Abstract
Members of the AraC/XylS family of transcriptional regulators are usually organized in two domains: a conserved domain made up of 100 amino acids and frequently located at the C-terminal end, involved in DNA binding; and an N-terminal nonconserved domain involved in signal recognition, as is the case for regulators involved in the control of carbon metabolism (R. Tobes and J. L. Ramos, Nucleic Acids Res. 30:318-321, 2002). The XylS protein, which is extremely insoluble, controls expression of the meta-cleavage pathway for alkylbenzoate metabolism. We fused the N-terminal end of XylS to the maltose-binding protein (MBP) in vitro and found in glutaraldehyde cross-linking assays that the protein dimerized. Experiments with a chimeric N-terminal XylS linked to a ′LexA protein showed that the dimer was stabilized in the presence of alkylbenzoates. Sequence alignments with AraC and UreR allowed us to identify three residues, Leu193, Leu194, and Ile205, as potentially being involved in dimerization. Site-directed mutagenesis of XylS in which each of the above residues was replaced with Ala revealed that Leu193 and Leu194 were critical for activity and that a chimera in which LexA was linked to the N terminus of XylSLeu193Ala or XylSLeu194Ala was not functional. Dimerization of the chimeras MBP-N-XylSLeu193Ala and MBP-N-XylSLeu194Ala was not observed in cross-linking assays with glutaraldehyde.
The TOL plasmid pWW0 of Pseudomonas putida encodes information for the catabolism of benzoate and alkylbenzoates through a meta-cleavage pathway. In this pathway, the aromatic carboxylic acids are first oxidized to the corresponding catechols, which undergo meta-cleavage fission to yield a derivative of muconic acid semialdehyde, which in turn is further metabolized to Krebs cycle intermediates (32). The genes that encode the enzymes of the meta-cleavage pathway form an operon in pWW0. The xylS gene, which encodes the regulator of the meta-cleavage pathway, is located at 3′ end with respect to the meta operon, and it is transcribed convergently with this operon (10). The XylS protein is synthesized constitutively at a low level and becomes transcriptionally active when a benzoate effector such as 3-methylbenzoate (3MBz) is added to the culture medium (4, 7, 34). However, when xylS is overproduced, it stimulates transcription from Pm (the promoter of the meta cleavage pathway) in the absence of effectors, albeit at a rate lower than that achieved with 3MBz.
The XylS protein binds at the −34 to −68 region of the Pm promoter and contacts the α subunit of RNA polymerase (6, 16, 36, 37), which uses the σ32 factor for transcription in the early logarithmic phase of growth and the σ38 factor thereafter to mediate transcription from the Pm promoter (23, 36).
The XylS protein is a member of the AraC/XylS family of positive transcriptional regulators, which includes more than 270 different bacterial proteins involved in the control of processes related to carbon metabolism, stress response, and pathogenesis (3, 5, 14, 21, 24, 27, 42). Many of the proteins in this family are about 300 amino acids long and are made of two domains, a nonconserved domain which seems to be involved in effector/signal recognition and dimerization, and a conserved domain characterized by significant amino acid sequence homology, which extends over 100-residue stretches and contains the bipartite DNA binding domain, made of two α-helix-turn-α-helix motifs. Some proteins in the family are extremely insoluble, although others, such as MarA and Rob, have been purified in soluble form and crystallized and their three-dimensional structure has been resolved (17, 35).
The N-terminal domain of XylS seems to be involved in effector recognition and XylS activation, as deduced from the isolation of XylS mutants with altered effector specificity (i.e., Arg45→Thr, Cys41→Gly, Asp137→Glu, and His153→Gln) or impaired effector recognition (i.e., Arg41→Leu and Asp137→Leu) (26, 33, 34). This has been taken as evidence that the XylS binding pocket for aromatic carboxylic acids consists of patches along the primary sequence of XylS.
