Abstract
Human cytomegalovirus (HCMV) is the major viral cause of birth defects and a serious problem for immunocompromised individuals. Here we show that infection of cells with HCMV during the S-phase of the cell cycle results in two specific chromosome 1 breaks at positions 1q42 and 1q21. We demonstrate that purified virions, and not infected cell supernatant alone, are responsible for the damage. In addition, we show that the specific breaks occur when different sources of fibroblasts and strains of HCMV are used. Incubation of the virus with neutralizing antibody prevents the induction of breaks. However, UV-inactivated virus is as efficient as untreated virus in inducing specific damage to chromosome 1. Thus, there is a requirement for viral adsorption/penetration, but not new viral gene expression. This HCMV-mediated induction of site-specific damage in actively dividing cells may provide clues for the development of neurological defects in the congenitally infected infant.
There is a great deal of evidence that nonspecific chromosomal aberrations and damage to the mitotic apparatus are caused by many human viruses, including adenovirus, herpes simplex types 1 and 2, herpes zoster, Epstein–Barr, human cytomegalovirus (HCMV), hepatitis B, mumps, measles, rubella, poliovirus, and papilloma virus (1–4). Chromatid breaks and chromosome pulverizations are the most frequently observed anomalies, although translocations, coiling deficiencies, and overcondensation have also been reported. Most of the damage appears to be random, and with the exception of the oncogenic adenoviruses, there is no evidence for specific site-directed breakage. In the case of adenovirus type 12, infection of human cells at low multiplicity induces specific fragility at four distinct loci (5, 6). The three most common sites, 17q21–22, 1p36, and 1q12–22, coincide with the major small nuclear RNA loci, while the fourth site at 1q42–43, which is only observed at a low frequency in infected primary human cells, lies adjacent to the 5S rRNA locus (5–10).
The above observations raise key questions regarding the role of virus-mediated mutagenesis in human disease. In this paper, we focus on HCMV, a herpesvirus that is the major viral cause of birth defects. Each year, approximately 1% of all newborns are congenitally infected, and of these infants, 5–10% manifest signs of serious neurological defects at birth, which can include deafness, mental retardation, blindness, microcephaly, and cerebral calcification (11–14). In addition, 10–15% of infected infants who are asymptomatic at birth subsequently develop varying levels of sensorineural hearing loss and/or learning disabilities. HCMV infection is also a major medical problem in immunocompromised individuals (11).
In the past few years, it has become apparent that HCMV markedly dysregulates host cell functions and can inhibit cell cycle transit (15–18). Initial entry of HCMV into the cell leads to a second messenger-type response similar to that which occurs during regulation by means of hormones and growth factors (19). The productive HCMV infection stimulates the expression of genes encoding several proteins involved in preparing the cell for DNA replication, thereby leading to a fully “activated” state in the permissive cell (20–24). Although HCMV induces elevated steady state levels of p53 (15, 25, 26), there does not appear to be signaling to its downstream damage response targets, p21 and MDM2 (refs. 17 and 27, and E.A.F., unpublished results). Whether this block in signaling is because of viral protein binding and inhibition (25, 28, 29) or sequestration of p53 (30), it suggests a mechanism by which HCMV may fully “activate” the infected cell and precipitate genotoxic effects without triggering cell death.
Several investigators have previously noted a significant increase in the number of randomly distributed chromatid breaks and gaps in HCMV-infected cultures, but no specific breaks were reported (2–4). However, aspects of the experimental design of these early studies may well have obscured the observance of specific chromosomal aberrations. To detect this type of DNA damage, mitotic chromosomes must be examined. In some of the earlier experiments, the cells were synchronized in Go-phase and then infected. As noted above, these conditions inhibit cellular DNA synthesis and division, producing negligible metaphase cells for analysis (15–18). In addition, most of the previous experiments were performed at low multiplicities of infection (moi < 1), complicating the initial round of infection with unequal viral distribution. Finally, the cells were harvested for analysis at late times post infection (pi) (48 h or later), when viral DNA synthesis was at its peak and the cells could no longer cycle and display mitotic figures.
