Abstract
Studies of naturally occurring polymorphisms of the CCR5 gene have shown that deletion of the functional receptor or reduced expression of the gene can have beneficial effects in preventing HIV-1 infection or delaying disease. Because these polymorphisms are found in otherwise healthy people, strategies that aim to prevent or limit expression of CCR5 should be beneficial in the treatment of HIV-1 disease. To test this approach we have developed a CCR5-specific single-chain antibody that was expressed intracellularly and retained in the endoplasmic reticulum. This CCR5-intrabody efficiently blocked surface expression of human and rhesus CCR5 and thus prevented cellular interactions with CCR5-dependent HIV-1 and simian immunodeficiency virus envelope glycoprotein. Intrabody-expressing cells were shown to be highly refractory to challenge with R5 HIV-1 viruses or infected cells. These results suggest that gene therapy approaches that deliver this intracellular antibody could be of benefit to infected individuals. Because the antibody reacts with a conserved primate epitope on CCR5 this strategy can be tested in nonhuman lentivirus models of HIV-1 disease.
The molecular mechanism of human immunodeficiency virus type 1 (HIV-1) entry into cells involves specific interactions between the viral envelope glycoproteins (env) and two target cell proteins, CD4 and a chemokine receptor. HIV-1 cell tropism is determined by the specificity of the env for a particular chemokine receptor. Macrophage (M)-tropic viruses require the chemokine receptor CCR5 for entry, and these viruses are designated as R5 viruses. T-cell-line (TCL)-tropic viruses use CXCR4 for entry and are designated as X4 viruses (1). While a multiplicity of coreceptors have been shown to facilitate HIV-1 entry in vitro, only CCR5 and CXCR4 have been convincingly demonstrated to be relevant in vivo (2, 3).
Several findings suggest that CCR5-positive cells are typically the critical first targets in HIV-1 infection and that CCR5 expression levels are key in disease progression. Individuals with a homozygous deletion (Δ32) in their CCR5 gene lack functional CCR5 expression and are highly protected against transmission, which usually involves R5 viruses (2). Individuals that are heterozygous for this mutation express reduced levels of CCR5 and are delayed in their progression to AIDS by 1–2 years (4). Furthermore, the 59029 G/A polymorphism reduces the activity of the CCR5 promoter by ≈45%; individuals with this mutation are delayed in their progression to AIDS by ≈4 years (5). Significantly, these natural polymorphisms are not known to be associated with any detrimental phenotype. Therefore, intervention strategies aimed at blocking CCR5 expression should be beneficial for cellular protection against viral infection and may provide a clinical benefit.
In attempts to disrupt HIV-1 replication, intracellular immunization strategies based on the expression of trans-dominant mutants, ribozymes, and intracellular antibodies (“intrabodies”) have been studied (6–8). Approaches that aim to prevent viral entry should have advantages over strategies that target postentry steps of the HIV-1 life cycle. In this direction, intracellular expression of chemokines has shown some promise in limiting, to some extent, viral entry (9–11).
The study presented here describes the development and characterization of a CCR5-specific intrabody, ST6. We show that intracellular expression of ST6 with an endoplasmic reticulum (ER)-retention signal efficiently blocks surface expression of CCR5. A CCR5+ T cell line, PM1, was transduced to express the intrabody. No CCR5 surface expression was detected in the transduced PM1 cells, and they were protected from both direct and cell-to-cell infection with R5 virus strains. Our results suggest that the introduction of a CCR5-specific intrabody into hematopoietic stem cells is a plausible strategy for the generation of a cell pool in infected individuals that is protected from R5-HIV-1 infection.
Materials and Methods
Cells, Viruses, and Reagents.
Cells.
PM1 cells were grown in RPMI medium 1640 containing 10% FBS and antibiotics. Transduced PM1 cells were usually maintained in the presence of puromycin (0.5 μg/ml) except during cell–cell fusion assays and infection assays. COS7 cells and PA317 (both from the American Type Culture Collection) and 293T cells (obtained from R. W. Doms, University of Pennsylvania) were maintained in DMEM containing 10% FBS and antibiotics. Tissue culture media and reagents were from GIBCO/BRL.
Viruses.
The following vaccinia recombinants were used: vCB-21R (lacZ gene) (12), vTF7–3 (T7 RNA polymerase) (13), vCB-28 (JR-FL env) (14), vCB-32 (SF162 env) (15), vCB-43 (Ba-L env) (16, 17), vBD3 (89.6 env) (18), and vCB 74 [simian immunodeficiency virus (SIV) mac 239 env] (19). Infection and further treatment of the effector cells were done as described (20). The reporter R5 HIV-1 virus construct, NFN-SX-r-HSAS, was obtained from B. D. Jamieson and J. A. Zack (UCLA School of Medicine).
Plasmids.
Plasmids encoding human CCR5 and CXCR4 (21) and rhesus CCR5 and CD4 (22) were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. A plasmid encoding human CD4 was obtained from B. J. Doranz (University of Pennsylvania). A reporter plasmid containing the luciferase gene under the control of the T7 RNA polymerase promoter was purchased from Promega, and plasmid pcDNA3.1/Zeo was purchased from Invitrogen.
Antibodies.