AraC is the best-characterized protein in this family, and its N-terminal domain has been resolved by X-ray diffraction (38, 40). This domain is now known to be involved in effector recognition and dimerization. Three critical leucines (Leu150, Leu151, and Leu161) are involved in AraC dimerization (19, 40). UreR is another member of the AraC/XylS family, and it also contains three conserved leucine residues in the same relative locations with the same spatial distance relative to each other as in AraC (Leu147, Leu148, and Leu158) (31). Ramos et al. (33) noticed some sequence conservation at the N-terminal end of the XylS protein with respect to AraC and observed that XylS also exhibited a set of leucines conserved near the linker of the N- and C-terminal domains, corresponding to Leu193 and 194, and Ile205 (Fig. 1). Given the intrinsic insolubility of XylS, it is still unknown whether the protein is a dimer or not and, if it is a dimer, whether these three leucines play a role in dimerization.
FIG. 1.
Partial alignment of AraC, XylS, and UreR. Leucine residues 150, 151, and 161 in AraC are critical for AraC dimerization (39). Leucines 147, 148, and 157 in UreR are critical for dimerization of UreR (30). Leucines 193 and 194 and isoleucine 205 in XylS align with the leucine residues of AraC and UreR that are critical for dimerization.
In this study, we constructed fusion proteins to determine whether the N-terminal end of XylS also dimerizes and to determine whether the two leucine and the isoleucine residues of XylS that align with the leucines of AraC and UreR are involved in XylS dimerization and activity.
MATERIALS AND METHODS
Bacterial strains, culture medium, and cloning vectors.
The bacterial strains used in this study are shown in Table 1. All strains were grown in Luria-Bertani (LB) medium at 30°C with shaking (200 strokes per min in a Kühner incubator). Relevant characteristics of the cloning vectors used for subcloning are shown in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strains or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 gyrA96 relA1 | 8 |
| JL1436 | deoC1 ptsF25 rbsR PsulA::′lacZ | 1 |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 | 18 |
| Plasmids | ||
| pCMX2 | pSELECT-1, xylS ori F1, ColE1, Tcr | 22 |
| pERD100 | pMP220, Pm:′lacZ, IncP1, Tcr | 33 |
| pGB002 | pSE380, LexA DNA binding domain (amino acids 1-87) | 1 |
| pLOW2 | pACYC177, p15A, Kmr | 9 |
| pLRRA1 | pLOW2 derivative bearing xylS mutant allele encoding XylSL193A | This study |
| pLRRA2 | As pLLAR1 but encoding XylSL194A | This study |
| pLRRA3 | As pLLAR1 but encoding XylSI205A | This study |
| pLRRA4 | As pLLAR1 but encoding XylSL193A, L194A | This study |
| pLRRA5 | As pLLAR1 but encoding XylS193A, 1205A | This study |
| pLRRA6 | As pLLAR1 but encoding XylSL194A, 1205A | This study |
| pLRRA7 | As pLLAR1 but encoding XylSL193A, L194A, I205A | This study |
| pLRRA8 | pGB002 derivative encoding chimeric N-XylS-LexA protein | This study |
| pLRRA9 | As pLRRA8 but encoding N-XylS L193A-LexA protein | This study |
| pLRRA10 | As pLRRA8 but encoding N-XylS L194A-LexA protein | This study |
| pLRRA11 | As pLRRA8 but encoding N-XylS I205A-LexA protein | This study |
| pLRRA12 | As pLRRA8 but encoding N-XylS L193A, L194A-LexA protein | This study |
| pLRRA13 | As pLRRA8 but encoding N-XylS L193A, I205A-LexA protein | This study |
| pLRRA14 | As pLRRA8 but encoding N-XylS L194A,I205A-LexA protein | This study |
| pLRRA15 | As pLRRA8 but encoding N-XylS L193A,L194A,I205A-LexA protein | This study |
| pMAL-pV | pMAL-C2, PtalmalE lacIq | 28, 29 |
Apr, Kmr, Smr, and Tcr stand for resistance to ampicillin, kanamycin, streptomycin, and tetracycline, respectively.
Construction of xylS mutants by PCR.