Unlike infection on release from Go, human fibroblasts infected in S-phase are refractory to viral protein expression (27). The majority of the S-phase-infected cells move through mitosis and by 24 h pi (hpi) are back in G1, where viral gene expression can then initiate. If DNA damage occurred in this cycling population, the cells could divide before virus replication, resulting in one of the daughter cells being free of viral genomes. This daughter cell could be a reservoir for genetic damage, opening up the possibility that disease syndromes might stem from this early damage rather than be because of active viral replication and cell lysis in the infected individual. As the target cells in the developing fetus are likely still dividing, we have revisited the question of HCMV-induced damage by using S-phase infected cells as a model system.
Materials and Methods
Cells and Virus.
Primary human foreskin fibroblasts (FFs) were obtained from the University of California, San Diego Medical Center, and human embryonic lung cells were obtained from the American Type Culture Collection (ATCC) (no. CCL 137). Both cell types were propagated in MEM Earle media in incubators maintained at 37°C and 5% CO2. Media was supplemented with 10% heat inactivated FBS, l-glutamine (2 mM), penicillin (200 units/ml), streptomycin (200 μg/ml), amphotericin B (1.5 μg/ml), and gentamycin sulfate (50 μg/ml). The Towne (no. VR 977) and AD169 (no. VR 538) strains of HCMV were obtained from the ATCC, and the Toledo strain was a gift from Stephen Spector (University of California, San Diego). HCMV strains were propagated as described earlier (31). Murine cytomegalovirus strain K181 was obtained and propagated as described (32). All viruses were used at an moi of 5 to ensure synchronous infection of all cells.
Cell Cycle Synchronization and Infection Conditions.
All experiments were performed under S-phase infection conditions (27). Cells were seeded into flasks and allowed to become confluent. After 2–3 days at confluence, cells were reseeded onto 10-cm dishes at 0.75 × 106 cells/dish. Approximately 24 h after plating, media was removed and the infection inoculum (either virus or an equivalent amount of mock conditioned media, diluted in fresh culture media) was added to the cells. At 2 hpi, the inoculum was removed, cells were washed, and fresh culture media was added. At 12 hpi, cells were harvested for metaphase chromosome analysis.
Pretreatment of Cells in Preparation for Mitotic Analysis.
Eleven hours after infection, 5 μg/ml ethidium bromide was added to the cells for 20 min to prevent overcondensation of chromosomes. Fresh media containing 0.1 μg/ml demecolcine was added for an additional 20 min to block microtubule polymerization, and then cells were trypsinized and collected for mitotic analysis. Cells were swollen in a hypotonic buffer containing 75 mM KCl and 10 mM EDTA (to prevent nuclease activity) for 20 min at room temperature. They were then pelleted, fixed using a series of incubations in 3:1 MeOH/acetic acid, and metaphase spreads were prepared and G-banded by standard cytogenetic methods. One hundred metaphase cells per sample were analyzed, unless otherwise noted.
Partial Purification of Virions.
Four milliliters of viral inoculum was placed in an ultracentrifuge tube, underlayed with 1.2 ml of a 25% sucrose solution (in PBS), and then spun at 40,000 rpm for 60 min at 4°C in a Beckman SW55 rotor. The top 3 ml were removed and considered the supernatant fraction. The remaining liquid above the pellet was discarded. The pelleted virus was then washed in 5 ml of cold PBS and repelleted as described above. The PBS was removed, and the pellet was resuspended in 4 ml of cold media. The pellet fraction (and the supernatant from spin one) was then sonicated for 1 min to ensure thorough resuspension. Equivalent amounts of either spun supernatant or resuspended virions were then used to infect cells. Mock and viral control samples were kept on ice for a period equivalent to the 2 spins to control for variability resulting from 4°C incubations.
UV Irradiation of Virus.