Antibodies specific to human CCR5, CXCR4, CD4, and RANTES were purchased from PharMingen. FITC- or phycoerythrin (PE)-conjugated secondary antibodies were purchased from Jackson ImmunoResearch except for the anti-rat-FITC conjugate, which was obtained from PharMingen. A high-affinity hemagglutinin epitope (HA)-tag-specific monoclonal rat antibody was purchased from Roche Molecular Biochemicals. CCR5-specific antibody 5C7 (23) was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program.
Conversion of a CCR5-Specific Fab Clone into a Single-Chain Antibody Fragment (scFv).
Specific oligonucleotide primers were used to amplify VH and VL gene segments from purified phagemid DNA isolated from ST6, a Fab fragment specific for the N-terminal extracellular domain of CCR5. This Fab was isolated from an immunized rabbit by using the phage display approach (24). The following primers were used: VL, RSCVK1 5′-GGGCCCAGGCGGCCGAGCTCGTGMTGACCCAGACTCCA-3′ and RKB9J0-B 5′-GGAAGATCTAGAGGAACCACCTAGGATCTCCAGCTCGGTCCC-3′; VH, RSCVH3 5′-GGTGGTTCCTCTAGATCTTCCCAGTCGYTGGAGGAGTCCGGG-3′ and HSCG1234-B 5′-CCTGGCCGGCCTGGCCACTAGTGACCGATGGGCCCTTGGTGGARGC-3′. The purified PCR products were assembled by another PCR using the following primers: RSC-F 5′-GAGGAGGAGGAGGAGGAGGCGGGGCCCAGGCGGCCGAGCTC-3′ and RSC-B 5′-GAGGAGGAGGAGGAGGAGCCTGGCCGGCCTGGCCACTAGTG-3′. The resulting overlap-PCR product encodes an scFv in which the N-terminal VL region is linked with the VH region through a 7-aa peptide linker (GGSSRSS). The DNA fragment was gel-purified, digested with the restriction endonuclease SfiI, and cloned into the appropriately cut phagemid vector pComb3X, a variant of pComb3H (24). Binding activity of the expressed scFv was confirmed, and the gene encoding the scFv was transferred to pcDNA3.1/Zeo and pBabe Puro vectors.
Generation of pcDNA3.1/Zeo and pBabe Puro Intrabody and Intrakine Constructs.
Both pcDNA3.1/Zeo and pBabe Puro (25) were modified by introducing two SfiI sites into their multiple cloning sites. A human κ leader sequence was cloned into the vectors upstream of the 5′ SfiI sites. Downstream of the 3′ SfiI site, a sequence encoding the HA-tag sequence (YPYDVPDYA) (26) and an endoplasmic reticulum (ER) retention signal (KDEL) followed by a stop codon was introduced. The ST6 scFv, as well as control scFv encoding DNA fragments were cloned into the appropriately digested vector DNAs. The modified pcDNA3.1/Zeo plasmid encoding ST6 was designated pIB6.
For control purposes an intrakine construct for intracellular expression of the CCR5 binding chemokine RANTES (9) was also made: Human RANTES cDNA was PCR amplified by using the primers RANF 5′-GAGGAGGAGGAGGAGGCTAGCATGAAGGTCTCCGCGGCAC-3′ and RAN-B-SFII 5′-CGGAACGTCGTACGGGTACTGGCCGGCCTGGCCGCTCATCTCCAAAGAGTTGATGTACTCCCG-3′. The PCR product was digested with NheI and SfiI and was gel-purified. The RANTES-encoding DNA insert was cloned into the modified and appropriately cut pcDNA 3.1/Zeo DNA. The resulting plasmid encoding the RANTES intrakine without the κ leader sequence was designated pRAN. The sequence of the intrakine insert was confirmed by DNA sequence analysis.
Cotransfection of 293T Cells by Using Chemokine Receptor-Encoding Plasmid and Intrabody- or Intrakine-Encoding Plasmid.
293T cells were transfected by using Lipofectamine (GIBCO/BRL) according to the manufacturer's protocol with plasmids containing coreceptor genes. At the same time, cells were cotransfected with 2-fold molar excess of plasmid encoding CCR5-specific intrabody (pIB6) or intrakine (pRAN) or with control plasmid, pcDNA3.1/Zeo containing no insert.
Flow Cytometric Analysis of Cotransfected 293T Cells and Transduced PM1 Cells.
For surface staining, cells were incubated with primary antibodies for 30 min, washed, and stained with appropriate FITC or PE conjugates. For intracellular staining, cells were permeabilized with PBS containing 4% paraformaldehyde (Sigma) and 0.1% saponin (Sigma) for 10 min and washed. Cells were then incubated with primary antibodies for 30 min, washed, and stained with appropriate FITC or PE conjugates. Throughout the staining, the washing and staining buffers contained saponin (0.1%). After staining and washing, the cells were resuspended in PBS without saponin. Cells were analyzed on Becton Dickinson flow cytometers (FACScan, FACSort, or FACSCalibur) using CellQuest software.
Reporter Gene Fusion Assay.