The xylS mutants were generated by overlap extension PCR mutagenesis (12, 13) with internal oligonucleotide primers that exhibited one or more mismatches with respect to the wild-type sequence. The forward and reverse primers were 5′-GCTATCTCAGTTATACTACG-3′ and 5′-CGAGAAATTTATCGTTAAATTGCC-3′, respectively. After DNA amplification, the resulting DNA was digested with XhoI and MfeI, and the 379-bp XhoI-MfeI xylS mutant fragments were inserted between the XhoI and MfeI sites of pCMX2 (22) to yield plasmids pCMX2::xylS* (the asterisk indicates that one or more of the amino acids in the wild-type protein have been changed). All the xylS mutant alleles generated in this study were verified by DNA sequencing. Plasmids bearing the xylS mutant alleles were digested with EcoRI and XbaI, and the 1,609-bp EcoRI-XbaI fragments, which contained the entire set of xylS mutant alleles, were subcloned between the EcoRI and XbaI sites of pLOW2 to generate plasmids pLRRA1 through pLRRA7, which encoded the mutant XylS proteins shown in Table 2.
TABLE 2.
β-Galactosidase activity of E. coli JL1436 expressing the N-terminal domain of XylS and the XylS mutants in the LexA-based two-hybrid systema
| XylS protein | β-Galactosidase (Miller units)
|
|
|---|---|---|
| Without 3MBz | With 3MBz | |
| None | 4,280 ± 400 | 4,380 ± 400 |
| N-XylS (wild type) | 4,100 ± 300 | 1,300 ± 100 |
| N-XylSL193→A | 4,100 ± 300 | 3,000 ± 250 |
| N-XylSL194→A | 3,950 ± 200 | 3,800 ± 30 |
| N-XylSI205→A | 3,890 ± 284 | 1,250 ± 80 |
| N-XylSL193,L194→A,A | 4,450 ± 350 | 4,750 ± 100 |
| N-XylSL193,I205→A,A | 4,200 ± 280 | 4,050 ± 300 |
| N-XylSL194,I205→A,A | 4,150 ± 300 | 4,000 ± 300 |
| N-XylSL193,L194,I205→A,A,A | 4,200 ± 400 | 4,400 ± 350 |
E. coli Jl1436 bearing a plasmid encoding LexA, LexA-N-Xyls, or LexA-N-XylS* protein was grown, and β-galactosidase was assayed as described in Materials and Methods. Data (values are rounded) are the averages and standard deviations of at least four independent assays performed in triplicate.
Chimeric fusions of N-terminal domain of XylS* to MalE protein or LexA protein.
All cloning steps were performed in Escherichia coli DH5α (8). The fragment of the gene that encodes the N-terminal half of XylS or XylS* (amino acids 1 to 213) was amplified from plasmid pCMX2 or its derivatives generated in this study by PCR with the forward primer 5′-GAACCGGGATCCATGGATTTTTGC-3′ and the reverse primer 5′-GAATTGGTCGACTCACTAGAAAGACG-3′. The PCR product was digested with BamHI and SalI and ligated into the expression vector pMAL-pV (29) cut with the same enzymes to obtain plasmids pMAL-NXylS and pMAL-NXylS*, respectively. Alternatively, we used 5′-GAACCGGAATTCATGGATTTTTGC-3′ as the forward primer and 5′-GAATTGAACGACGGATCCGAAAGACG-3′ as the reverse primer for PCR amplification, and upon digestion of the PCR product with BamHI and EcoRI, it was ligated into the pGB002 vector digested with the same enzymes to produce plasmids pLRRA8 through to pLRRA15 and yield the N-XylS-LexA and N-XylS*-LexA chimeric proteins. The identity of the insert in all resulting plasmids described above was confirmed by DNA sequencing.
Purification of MalE-N-XylS* proteins.
Overnight cultures (10 ml) of E. coli DH5α(pMAL-NXylS) and E. coli DH5α(pMAL-NXylS*) were inoculated into 1 liter of LB medium supplemented with glucose and 100 μg of ampicillin per ml and grown at 30°C. When the cultures reached a turbidity at 600 nm of 0.6, isopropyl-β-d-galactopyranoside (IPTG) was added to reach a concentration of 1 mM, and incubation was continued for 4 h. Cells were then harvested by centrifugation (5,000 × g for 10 min) and stored at −20°C until use.
