, M. Braster, and A. H. Stouthamer. 1984. Reconsideration of the efficiency of energy transduction in Paracoccus denitrificans during growth under a variety of culture conditions. Arch. Microbiol. 139:344-350. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Danneberg, G., W. Zimmer, and H. Bothe. 1989. Energy transduction efficiencies in nitrogenous oxide respirations of Azospirillum brasilense Sp7. Arch. Microbiol. 151:445-453. [Google Scholar]
- 6.De Groot, G. N., and A. H. Stouthamer. 1969. Regulation of reductase formation in Proteus mirabilis. I. Formation of reductases and enzymes of formic hydrogenase complex in the wild type and in chlorate-resistant mutants. Arch. Microbiol. 66:220-233. [PubMed] [Google Scholar]
- 7.Hagen, W. R., M. A. Vanoni, K. Rosenbaum, and K. D. Schnackerz. 2000. On the iron-sulfur clusters in the complex redox enzyme dihydropyrimidine dehydrogenase. Eur. J. Biochem. 267:3640-3646. [DOI] [PubMed] [Google Scholar]
- 8.Hille, R. 1996. The mononuclear molybdenum enzymes. Chem. Rev. 96:2757-2816. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, H. A., D. A. Pelletier, and M. Spormann. 2001. Isolation and characterization of anaerobic ethylbenzene dehydrogenase, a novel Mo-Fe-S enzyme. J. Bacteriol. 183:4536-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kengen, S. W. M., G. B. Rikken, W. R. Hagen, C. G. van Ginkel, and A. J. M. Stams. 1999. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J. Bacteriol. 181:6706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwan, H. S., and E. L. Barrett. 1983. Purification of trimethylamine oxide reductase from Salmonella typhimurium. J. Bacteriol. 155:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed]
- 13.Logan, B. E. 1998. A review of chlorate- and perchlorate-respiring microorganisms. Bioremed. J. 2:69-79. [Google Scholar]
- 14.McDevitt, C. A., P. Hugenholtz, G. R. Hanson, and A. G. McEwan. 2002. Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes. Mol. Microbiol. 44:1575-1587. [DOI] [PubMed] [Google Scholar]
- 15.Oltmann, L. P., W. N. M. Reijnders, and A. H. Stouthamer. 1976. The correlation between the protein composition of cytoplasmic membranes and the formation of nitrate reductase A, chlorate reductase C and tetrathionate reductase in Proteus mirabilis wild type and some chlorate resistant mutants. Arch. Microbiol. 111:37-43. [DOI] [PubMed] [Google Scholar]
- 16.Pierik, A. J., and W. R. Hagen. 1991. S=9/2 electron paramagnetic resonance signals are evidence against coupling between the siroheme and the Fe/S cluster prosthetic groups in Desulfovibrio vulgaris (Hildenborough) dissimilatory sulfite reductase. Eur. J. Biochem. 195:505-516. [DOI] [PubMed] [Google Scholar]
- 17.Rikken, G. B., A. G. M. Kroon, and C. G. van Ginkel. 1996. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45:420-426. [Google Scholar]
- 18.Schiltz, E., A. Kreusch, U. Nestel, and G. E. Schulz. 1991. Primary structure of porin from Rhodobacter capsulatus. Eur. J. Biochem. 199:587-594. [DOI] [PubMed] [Google Scholar]
- 19.Schröder, I., S. Rech, T. Krafft, and J. M. Macy. 1997. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 272:23765-23768. [DOI] [PubMed] [Google Scholar]
- 20.Sebban, C., L. Blanchard, L. Bruschi, and F. Guerlesquin. 1995. Purification and characterization of the formate dehydrogenase from Desulfovibrio vulgaris Hildenborough. FEMS Microbiol. Lett. 133:143-149. [DOI] [PubMed] [Google Scholar]
- 21.Shimokawa, O., and M. Ishimoto. 1979. Purification and some properties of inducible tertiary amine N-oxide reductase from Escherichia coli. J. Biochem. 86:1709-1717. [DOI] [PubMed] [Google Scholar]
- 22.Wolterink, A. F. W. M., A. B. Jonker, S. W. M. Kengen, and A. J. M. Stams. 2002. Pseudomonas chloritidismutans sp. nov., a nondenitrifying, chlorate-reducing bacterium. Int. J. Syst. E vol. Microbiol. 52:2183-2190. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimatsu, K., T. Sakurai, and T. Fujiwara. 2000. Purification and characterization of dissimilatory nitrate reductase from a denitrifying halophilic archaeon, Haloarcula marismortui. FEBS Lett. 470:216-220. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimatsu, K., T. Iwasaki, and T. Fujiwara. 2002. Sequence and electron paramagnetic resonance analyses of nitrate reductase NARGH from a denitrifying halophilic euryarchaeote, Haloarcula marismortui. FEBS Lett. 516:145-150. [DOI] [PubMed] [Google Scholar]
- 25.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]