A modified reporter gene assay was used to quantify cell–cell fusion events (20, 27). Briefly, T7 RNA polymerase and HIV-1 or SIV env were introduced into 293T effector cells by using vaccinia virus recombinants. COS7 cells, used as target cells, were transfected with a plasmid encoding luciferase under the control of a T7 promoter and plasmids containing human or rhesus CCR5 genes, and human or rhesus CD4 genes by using Lipofectamine. Target cells were cotransfected with a plasmid encoding ST6 intrabody (pIB6), a plasmid encoding RANTES intrakine (pRAN), or the control plasmid pcDNA3.1/Zeo containing no insert. To assure cotransfection, the plasmids encoding intrabody or control plasmids were introduced in 2-fold molar excess over the plasmids encoding the coreceptors. To assess background luciferase activity, a set of target cells was transfected with luciferase- and CD4-encoding plasmids but not CCR5-encoding plasmid. Duplicate transfection mixes were set up for each kind of target cell, and each transfection mix was distributed to two wells. After overnight incubation, effector cells were added to wells containing target cells and cocultured for 8–10 hr in the presence of rifampicin (100 μg/ml, Sigma) and araC (cytosine β-d-arabinofuranose, 10 μM, Sigma). Cells were then lysed and assayed for luciferase activity. When transduced and untransduced PM1 cells were used as target cells in a reporter gene fusion assay, the cells were infected with a vaccinia recombinant encoding the T7 RNA polymerase. 293T cells that were used as effector cells were transfected with luciferase reporter plasmid and infected with vaccinia virus recombinants encoding HIV-1 env. To assess background luciferase activity, a set of target cells was infected with a control vaccinia recombinant containing the lacZ gene.
Retroviral Gene Transfer: Generation of Transduced PM1 cells.
The amphotropic packaging cell line PA317 (28) was transfected with pBabe Puro plasmids encoding the ST6 scFv insert. For control purposes pBabe Puro plasmids encoding scFv specific to glutathione S-transferase (GST) or human integrin αvβ3 (RAI3) were also used. Producer lines were selected by adding 2 μg/ml puromycin to the cultures. These stable lines were used to generate virus-containing medium for two rounds of infection of PM1 cells in the presence of 8 μg/ml Polybrene (Sigma). Two days after the last infection, transduced PM1 cells were selected in puromycin (0.5 μg/ml). After 14 days of selection, analysis of cells for CCR5 expression and infectability was started. The untransduced parental PM1 cell line was named PM1-P, and the PM1 cells transduced to express ST6 intrabody were named PM1–6. The PM1 cell lines transduced to express the control intrabodies RAI3 and a GST-specific intrabody were named PM1-RAI3 and PM1-GST, respectively. For some experiments transduced intrabody-expressing PM1 clones obtained from limiting dilution cultures were used. PM1–6-A2 and PM1–6-G were cloned from the PM1–6 cell line and PM1-RAI3–5 was cloned from the PM1-RAI3 cell line.
HIV-1 Infection of the PM1 Cells.
Transduced and untransduced PM1 cells were infected at an multiplicity of infection of 0.01 for 5 hr at 37°C, washed, and then cultured for up to 16 days. To monitor infection, aliquots were taken from the cultures at the indicated time points and p24 levels were determined in an HIV-1 ELISA (NEN Life Sciences).
Cocultivation of PM1 Cells with Infected Parental PM1 Cells.
Parental PM1 cells were infected with the NFN-SX-r-HSAS reporter virus. In this virus the HIV-1 vpr is replaced with murine heat-stable antigen (HSA, CD24), allowing infected cells to be monitored by flow cytometry. Here, a virus was constructed by replacing the env of NFN-SX-r-HSAS (29) with the env sequence of the CCR5-using JR-FL (B. D. Jamieson and J. A. Zack, personal communication). When about 5% of the cells were infected, three cocultures were initiated: A 2-fold excess of cells from the infected PM1-P culture were mixed with PM1-P cells and with the transduced PM1 cell clones PM1-RAI3–5 and PM1–6-G. Cocultures were monitored for intrabody and HSA expression by flow cytometry. To reduce false positives, doublets or larger aggregates of PM1 cells were excluded from analysis.
Results
Transfection of an ST6-Encoding Plasmid (pIB6) Blocks Surface Expression of CCR5.
An antibody fragment, ST6, which binds the N-terminal extracellular domain of CCR5, was originally derived from an Fab phage display library (P.S. and C.F.B.). ST6 was converted into an scFv in which the VL and VH fragments were covalently linked with a peptide linker consisting of seven amino acids. Upon expression, use of this short peptide linker results in dimeric scFv proteins (30). To retain the antibody fragment in the ER, an ER retention peptide (KDEL) was appended to the C terminus of the protein (31). We anticipated that ST6 scFv dimers expressed within cells as intrabodies possessing two functional CCR5 binding sites and two ER retention signals would efficiently trap CCR5 proteins en route to the cell surface via their natural ER trafficking pathway. As a control protein, the C-C-chemokine RANTES was cloned into the same expression vector as a fusion with the ER retention sequence as described (9). RANTES expressed in this manner has been shown to be retained predominately in the ER and has been termed an “intrakine.”