Frozen cells were thawed and suspended in 50 ml of column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA) supplemented with complete mini-protease inhibitor cocktail (Roche Molecular Biochemical) and then disrupted by passing cells at 10,000 lb/in2 through a French press. All steps were performed at 4°C. The cell lysate was centrifuged at 9,000 × g for 30 min, and the supernatant was collected and diluted fivefold in column buffer and then loaded onto an XK16 column (Amersham Pharmacia Biotech) with 15 ml of amylose resin equilibrated with column buffer. The column was washed with 180 ml of column buffer. MalE-N-XylS and MalE-N-XylS* proteins were eluted with column buffer and 10 mM maltose. The procedure yielded almost homogenous proteins, as judged by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and staining of proteins with Coomassie brilliant blue R-250.
Glutaraldehyde cross-linking assays.
In vitro cross-linking assays were done as described by Lu and Abdelal (19). Aliquots of 300 pmol of MalE, MalE-N-XylS, or MalE-N-XylS* were incubated for 1 h at room temperature with and without 100 μM 3MBz. Each aliquot was then divided in two; one served as a control, and glutaraldehyde was added to the other to reach a final concentration of 0.005% (vol/vol). The final volume of each sample was adjusted to 20 μl with column buffer. After 2 h at room temperature 10-μl aliquots of the reaction mixtures were mixed with 10 μl of loading buffer (100 mM Tris-HCl [pH 6.8], 0.4% [wt/vol] SDS, 20% [vol/vol] glycerol and 0.001% [wt/vol] bromophenol blue) and subjected to boiling in a water bath for 5 min. Then, the proteins were separated by SDS-8% (wt/vol) PAGE, and the products were stained with Coomassie brilliant blue R-250.
β-Galactosidase expression assays.
To determine expression from the PsulA promoter in E. coli JL1436 bearing plasmids encoding the different XylS-LexA fusions constructed in this study (see Table 1), fresh medium was inoculated with a single colony from LB-agar plates containing the appropriate antibiotics and cultured at 30°C overnight. These cultures were diluted 100-fold in the same medium supplemented or not with 1 mM 3MBz, and cell growth was monitored over time. When the cultures reached an optical density at 600 nm of ≈0.4 to 0.6, IPTG was added to reach a concentration of 2 mM, and incubation was continued for 1 h. β-Galactosidase activity was then determined as described by Platt et al. (30).
To determine the induction capacity of the XylS mutants, E. coli MC4100(pERD100) was transformed with plasmids bearing the wild-type and mutant xylS alleles in pLRRA1 to pLRRA7 (pERD100 is an IncQ group plasmid that carries a fusion of Pm to a promoterless ′lacZ gene and encodes resistance to tetracycline [33]). These cells were grown overnight at 30°C in LB medium containing the appropriate antibiotics. Cultures were diluted 100-fold in the same medium supplemented or not with 1 mM 3MBz. After 5 h of incubation, β-galactosidase activity was assayed in permeabilized whole cells. All constructs were assayed in triplicate in three or more independent assays.
RESULTS AND DISCUSSION
Construction of chimeric fusion proteins containing the N-terminal domain of XylS.
Based on the crystallographic data for the MarA and Rob proteins (17, 35) and the alignment of the 270 AraC/XylS family members (42), the first 213 amino acids were considered to constitute the N-terminal region of XylS (N-XylS). On the basis of the biochemical analysis and the crystallographic data for the N-terminal end of the AraC protein, we deduced that the XylS stretch includes the linker arm between the N-terminal and C-terminal domains (11, 40, 43).
Using the appropriate primers, we amplified the segment of the xylS gene that would give rise to the N-XylS′ polypeptide by PCR. This amplified fragment was subcloned in different vectors to provide either a hexahistidine tail or fusions to thioredoxin, glutathione S-transferase, or the maltose-binding protein (MBP). Of all these constructions, the only one that yielded a partially soluble chimera was N-XylS-MBP, in agreement with the finding of Kapust and Waugh (15) that MBP is uncommonly effective in promoting the solubility of polypeptides to which it is fused. We therefore concentrated our efforts on characterizing the N-terminal end of XylS in this chimera. Induction of the expression of N-XylS-MBP in E. coli(pMAL-NXylS) yielded a 65-kDa fusion protein that represented almost 10% of the total protein in the extract, with one-third of it being soluble under the best production conditions described in Materials and Methods. The soluble protein was purified to apparent homogeneity by α-amylose affinity chromatography (not shown).