HCMV was UV-inactivated either by exposure to 6,000 J/m2 of irradiation in a Stratalinker 1800 (Exp. 9) or by exposure to the UV light within a bioguard tissue culture hood for 1 h (Exp. 10). Sodium pyruvate (5 mM final concentration) was added to the inoculum immediately after irradiation.
Virus Neutralization Assay.
Stocks of AD169 were preincubated for 1 h at 37°C with mAb to glycoprotein B, CH253 (a gift from Lenore Periera, University of California, San Francisco), Cytogam polyclonal hyperimmune globulin (a gift from MedImmune), control rabbit anti-mouse IgG (Jackson ImmunoResearch), or sterile PBS before addition to the cells. Abs were added at a concentration of 0.5 ng Ab/plaque forming unit. Volumes were adjusted with PBS to insure equivalent dilution. After 4 h, cells were washed, refed with fresh media, then harvested as described above for mitotic analysis.
Immunofluorescence Analysis.
Coverslips were included when cells were seeded and harvested at the indicated times pi. All coverslips were simultaneously fixed and permeabilized by incubation for 10 min with 100% MeOH at −20°C. Coverslips were blocked with normal goat sera (Jackson ImmunoResearch), then incubated with either mAb to pp65 or to IE1 72 and IE2 86 (CH 16.0) (both from the Goodwin Institute, Plantation, FL). Primary Abs were detected either with goat anti-mouse IgG (Jackson ImmunoResearch) or goat anti-mouse IgG1 (Southern Biotechnology Associates) coupled to fluorescein. All samples were counterstained with Hoechst dye (Sigma) to visualize the DNA. Slides were analyzed with a Zeiss Axiophot microscope fitted with a charge-coupled device camera and National Institutes of Health image software to capture images.
Results
The Towne Strain of HCMV Causes Specific Chromosomal Breaks in Primary Fibroblasts.
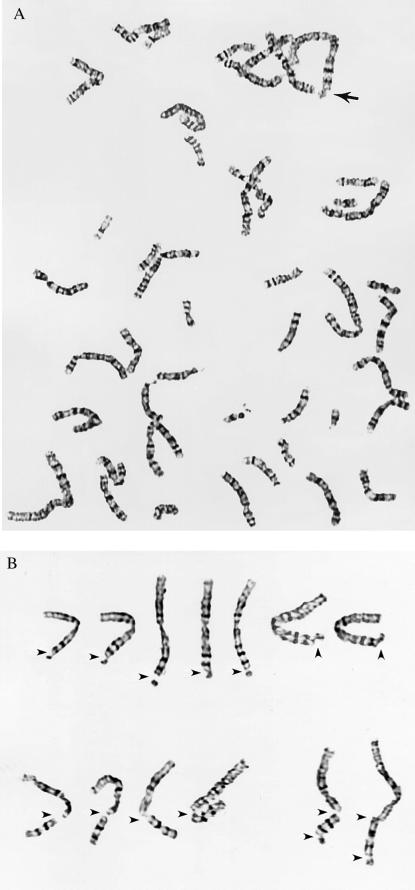
Analysis of primary FFs infected in S-phase with the Towne strain of HCMV revealed a dramatic increase in the incidence of specific breakage on the long arm of chromosome 1. Approximately 16% of the infected cells showed a specific break at either position 1q42 or 1q21, and this was virtually never seen in the mock-infected samples. Fig. 1 shows examples of the typical damage observed in HCMV-infected cells at metaphase (including breaks/gaps in band 1q42, breaks in 1q21, and 2 examples of double 1q21/1q42 breaks).
Figure 1.
Infection with HCMV produces specific breaks in chromosome 1. (A) A typical virally infected cell at metaphase, with the break at 1q42 indicated by an arrow. (B) Several different examples of the types of breaks observed in chromosome 1 at either position 1q42 alone (Upper), at position 1q21 alone (Lower Left), or a double break at positions 1q21 and 1q42 (Lower Right).