The effect of intrabody and intrakine coexpression on the surface expression of human and rhesus CCR5 was examined by flow cytometry. Upon transfection with a CCR5-encoding plasmid, 293T cells expressed high levels of CCR5. To study the effect of expression of the intrabody and intrakine, cotransfections were performed with a 2-fold molar excess of control plasmid (pcDNA3.1/Zeo) or plasmid encoding ST6 (pIB6) or RANTES (pRAN). Upon cotransfection with pIB6-DNA, no surface expression of human CCR5 was detected by flow cytometry, whereas cotransfection with pRAN resulted in only a slight reduction of CCR5 expression. Intracellular staining using an antibody specific to the HA-tag encoded in the expression plasmids upstream of the KDEL sequence showed that the expression levels for the intrabody and the intrakine were comparable. Intracellular expression of RANTES in cells cotransfected with pRAN was also confirmed by incubating permeabilized cells with a RANTES-specific antibody (Fig. 1A). The ability of ST6 intrabody to block rhesus CCR5 expression was studied to assess whether it could be used in nonhuman lentivirus models. CCR5 is the primary coreceptor for SIV, and the N-terminal extracellular domain sequence of rhesus CCR5 has only two amino acid substitutions as compared with the human CCR5 sequence (32). Transfection studies of 293T cells were performed as described above with a plasmid encoding rhesus CCR5 replacing that used for human CCR5 expression. Again cotransfection with ST6 scFv-encoding plasmid pIB6 completely blocked transient rhesus CCR5 expression, whereas pRAN had little effect (Fig. 1B). Note that rhesus RANTES is identical in sequence to the human chemokine (32). Cotransfection studies using plasmids encoding human CXCR4 with pIB6 or pRAN demonstrated that neither pIB6 nor pRAN affected transient CXCR4 expression (Fig. 1C). Intracellular HA and RANTES staining confirmed that both the intrabody and the intrakine were expressed at similar levels in these experiments (not shown). Cotransfection studies of the CCR5-expressing plasmid with plasmid encoding an irrelevant intrabody had no effect on CCR5 surface expression (results not shown).
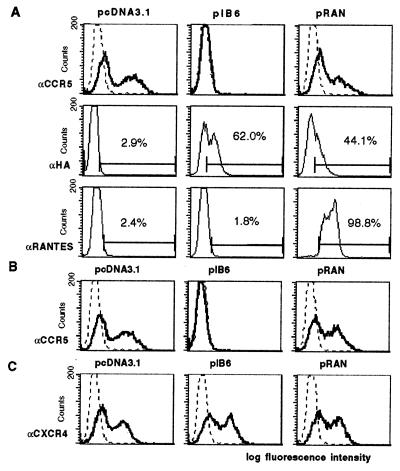
Figure 1.
Inhibition of transient CCR5 surface expression upon cotransfection with ST6 intrabody-encoding plasmid, pIB6. (A) 293T cells were transiently transfected with plasmid encoding human CCR5 and were cotransfected with the indicated plasmids: control plasmid, pcDNA3.1/Zeo, plasmid encoding ST6, pIB6, or with plasmid encoding RANTES, pRAN. (Top) Cell surface staining using a CCR5-specific antibody (clone 5C7, bold lines) or with an irrelevant mouse antibody for control purposes (dashed lines). (Middle) Staining of permeabilized cells with an HA-tag-specific antibody to detect intrabody or intrakine. (Bottom) Staining of permeabilized cells for intracellular RANTES expression with an anti-human RANTES antibody. (B) 293T cells were transiently transfected with plasmid encoding rhesus CCR5 and were cotransfected with plasmids as indicated. Cells were surface stained for CCR5 expression as described above. (C) 293T cells were transiently transfected with plasmid encoding human CXCR4 and were cotransfected with plasmids as indicated. Transfected cells were surface stained with a CXCR4-specific antibody (bold lines), or control stained with an irrelevant mouse antibody (dashed lines).
ST6-Encoding Plasmid (pIB6) Prevents CCR5-Dependent Cell–Cell Fusion.
The effect of ST6 intrabody expression on CCR5-dependent cell–cell fusion was investigated by using a reporter gene assay. Plasmids encoding luciferase under the control of the T7 promoter (reporter plasmid) and human or rhesus CD4 and CCR5 were introduced into COS7 cells to generate two target cell populations. These cells were also cotransfected with ST6 plasmid pIB6 or with control plasmid. In some experiments, target cells were cotransfected with RANTES intrakine-encoding plasmid pRAN. Effector cells of five types were prepared that expressed T7 RNA polymerase and env derived from any one of three different R5 HIV-1 variants, the R5X4 HIV-1 strain 89.6, or an SIV strain. Effector cells were then cocultured with the target cells. In this assay, measurement of luciferase activity allows cell–cell fusion activity to be quantified (20). As shown in Fig. 2, cotransfection with pIB6 reduced CCR5-dependent cell fusion to background levels. Fusion assays were repeated at least twice with similar outcomes. Cotransfection with plasmid encoding the RANTES intrakine, pRAN, produced only a slight reduction of cell fusion activity. Cotransfection of pIB6 DNA with CXCR4-encoding plasmid did not affect CXCR4-dependent cell fusion (data not shown).