Glutaraldehyde cross-linking in vitro.
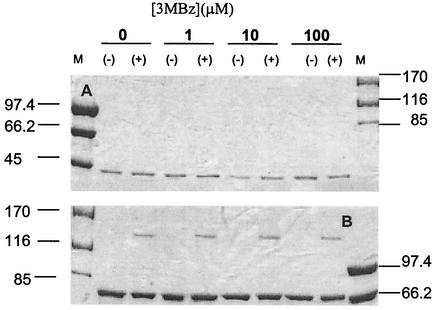
To determine whether the chimeric N-XylS-MBP protein forms dimers, we carried out in vitro cross-linking assays in the absence and presence of 3MBz. In the absence of glutaraldehyde, the N-XylS-MBP protein appeared as a single band, in agreement with the fact that MBP is a monomer (39, 41). However, in the presence of glutaraldehyde, a fraction of the chimeric protein was shifted, with a molecular weight corresponding to a dimer (Fig. 2). A control with MBP without N-XylS did not produce dimers, which suggests that the XylS N-terminal domain in the chimera is responsible for the dimerization observed. The fact that the dimer formed in the absence of 3MBz suggests that XylS may form a dimer in the absence of effector. This has also been observed with AraC (1) and MelR (2), two other members of the AraC/XylS family.
FIG. 2.
Cross-linking of MBP-N-XylS. Top panel: SDS-PAGE (8% [wt/vol]) of MBP. Bottom panel: SDS-PAGE (6% [wt/vol]) of MBP-N-XylS. Lane M, molecular size markers, with sizes shown on the left or the right (in kilodaltons). The concentrations of 3MBz are indicated along the top. The + and − symbols indicate whether the samples were incubated or not with glutaraldehyde, as described in Materials and Methods.
Two-hybrid system confirms that the N-terminal domain of XylS dimerizes.
Transcription activation by AraC has been studied intensively (38) and serves as a model for activation of transcription mediated by other proteins in the family. AraC is the activator of the ara regulon, which is essential for arabinose transport and metabolism. In the absence of the sugar arabinose, it binds to two 16-bp sites (denoted O2 and I1) 200 bp apart at the araBAD promoter, forming a repression loop. In the presence of arabinose, AraC binds to I2, which is adjacent to I1, rather than to O2. This breaks the repression loop, and the presence of the activator at I2 next to the RNA polymerase activates transcription. AraC-dependent transcription initiation at the araBAD promoter is increased by catabolite repression protein, which binds to a single DNA site upstream of I1 and I2 (44).
The LexA protein controls expression of the PsulA promoter and, by binding to the promoter region, downregulates expression (1). Such repression requires dimerization of the LexA DNA binding domain. Previously, chimeric proteins containing the N-terminal domain of AraC, predicted to be involved in AraC dimerization, were fused to the LexA DNA binding domain (1). The chimera was shown to behave like a full-length LexA protein, because transcription of PsulA::lacZ was repressed in the presence of the chimeric AraC-LexA fusion protein. To obtain further insights on the possibility that N-XylS is a dimerization domain for XylS, we used the system described above. We used vector pGB002, engineered to encode only the DNA-binding domain of LexA′, and we cloned part of the xylS gene that, when translated, will produce a chimera of the N-XylS′ domain to LexA. The construct was transformed into the E. coli JL1436 reporter strain with a lacZ fusion under the control of PsulA and screened for repression of PsulA::lacZ (lower levels of β-galactosidase).
The pGB002 vector provides a positive control for the system, and β-galactosidase levels were around 4,000 Miller units (Table 2). When we used a LexA′ fusion to the N-terminal end of XylS, activity decreased to 25% of that of the control strain only in the presence of 3MBz in the culture medium (Table 2). These results are in apparent contradiction with the cross-linking assays, but differences could be attributed either to the monomeric nature of the MBP protein versus the dimeric nature of the LexA protein or to the fact that the dimer may be stabilized in the presence of the effector. Another possibility is that the dimer in the presence of effector links to the target DNA and is the stable form of the protein in vivo.