Exps. 1 and 2 of Table 1 summarize the data obtained with the Towne strain of HCMV. Several important features (that also hold true for the results presented in the subsequent tables) should be pointed out. In both mock- and HCMV-infected samples, cells were scored as aberrant if they possessed one or more breaks, gaps, or deletions. At the 1q42 position, the vast majority of damage was in the form of single chromatid breaks, while damage at the 1q21 position was almost equally divided between chromatid and chromosome breaks. The 1q42 break was observed in all viral samples. However, the 1q21 break, although reproducible, was not detected in every trial. Although the preponderance of cells incurred a break in only one copy of chromosome 1, breakage of both copies was present at a low frequency. The dual combination of 1q21 and 1q42 breaks within the same cell (and even the same chromosome) was also sporadically observed. Although aberrations were observed in the mock samples, the damage never included the specific 1q42 break and only once included the 1q21 break (Exp. 9).
Table 1.
The Towne strain of HCMV produces specific chromosomal damage in primary FFs
| Sample | No. aberrant cells/100 | No. of cells with specific breaks
|
||||
|---|---|---|---|---|---|---|
| 1q42 (×1) | 1q42 (×2) | 1q21 (×1) | 1q21 (×2) | 1q42 + 1q21* | ||
| Exp. 1 | ||||||
| HCMV Towne | 30 | 14 | 2 | 5 | 0 | 2 |
| Mock | 11 | 0 | 0 | 0 | 0 | 0 |
| Exp. 2 | ||||||
| HCMV Towne | 19 | 9 | 2 | 0 | 0 | 0 |
| Mock | 7 | 0 | 0 | 0 | 0 | 0 |
| Exp. 3† | ||||||
| HCMV Towne | 24 | 14 | 0 | 0 | 0 | 0 |
| Mock | 2 | 0 | 0 | 0 | 0 | 0 |
| Pelleted virus | 26 | 12 | 0 | 6 | 0 | 2 |
| Supernatant | 0 | 0 | 0 | 0 | 0 | 0 |
Breaks in chromosome 1q42 and 1q21 were scored as occurring either in one (×1) or both (×2) copies of chromosome 1 within the same cell. The small number of gaps seen at these loci have also been represented in these totals. In addition, the frequency with which both breaks occurred within the same cell is presented.
Indicates that different combinations of these two breaks were seen (i.e., 1q42(×2) + 1q21(×1) or 1q42(×1) + 1q21(×2), etc.).
†Denotes that in Experiment 3, 50 metaphases were scored and each entry represents the no. of cells/50 metaphases (×2).
Purified Viral Particles Can Induce Chromosome 1 Breaks.
When conducting studies on virus/host cell interactions, it is important to determine whether the virus itself or some component of the infected cell supernatant is responsible for the observed result. To answer this question, we purified virions from the cell supernatant by using a series of high-speed centrifugation and washing steps (see Materials and Methods). Exp. 3 in Table 1 documents that purified virions, and not the infected cell supernatant alone, were capable of inducing the specific chromosome 1 breaks.
Chromosome 1 Damage Is Not Cell-Type-Specific.
Because primary cells must be used for HCMV infections, there is a risk that underlying fragility in a single individual's chromosomes will be revealed on exposure to a potentially damaging agent. We therefore tested a completely unrelated source of human embryonic lung cells, also of primary origin. The data in Table 2 clearly demonstrates that the 1q42 break was observed in both trials at comparable or higher levels than in FFs. In Exp. 4, there was also a high incidence of 1q21 breaks over background. These results eliminate the possibility of cell-type specificity.
Table 2.