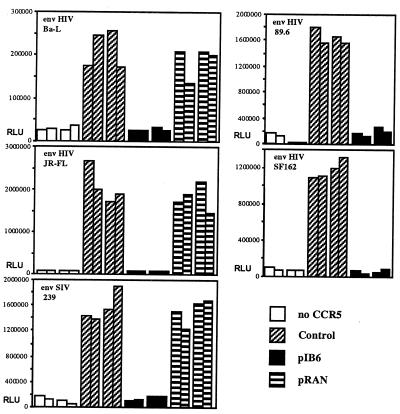
Figure 2.
Cotransfection of ST6-encoding plasmid (pIB6) inhibits CCR5-dependent cell fusion between target cells transiently transfected to express CCR5 and CD4 and effector cells expressing env derived from various HIV-1 or SIV strains. For determination of background luciferase activity, a set of target cells was cotransfected with plasmids encoding CD4 and luciferase but no CCR5-encoding plasmid (“no CCR5”). “Control” indicates fusion assays using target cells cotransfected with plasmids encoding CCR5, CD4, luciferase, and control plasmid pcDNA3.1/Zeo containing no insert. “pIB6” indicates luciferase activity where target cells were cotransfected with plasmids encoding CCR5, CD4, luciferase, and ST6. “pRAN” indicates luciferase activity, where target cells were cotransfected with plasmids encoding CCR5, CD4, luciferase, and RANTES. Relative light units (RLU) are shown on the y-axis. Each bar represents luciferase activity from one well of target cells.
Generation and Characterization of an ST6 scFv-Expressing PM1 Cell Line.
Recombinant retroviruses encoding intrabody ST6 or control intrabodies were used to transduce the CCR5+/CD4+-human lymphocyte cell line PM1. Transduced PM1 cell lines were established through puromycin selection. Parental cells, transduced cell lines, and clones were analyzed for CCR5 expression and intrabody expression by flow cytometry (Fig. 3). The untransduced parental cell line and PM1 cells transduced with retrovirus directing the expression of a control intrabody that does not bind CCR5 expressed CCR5 on their cell surface, as was reported previously (23). In contrast, no CCR5 surface-expression could be detected with PM1–6, the PM1 cell line transduced with ST6 intrabody encoding retrovirus (Fig. 3A). Intrabody could be detected by staining permeabilized transduced PM1 cells with an anti-HA antibody (Fig. 3B). The PM1–6 line showed homogeneous and stable expression of intrabody after puromycin selection, whereas only about 30% of PM1 cells transduced with control intrabody (PM1-RAI3) encoding retrovirus stained positively for intrabody expression. Therefore, limiting dilution cloned transduced PM1 lines and a control intrabody-expressing clone (PM1-RAI3–5) were used in some experiments. For comparative studies two clones derived from the PM-6 line, PM1–6-G and PM1–6-A2, were also isolated. No single-chain antibody was detected on the surface of unpermeabilized transduced PM1 cells by using the same primary and secondary antibodies used for intracellular detection (Fig. 3C). PM1–6 culture supernatants were also examined for the presence of CCR5-specific scFv by ELISA using purified ST6 scFv as a reference. In this assay (sensitivity ≈2 ng of scFv per ml) no scFv was detected (not shown).
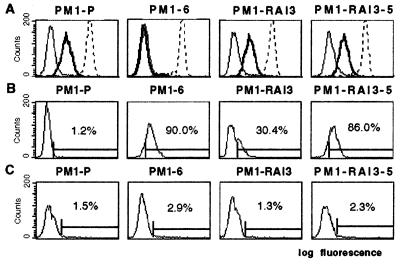
Figure 3.
Analysis of untransduced PM1 cells (PM1-P) and PM1 cell lines transduced to express ST6 intrabody (PM1–6) or control intrabody (PM1-RAI3) and a cell clone derived from the cell line PM1-RAI3 (PM1-RAI3–5) by flow cytometry. (A) PM1 cells were stained with CCR5-specific antibody (bold line), CD4-specific antibody (dashed line), or irrelevant negative control antibody (thin line). (B) PM1 cells were permeabilized and stained with an HA-tag-specific antibody. Percentages are number of cells staining positive. (C) Analysis of unpermeablized PM1 cells for cell surface scFv. PM1 cells were stained with an HA-tag-specific antibody.
PM1 Cells Expressing CCR5-Specific Intrabody ST6 Are Protected from env-Induced CCR5-Dependent Cell Fusion.
Parental PM1 cells (PM1-P) and transduced PM1 clones were analyzed for their interaction with HIV-1 env-expressing cells in a cell–cell fusion reporter assay. In this assay, transduced or untransduced PM1 target cells were infected with recombinant vaccinia virus expressing T7 RNA polymerase. These cells were subsequently cocultured with effector 293T cells that had been transfected with a luciferase reporter plasmid and infected with recombinant vaccinia virus that directs the expression of env derived from the R5 HIV-1 strain JR-FL. The background level of luciferase activity was established by using target cells infected with recombinant vaccinia virus expressing β-galactosidase instead of T7 RNA polymerase and is shown in Fig. 4 (PM1-P-lacZ). Cell–cell fusion that resulted from the interaction of env expressing effector cells with the CCR5+/CD4+-untransduced line, PM1-P, and the transduced control PM1 cells (PM1-RAI3–5) was quantified by luminometry. As shown in Fig. 4, no cell–cell fusion above background was detected after incubation of PM1–6-G cells with effector cells. In contrast, incubation of PM1–6-G cells with effector cells expressing env protein derived from 89.6, an HIV-1 strain that can also use CXCR4 as a coreceptor, led to cell fusion (Fig. 4).