The crystal structure of AraC reveals that a set of leucines corresponding to positions 150, 151, and 161 are important for dimerization (40). This arrangement of leucines has also been shown to occur in UreR (another AraC/XylS family member) (Fig. 1), and site-directed mutagenesis revealed that these leucines are critical for the dimerization and transcriptional activity of these two regulators (19, 31). A similar organization of residues is seen in XylS except that the furthermost C-terminal leucine is replaced by an isoleucine and the overall length is one amino acid longer (Fig. 1). We therefore decided to mutate these residues and replace them with alanine. The mutant alleles were present in the plasmid series pLRRA1 through pLRRA7. These plasmids were transformed into E. coli MC4100(pERD100), and β-galactosidase was measured (Table 3). Our results show that XylSI205A was as active as the wild-type protein, but the replacement of leucine 193 or 194 with alanine resulted in a marked decrease in activity. In particular, activation was not observed with XylSL194A. This suggests that L194 is a critical residue for XylS activity.
TABLE 3.
Induction of Pm by variants of XylS proteina
| N-XylS protein | β-Galactosidase (Miller units)
|
Induction ratio, +3MBz/−3MBz (fold increase) | |
|---|---|---|---|
| Without 3MBz | With 3MBz | ||
| Wild type | 50 ± 2 | 1,510 ± 80 | 30 |
| N-XylSL193A | 45 ± 1 | 450 ± 30 | 10 |
| N-XylSL194A | 50 ± 1 | 50 ± 5 | 1 |
| N-XylSI205A | 45 ± 3 | 1,650 ± 60 | 37 |
| N-XylSL193A,L194A | 45 ± 4 | 50 ± 8 | 1 |
| N-XylSL193A,I205A | 45 ± 2 | 60 ± 2 | 1 |
| N-XylSL194A,I205A | 45 ± 2 | 50 ± 1 | 1 |
| N-XylSL193A,L194A,I205A | 45 ± 2 | 45 ± 2 | 1 |
E. coli MC4100(pERD100) bearing a derivative of pLOW2 that encodes the indicated XylS protein was cultured as indicated in Materials and Methods. Data (values are rounded) are the averages of at least three independent assays performed in triplicate.
Combination of L194A with any of the other two mutations resulted in a mutant protein that was unable to activate transcription, as expected. The combination of L193A with I205 yielded a mutant that was unable to stimulate transcription, in contrast to the single-parent mutants. This suggests that these residues in XylS might work additively, although we cannot rule out that multiple amino acid substitutions alter either the secondary or tertiary structure of the protein, which would account for the loss of activity.
To determine whether the decrease in activity of the XylS mutant was due to dimerization defects, we used the variants of the N-terminal end with the L193→A, L194→A, I205→A, L193,L194→A,A, L193,I205→A,A, and L194,I205→A,A substitutions and the triple mutant in the PsulA system after fusion of the N-XylS* mutant ends to LexA′ (Table 2). We found that the single mutants L193→A and L194→A, the double mutant involving these two residues, and the triple mutant did not inhibit expression of PsulA regardless of the presence of 3MBz, whereas the single I205→A mutant did inhibit expression at a level similar to that seen with the wild type when 3MBz was added to the culture medium (Table 2). These results suggest that residues 193 and 194 are critical for dimerization of the N-terminal region of XylS.
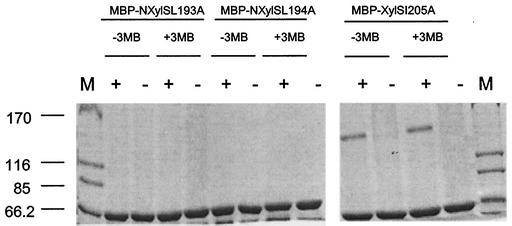
To further investigate the dimerization of XylS, we constructed chimeric fusions of the N-XylS* variants to MBP, and the chimeric N-XylS*-MBP proteins were purified as described above for glutaraldehyde cross-linking assays. Our results revealed that the MBP-N-XylSI205A protein dimerized (Fig. 3), but when the N-XylS domain contained the L193→A or L194→A change, dimerization did not occur (Fig. 3). This set of results further supports the idea that these two leucine residues are critical for dimerization or folding of the chimeric proteins.
FIG. 3.