The HCMV-induced breaks at 1q42 and 1q21 are not cell-type specific
| Sample | No. aberrant cells/100 | No. of cells with specific breaks
|
||||
|---|---|---|---|---|---|---|
| 1q42 (×1) | 1q42 (×2) | 1q21 (×1) | 1q21 (×2) | 1q42 + 1q21* | ||
| Exp. 4 | ||||||
| FFs | ||||||
| HCMV Towne | 22 | 9 | 1 | 7 | 0 | 4 |
| Mock | 3 | 0 | 0 | 0 | 0 | 0 |
| HELs | ||||||
| HCMV Towne | 53 | 36 | 8 | 4 | 0 | 4 |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 |
| Exp. 5 | ||||||
| FFs§ | ||||||
| HCMV Towne | 6 | 6 | 0 | 0 | 0 | 0 |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 |
| HELs | ||||||
| HCMV Towne | 14 | 13 | 0 | 0 | 0 | 0 |
| Mock | 1 | 0 | 0 | 0 | 0 | 0 |
The Towne strain of HCMV was used to infect FFs or human embryonic lung (HEL) cells in parallel to assess the specificity of the 1q42 and 1q21 breaks. See legend to Table 1 for description of *.
§Denotes that the FF samples in experiment 5 represent the same samples labeled in experiment 7, Table 3.
Chromosome 1 Damage Is Not Strain-Specific.
HCMV strains vary considerably in the length of time that they have been continuously cultured in the laboratory and to a lesser extent in their genotype. To assess the role of viral passage number and strain type in the occurrence of the induced 1q42 and 1q21 breaks, we compared the high passage Towne and AD169 strains to the Toledo strain of HCMV. Although slightly adapted to tissue culture, the Toledo strain is genotypically and phenotypically much closer to standard clinical isolates, as its genome possesses a 13-kbp region absent from the high passage laboratory strains (33). As shown in Table 3, both the AD169 and Toledo strains produced results identical to the Towne strain with respect to the incidence of the specific chromosomal breakage in region 1q42, thus discounting any tissue culture/strain-specific phenomena in the induction of this break. The AD169 strain also induced the break at 1q21 in both trials performed.
Table 3.
The 1q42 and 1q21 breaks are observed with multiple HCMV strains, but not with murine CMV (MCMV)
| Sample | No. aberrant cells/100 | No. of cells with specific breaks
|
||||
|---|---|---|---|---|---|---|
| 1q42 (×1) | 1q42 (×2) | 1q21 (×1) | 1q21 (×2) | 1q42 + 1q21* | ||
| Exp. 6 | ||||||
| HCMV Towne | 19 | 10 | 0 | 2 | 0 | 1 |
| HCMV AD169 | 18 | 7 | 0 | 3 | 1 | 2 |
| Mock | 3 | 0 | 0 | 0 | 0 | 0 |
| Exp. 7 | ||||||
| HCMV Towne§ | 6 | 6 | 0 | 0 | 0 | 0 |
| HCMV AD169 | 10 | 7 | 0 | 1 | 1 | 0 |
| HCMV Toledo | 14 | 13 | 0 | 0 | 0 | 0 |
| Mock§ | 0 | 0 | 0 | 0 | 0 | 0 |
| Exp. 8† | ||||||
| HCMV Towne | 18 | 10 | 0 | 0 | 0 | 6 |
| MCMV K181 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 |
In a related experiment, we tested murine cytomegalovirus (MCMV), which has the ability to infect human cells and initiate viral protein synthesis (34), to see whether induction of damage was a virus-specific phenomenon. As shown in Table 3, Exp. 8, MCMV was incapable of causing chromosomal damage in human cells.
Viral Protein Expression Is Not Required for Induction of Damage.