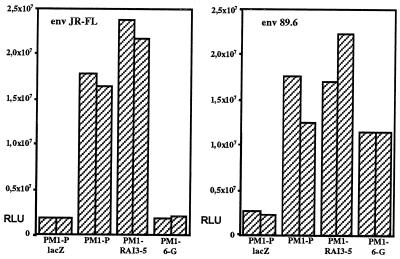
Figure 4.
Cell–cell fusion assay using parental PM1 cells (PM1-P) and clones derived from the control transduced PM1 cells (PM1-RAI3–5) and from PM1 cells transduced with ST6-encoding retrovirus (PM1–6-G). PM1 cells were infected with a vaccinia recombinant encoding T7 RNA polymerase. To assess background luciferase activity, parental PM1 cells were infected with vaccinia virus encoding β-galactosidase instead (PM1-P-lacZ). Reporter gene activity upon coculturing of PM1 cells with 293T cells expressing HIV-1 env and transfected with reporter plasmid is shown as relative light units (RLU) on the y-axis. Pairs of individual experiments are shown.
PM1 Cells Expressing CCR5-Specific Intrabody ST6 Are Protected from R5 HIV-1 Infection.
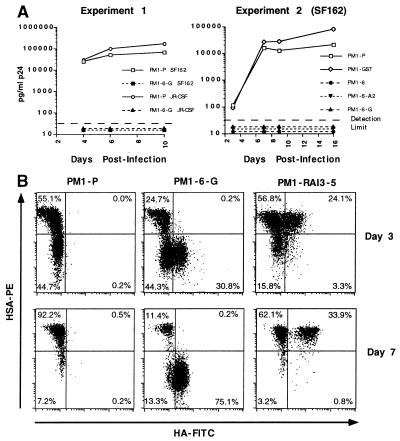
To verify the results of the fusion experiments, we challenged the intrabody-expressing PM1 cell lines with R5 HIV-1 isolates. Fig. 5A shows that the parental PM1 cell line, PM1-P, was readily susceptible to infection with the R5 strains SF162 and JR-CSF, as demonstrated by increasing p24 levels in the tissue culture supernatant (experiment 1). In contrast, p24 levels in cultures of ST6 intrabody-expressing PM1 cells (PM1–6-G) remained below the detection limit of 20 pg of p24 per ml over the 10-day course of the experiment. As a control for nonspecific intrabody effects, a PM1 line expressing an anti-GST intrabody was included in the second experiment (Fig. 5A). This control line was as readily infected with SF162 as were the parental PM1 cells, and the p24 protein production that resulted from this infection closely tracked that observed with the parental PM1 line. The intrabody-expressing line PM-6 as well as two clones derived from this line, PM1–6-G and PM1–6-A2, did not show any detectable p24 at any time during the 16-day time course of this experiment.
Figure 5.
ST6 intrabody-expressing PM1 cells are resistant to challenge by R5 HIV-1 viruses and infected cells. (A) ST6 intrabody-expressing PM1 cells are resistant to cell-free infection with the R5 HIV-1 isolates SF162 and JR-CSF. ST6-expressing PM1 cell lines and clones (dashed lines), parental PM1 cells, and transduced control lines (solid lines) were infected and monitored for p24 levels at indicated times. In experiment 1 the parental PM1 cell line (PM1-P) and the ST6-expressing PM1–6 line were infected with the isolates SF162 and JR-CSF. In experiment 2, HIV-1 SF162 was used to infect PM1 cell lines and clones as indicated. (B) ST6 intrabody-expressing PM1 cells are resistant to cell-mediated R5 HIV-1 infection. PM1 cell cultures infected with the R5-HIV-1 reporter virus NFN-SX-r-HSAS were cocultured with the parental PM1 cell line (PM1-P) and the transduced PM1 clones PM1–6-G and PM1-RAI3–5. Double staining of cells for intrabody expression (HA-FITC) and reporter virus infection (HSA-PE) after 3 and 7 days of coculturing is shown.
Independent studies of transduced PM1 and Jurkat lines expressing RANTES or SDF-1α intrakines, respectively, have shown that they are susceptible to low level infection and viral replication (9, 11). To challenge the intrabody-expressing PM1 cell lines with high amounts of virus under more stringent conditions, we cocultivated infected parental PM1 cells with the ST6-expressing PM1 cell clone PM1–6-G. To monitor infection of cells, a reporter virus, NFN-SX-r-HSAS, was used. The reporter virus construct encodes JR-FL env and carries murine HSA as a vpr replacement. Cells infected with this virus can be detected by surface staining for HSA. In this experiment a 2-fold excess of PM1-P cells infected at the level of 5% were added to uninfected PM1 cells and transduced PM1 cell clones.
Cells were stained for intrabody expression and for reporter virus infection by using HA- and HSA-specific antibodies, respectively. Flow cytometry results obtained after 3 and 7 days of cocultivation are shown in Fig. 5B.