Cross-linking of MBP-N-XylS*. E. coli bearing plasmids that will produce MBP-N-XylSL193A, MBP-N-XylSL194A, and MBP-N-XylSI205A were grown in the absence and in the presence of 3MBz. Proteins were purified as described in Materials and Methods, and samples were incubated in the presence (+) and in the absence (−) of 0.005% (vol/vol) glutaraldehyde.
Previous studies with XylS mutants revealed intra-allelic dominance of mutations of the C-terminal domain over those in the N-terminal domain and vice versa (25), and this led those authors to propose that the N- and C-terminal domains of XylS interact with each other. Recent evidence has supported the idea that transcription from PmelAB by MelR requires both the C-terminal DNA-binding domain and the N-terminal domain involved in melibiose recognition (14). Therefore, transcriptional regulators in the AraC/XylS family involved in the control of carbon metabolism seem to acquire a conformational form in the presence of effectors which stabilizes dimers and facilitates subsequent contacts with the target DNA promoter.
Acknowledgments
Work in our laboratory was supported by a CICYT grant to S.M. (BIO 2000-0964) and a grant from the European Commission (QLK3-CT-2000-0170).
We thank Robert Schleif for supplying strains and plasmids and José A. Paz for technical assistance. We thank Mai Fandila and Carmen Lorente for secretarial assistance and K. Shashok for reading the manuscript and improving the language.
REFERENCES
- 1.Bustos, S. A., and R. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 90:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caswell, R., J. Williams, A. Lyddiatt, and S. Busby. 1992. Overexpression, purification and characterization of the Escherichia coli MelR transcription activator. Biochem. J. 287:493-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallegos, M. T., S. Marqués, and J. L. Ramos. 1996. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a σ70-dependent promoter or from σ70- and σ54-dependent tandem promoters according to the (aromatic) compound used for growth. J. Bacteriol. 178:2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González-Pérez, M. M., S. Marqués, P. Domínguez, and J. L. Ramos. 2002. XylS activator and RNA polymerase binding sites at the Pm promoter overlap. FEBS Lett. 519:117-122. [DOI] [PubMed] [Google Scholar]
- 7.González-Pérez, M. M., J. L. Ramos, M. T. Gallegos, and S. Marqués. 1999. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J. Biol. Chem. 274:2286-2290. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, L. H., S. J. Sorensen, and L. B. Jensen. 1997. Chromosomal inscription of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene 186:167-173. [DOI] [PubMed] [Google Scholar]
- 10.Harayama, S., P. R. Lehrbach, and K. N. Timmis. 1984. Transposon mutagenesis analysis of meta cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J. Bacteriol. 160:251-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmer, T., and R. Schleif. 2001. The C-terminal end of AraC tightly binds to the rest of its domain. J. Biol. Chem. 276:4886-4888. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis et al. (ed.), PCR protocols: a guide to methods and applications. Academic Press. San Diego, Calif.
- 13.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 14.Howard, V., T. A. Belyaeva, S. J. W. Busby, and E. I. Hyde. 2002. DNA binding of the transcription activator protein MelR from Escherichia coli and its C-terminal domain. Nucleic Acids Res. 30:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapust, R. B., and D. S. Waugh. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8:1668-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-303. [DOI] [PubMed] [Google Scholar]
- 17.Kwon, H. J., M. H. J. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424-430. [DOI] [PubMed] [Google Scholar]
- 18.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 19.LaRonde-LeBlanc, N., and C. Wolberger. 2000. Characterization of the oligomeric states of wild type and mutant AraC. Biochemistry 39:11593-11601. [DOI] [PubMed] [Google Scholar]
- 20.Lu, C.-D., and A. T. Abdelal. 2001. The gdhB gene of Pseudomonas aeruginosa encodes an arginine-inducible NAD+-dependent glutamate dehydrogenase which is subject to allosteric regulation. J. Bacteriol. 183:490-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, Z., H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184:7001-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manzanera, M., S. Marqués, and J. L. Ramos. 2000. Mutational analysis of the highly conserved C-terminal residues of the XylS protein, a member of the AraC family of transcriptional regulators. FEBS Lett. 476:312-317. [DOI] [PubMed] [Google Scholar]