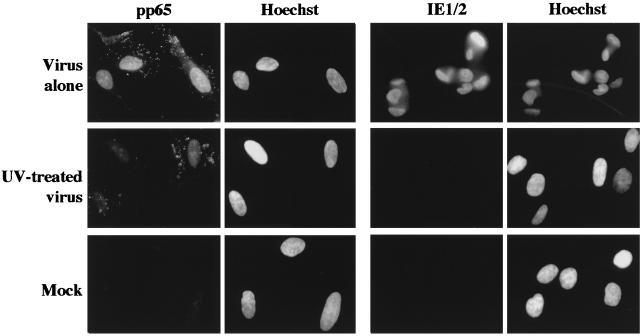
Next, we assessed the requirement for de novo viral gene expression in the induction of the specific breaks. Two parallel virus aliquots (equivalent to an moi of 5) were thawed; one was UV-inactivated, while the second was left untreated. Both UV irradiation procedures and doses (described in Materials and Methods) were predetermined to decrease expression of the HCMV genome, such that <1% of the cells synthesized viral immediate early proteins at 24 hpi (as assayed by immunofluorescence and demonstrated in Fig. 2). Under these conditions, viral entry was not affected, as measured by pp65 tegument protein staining in the nuclei of all cells, as was observed for cells incubated with untreated virus in this and all other experiments. The addition of sodium pyruvate to the media after irradiation was essential for survival of mitotic cells, as cultures with media lacking this supplement showed apoptotic destruction of all M-phase cells, most probably because of oxygen free radical production during irradiation. Mock supernatant was also treated to control for variability introduced by the irradiation procedure. As can be seen in Table 4, Exps. 9 and 10, both untreated virus and virus UV-inactivated with either irradiation procedure scored equally well with respect to induction of the specific 1q42 and 1q21 damage. Thus, new viral gene expression is not required for the induction of chromosome 1 breaks.
Figure 2.
UV inactivation does not inhibit virion entry, but does inhibit viral protein expression. Coverslips from Exp. 10 were harvested at either 4.5 hpi (for pp65 staining) or 24 hpi (for immediate early protein staining). Primary Abs to pp65 and to the immediate early proteins (CH 16.0) were both detected with goat anti-mouse IgG coupled with fluorescein. All cells were counterstained with Hoechst dye to visualize the DNA. (Magnification, ×240).
Table 4.
The 1q42 and 1q21 breaks do not require viral gene expression, but do require viral binding and/or penetration
| Sample | No. aberrant cells/100 | No. of cells with specific breaks
|
||||
|---|---|---|---|---|---|---|
| 1q42 (×1) | 1q42 (×2) | 1q21 (×1) | 1q21 (×2) | 1q42 + 1q21* | ||
| Exp. 9 | ||||||
| HCMV Towne | 18 | 11 | 0 | 4 | 1 | 2 |
| HCMV Towne + UV | 22 | 11 | 0 | 2 | 1 | 3 |
| Mock | 1 | 0 | 0 | 1 | 0 | 0 |
| Exp. 10† | ||||||
| HCMV Towne | 20 | 10 | 0 | 4 | 0 | 2 |
| HCMV Towne + UV | 14 | 12 | 0 | 2 | 0 | 0 |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 |
| Exp. 11† | ||||||
| HCMV AD169 | 6 | 6 | 0 | 0 | 0 | 0 |
| HCMV + CH253 | 0 | 0 | 0 | 0 | 0 | 0 |
| HCMV + cytogam | 0 | 0 | 0 | 0 | 0 | 0 |
| HCMV + control Ab | 8 | 6 | 0 | 2 | 0 | 0 |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 |
HCMV Towne was UV-irradiated to eliminate viral gene expression and then used to assay for damage in experiments 9 and 10. In addition, HCMV AD169 was incubated with either control or neutralizing Abs before exposure to cells in Experiment 11. See Table 1 for explanation of ∗ and †.
Virion Binding and/or Entry Is Required for Damage Induction.