In the coculture of parental PM1 cells with infected cells, the number of infected cells increased from 55% on day 3 to more than 92% on day 7. In contrast, when infected PM1 cells were added to the (HA+) ST6-expressing cell line, PM1–6-G, virtually all HA+ cells remained HSA− through the 20-day time course of the experiment. In this culture, the number of HSA+ cells on day 3 is higher (24%) than on day 7 (11%). This is likely to be because of a depletion of infectable PM1-P cells in the culture, because on day 7, 85% of the HSA− cells (75% of the total cell population) stain positive for HA. The very low number of HA+/HSA+ cells (0.2%) are probably false positives, because a similar number of cells stained HA+ in the coculture of infected cells with intrabody-negative (HA−) PM1-P cells. Furthermore, selection against untransduced PM1-P cells in the coculture using puromycin led to the loss of all HSA+ cells (not shown). Thus, even when exposed to R5 HIV-1 virus and infected cells for a prolonged period of time, PM1–6-G cells were completely resistant to infection. A transduced clone PM1-RAI3–5 expressing an irrelevant intrabody that was included as a control was readily infected by the reporter virus (Fig. 5B). PM1–6-G cells were susceptible to infection by an otherwise identical reporter virus expressing an X4 env (not shown).
Cocultivation of PM1-P cells with transduced PM1 cell lines demonstrated that the two cell types had similar growth rates, since the proportion of intrabody-expressing PM1 cells was found to be stable when analyzed by intracellular staining using an HA-tag-specific antibody (not shown). This result is consistent with previous studies that have shown that intracellular antibody expression has no obvious negative effects on cell viability or proliferation (33, 34).
Discussion
Primary HIV-1 infection typically occurs through a CCR5-dependent mechanism with R5 virus. After expansion in the host, the virus can evolve to use CXCR4 as a coreceptor, a shift that is characterized by a syncytium-inducing phenotype and accelerated destruction of CD4+ T cells (2). The phenotypic switch from R5 to X4 viruses is not an inevitable consequence of HIV-1 infection, and most individuals who die of HIV-1 disease do so without developing viruses of the X4 phenotype (35). An important question facing CCR5-targeted strategies is if they will encourage a phenotypic switch to the more virulent X4 virus (36). The genetic approach described here abrogates viral entry by functional deletion of the coreceptor. The selective pressure imposed on HIV-1 evolution by this strategy is different from that imposed by receptor antagonists. Antagonists typically function by blocking or otherwise modifying the surface of the receptor involved in env interaction. HIV-1 has already proven itself adept at rapidly evolving compensating env mutations in response to the receptor modification strategies that have been tested: SDF-1a, RANTES analogs, and the CXCR4 antagonist AMD3100 (37–39).
Another approach to produce phenotypic HIV-1-coreceptor knockouts has used chemokines as intrakines to inhibit, to some extent, the trafficking of the chemokine receptors (9–11). While there have been promising reports of the use of intrakines to enhance the resistance of cells to infection, it is anticipated that this approach may suffer from a lack of specificity because chemokines typically interact with multiple receptors. Furthermore, intrakine-expressing cells remain susceptible to low-level viral infection and replication. The most detailed intrakine studies have been performed with RANTES, and while the dispensable nature of CCR5 has been established, the RANTES intrakine strategy would be expected to affect the other three RANTES receptors, CCR1, CCR3, and CCR9 (40), and perhaps others that are not yet defined. The consequences of the functional deletion in stem cells of four or more chemokine receptors are difficult to predict but are unlikely to be favorable. Antibody ST6 recognizes a sequence in the first extracellular domain of CCR5 that is conserved in human and nonhuman primates and is not found in any other known protein. Thus the antibody-based strategy demonstrated here presents significant advantages in terms of receptor selectivity. In functional terms the ST6 intrabody has been shown to be superior to RANTES intrakine in blocking CCR5 surface expression and in preventing cell–cell fusion events.
The efficiency of the ST6 intrabody in terms of functional deletion of the coreceptor CCR5 is evident in the resistance it imparts to cells in the face of stringent viral and infected cell challenge. Extended in vitro challenge of a receptor-deleted cell line with infected cells resulted, in time, in a culture consisting of virtually only the receptor-deleted HIV-1-resistant cell line. It is our hope, that this outcome could be repeated in vivo, allowing for the establishment of an HIV-1-resistant cell pool in infected individuals. The advent of improved gene delivery systems shown to efficiently and stably transduce human hematopoietic stem cells (41) will allow this hypothesis to be tested. Because ST6 reacts with CCR5 from nonhuman primates, this strategy can be tested in SIV and chimeric simian-human immunodeficiency virus (SHIV) models of human AIDS, allowing the benefits as well as the potential drawbacks of this approach to be assessed.
Acknowledgments
We are very grateful to E. A. Berger, C. C. Broder, R. W. Doms, B. J. Doranz, B. D. Jamieson, and J. A. Zack for generous gifts of reagents. We thank Jorun K. Sutton and Kent A. Smith for technical assistance and Roger R. Beerli and Christoph Rader for helpful discussions. This work was supported by National Institutes of Health Grants AI41944 (C.F.B.) and DL49886 (B.E.T.). J.A.-W. was supported in part by National Institutes of Health Individual Research Service Award for Postdoctoral Fellows 5F32 AI04397-03. P.S. was supported by Postdoctoral Fellowship J01296-MED from the Austrian Science Foundation.