- 23.Marqués, S., M. Manzanera, M. M. González-Pérez, M. T. Gallegos, and J. L. Ramos. 1999. The XylS-dependent Pm promoter is transcribed in vivo by RNA polymerase with σ32 or σ38 depending on the growth phase. Mol. Microbiol. 31:1105-1113. [DOI] [PubMed] [Google Scholar]
- 24.Martin, R. G., and J. L. Rosner. 2000. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 25.Michán, C., B. Kessler, V. de Lorenzo, K. N. Timmis, and J. L. Ramos. 1992. XylS domain interactions can be deduced from intrallelic dominance in double mutants of Pseudomonas putida. Mol. Gen. Genet. 235:406-412. [DOI] [PubMed] [Google Scholar]
- 26.Michán, C., L. Zhou, M. T. Gallegos, K. N. Timmis, and J. L. Ramos. 1992. Identification of critical amino-terminal regions of XylS. J. Biol. Chem. 267:22897-22901. [PubMed] [Google Scholar]
- 27.Munson, G. P., L. G. Holcomb, and J. R. Scott. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Martín, J., and M. Espinosa. 1992. A genetic system to study the in vivo role of transcriptional regulators in Escherichia coli. Gene 116:75-80. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Martín, J., I. Cases, and V. de Lorenzo. 1997. Design of a solubilization pathway for recombinant polypeptides in vivo through processing of a bi-protein with a viral protease. Protein Eng. 10:725-730. [DOI] [PubMed] [Google Scholar]
- 30.Platt, T., B. Meueler-Hill, and J. Miller. 1972. Assays of β-galactosidase activity, p. 352-355. In J. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 31.Poore, C. A., C. Coker, J. D. Dattlebaum, and L. T. Mobley. 2001. Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 183:4526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos, J. L., S. Marqués, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 33.Ramos, J. L., C. Michán, F. Rojo, D. Dwyer, and K. N. Timmis. 1990. Signal-regulator interactions. Genetic analysis of the effector binding site of xylS, the benzoate-activated positive regulator of Pseudomonas TOL plasmid meta-cleavage pathway operon. J. Mol. Biol. 211:373-382. [DOI] [PubMed] [Google Scholar]
- 34.Ramos, J. L., A. Stolz, W. Reineke, and K. N. Timmis. 1986. New effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of catabolic pathways. Proc. Natl. Acad. Sci. USA 83:8467-8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure of an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruíz, R., and J. L. Ramos. 2002. Residues 137 and 153 at the N terminus of the XylS protein influence the effector profile of this transcriptional regulator and the sigma factor used by RNA polymerase to stimulate transcription from its cognate promoter. J. Biol. Chem. 277:7282-7288. [DOI] [PubMed] [Google Scholar]
- 37.Ruíz, R., J. L. Ramos, and S. Egan. 2001. Interactions of the XylS regulators with the carboxyl-terminal domain of the α subunit of RNA polymerase influence the expression level from the cognate Pm promoter. FEBS Lett. 491:207-211. [DOI] [PubMed] [Google Scholar]
- 38.Schleif, R. 2002. Regulation of the l-arabinose operon of Escherichia coli. Trends Genet. 16:559-565. [DOI] [PubMed] [Google Scholar]
- 39.Sharff, A. J., L. E. Rodseth, J. C. Spurlino, and F. A. Quiocho. 1992. Crystallographic evidence of a ligand-induced hinge-twist mutation between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry 31:10657-10663. [DOI] [PubMed] [Google Scholar]
- 40.Soisson, S. M., McDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligmerization of AraC. Science 276:421-425. [DOI] [PubMed] [Google Scholar]
- 41.Spurlino, J. C., G.-Y. Lu, and F. A. Quiocho. 1991. The 2.3-Å resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J. Biol. Chem. 266:5202-5219. [DOI] [PubMed] [Google Scholar]
- 42.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 29:72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, M., and R. Schleif. 2001. Strengthened arm-dimerization domain interaction in AraC. J. Biol. Chem. 276:2562-2564. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, X., and R. Schleif. 1998. Catabolite activator protein mutations affecting the activity of the araBAD promoter. J. Bacteriol. 180:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]