Abbreviations
- VH
heavy-chain variable region
- VL
light-chain variable region
- scFv
single-chain antibody fragment
- PE
phycoerythrin
- HA
hemagglutinin epitope
- HSA
heat-stable antigen
- env
viral envelope glycoprotein
- SIV
simian immunodeficiency virus
- GST
glutathione S-transferase
- ER
endoplasmic reticulum
Note Added in Proof
Antibody ST6 has now been humanized (P.S. and C.F.B., unpublished data).
References
- 1.Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. Nature (London) 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 2.Berger E A, Murphy P M, Farber J M. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y J, Moore J P. J Virol. 1999;73:3443–3448. doi: 10.1128/jvi.73.4.3443-3448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, et al. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 5.McDermott D H, Zimmerman P A, Guignard F, Kleeberger C A, Leitman S F, Murphy P M. Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 6.Malim M H, Bohnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 7.Sarver N, Cantin E M, Chang P S, Zaia J A, Ladne P A, Stephens D A, Rossi J J. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 8.Marasco W A, Haseltine W A, Chen S Y. Proc Natl Acad Sci USA. 1993;90:7889–7893. doi: 10.1073/pnas.90.16.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang A G, Bai X, Huang X F, Yao C, Chen S. Proc Natl Acad Sci USA. 1997;94:11567–11572. doi: 10.1073/pnas.94.21.11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang A G, Zhang X, Torti F, Chen S Y. Hum Gene Ther. 1998;9:2005–2018. doi: 10.1089/hum.1998.9.14-2005. [DOI] [PubMed] [Google Scholar]
- 11.Chen J D, Bai X, Yang A G, Cong Y, Chen S Y. Nat Med. 1997;3:1110–1116. doi: 10.1038/nm1097-1110. [DOI] [PubMed] [Google Scholar]
- 12.Alkhatib G, Broder C C, Berger E A. J Virol. 1996;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst T R, Niles E G, Studier F W, Moss B. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. Nature (London) 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 15.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 17.Broder C C, Berger E A. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 19.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, et al. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rucker J, Doranz B J, Edinger A L, Long D, Berson J F, Doms R W. Methods Enzymol. 1997;288:118–133. doi: 10.1016/s0076-6879(97)88011-1. [DOI] [PubMed] [Google Scholar]
- 21.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Gettie A, Ho D D, Marx P A. Virology. 1998;246:113–124. doi: 10.1006/viro.1998.9174. [DOI] [PubMed] [Google Scholar]
- 23.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rader C, Barbas C F., III Curr Opin Biotechnol. 1997;8:503–508. doi: 10.1016/s0958-1669(97)80075-4. [DOI] [PubMed] [Google Scholar]
- 25.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 27.Nussbaum O, Broder C C, Berger E A. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller A D, Buttimore C. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamieson B D, Zack J A. J Virol. 1998;72:6520–6526. doi: 10.1128/jvi.72.8.6520-6526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z, Zapata G, Shalaby R, Snedecor B, Chen H, Carter P. Bio/Technology. 1996;14:192–196. doi: 10.1038/nbt0296-192. [DOI] [PubMed] [Google Scholar]
- 31.Munro S, Pelham H R. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 32.Villinger F, Brice G T, Mayne A, Bostik P, Ansari A A. Immunol Lett. 1999;66:37–46. doi: 10.1016/s0165-2478(98)00183-7. [DOI] [PubMed] [Google Scholar]
- 33.Chen S Y, Bagley J, Marasco W A. Hum Gene Ther. 1994;5:595–601. doi: 10.1089/hum.1994.5.5-595. [DOI] [PubMed] [Google Scholar]
- 34.Beerli R R, Wels W, Hynes N E. J Biol Chem. 1994;269:23931–23936. [PubMed] [Google Scholar]
- 35.Richman D D, Bozzette S A. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 36.Michael N L, Moore J P. Nat Med. 1999;5:740–742. doi: 10.1038/10462. [DOI] [PubMed] [Google Scholar]
- 37.Schols D, Este J A, Cabrera C, De Clercq E. J Virol. 1998;72:4032–4037. doi: 10.1128/jvi.72.5.4032-4037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosier D E, Picchio G R, Gulizia R J, Sabbe R, Poignard P, Picard L, Offord R E, Thompson D A, Wilken J. J Virol. 1999;73:3544–3550. doi: 10.1128/jvi.73.5.3544-3550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Este J A, Cabrera C, Blanco J, Gutierrez A, Bridger G, Henson G, Clotet B, Schols D, De Clercq E. J Virol. 1999;73:5577–5585. doi: 10.1128/jvi.73.7.5577-5585.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung S, Littman D R. Curr Opin Immunol. 1999;11:319–325. doi: 10.1016/s0952-7915(99)80051-x. [DOI] [PubMed] [Google Scholar]
- 41.Miyoshi H, Smith K A, Mosier D E, Verma I M, Torbett B E. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]