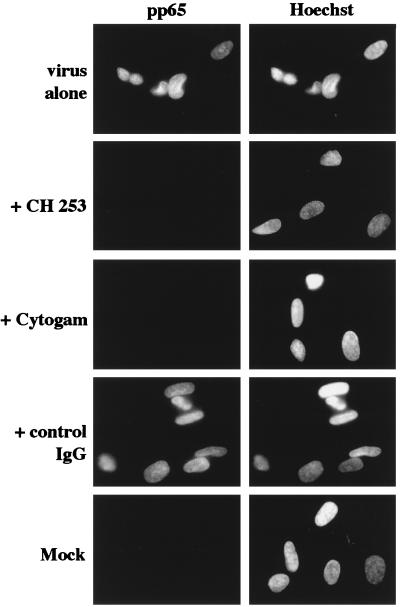
To determine the nature of the virus/host cell interaction required for the induction of chromosome 1 breaks, we utilized the following neutralizing Abs: a mAb (CH 253) specific for the viral envelope glycoprotein gB, which is a main component in viral attachment and penetration, and a purified IgG fraction of polyvalent hyperimmune sera isolated from HCMV-positive individuals (Cytogam). CH253 does not block viral binding to the cell surface, but does inhibit penetration of the virus into the host cell (35). As described above, viral penetration was assessed in this experiment by immunostaining with an Ab specific for the pp65 tegument protein. Fig. 3 shows that viral entry is blocked by incubation with the neutralizing Abs, but not with control IgG. As can be seen in Table 4, Exp. 11, both test Abs were equally potent at preventing the induction of chromosomal damage by strain AD169 (and the Towne strain; data not shown). These data suggest that virion penetration, and/or a specific receptor/virion protein interaction at the cell surface (blocked by the neutralizing Ab), are required for chromosome 1 breakage to occur in S-phase-infected fibroblasts.
Figure 3.
Neutralizing Abs prevent virus penetration. Coverslips from Exp. 11 were harvested at 12 hpi and stained with pp65 primary Ab, followed by detection with goat anti-mouse IgG1 coupled to fluorescein. Nuclei were counterstained with Hoechst dye. (Magnification, ×240).
Discussion
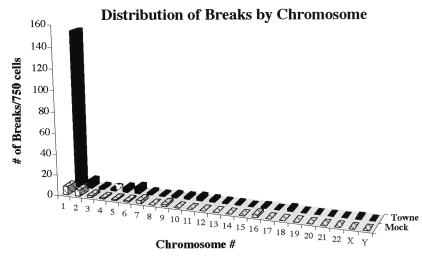
One might expect, in light of the wealth of data concerning other viruses, to see an increased incidence of breakage in all chromosomes in HCMV-infected cells. However, if one compiles the data from all experiments using untreated Towne virus on FFs and examines the distribution of breaks within the aberrant cells, a striking pattern is revealed. As illustrated in Fig. 4, there appears to be a nonspecific, random distribution of low background breaks/gaps occurring in the virally infected cells that is parallel to that seen in the mock cells. However, HCMV causes a dramatic increase in the number of breaks in chromosome 1, almost completely accounted for by specific breaks at 1q42 and 1q21. The data show that there is not an increased incidence in overall breaks spread throughout the genome in the HCMV-infected cells, but rather an enhanced fragility at specific loci on chromosome 1.
Figure 4.
HCMV does not lead to an increase in random breaks, but rather an increase in specific breaks on chromosome 1. Data from all experiments using FFs infected with untreated HCMV Towne (or their mock counterparts) were compiled (750 metaphase cells in total). Bars represent the total number of breaks per chromosome for those 750 cells. It should be noted that a single aberrant cell scored in Tables 1–4 could possess a break in more than one chromosome and could therefore be represented in more than one bar in this graph.
Why is this the first time that experiments using HCMV-infected cells have revealed specific damage to chromosome 1? First, our studies were performed at mois sufficient to ensure that all cells were infected simultaneously (as seen in Fig. 2). It may be that a higher concentration of viral particles is needed to observe specific damage. In fact, others have noted that not even nonspecific damage was significantly increased above background until mois greater than 1 were used (3). Secondly, and perhaps most importantly, the infections were carried out when a large proportion of cells (approximately 50–60%) were in the S- and G2/M-phases of the cell cycle. At these times, the chromosomes are likely most susceptible to damaging agents resulting from the partial unwinding that occurs during S-phase and the partial condensation events that occur during the G2/M-phases. The preponderance of chromatid vs. chromosome breaks at the 1q42 position would also argue that specific chromosome 1 damage is incurred after DNA replication rather than during G1 (36). In addition, because our experimental protocol resulted in a much larger proportion of mitotic cells at the time of observation, the probability of being able to detect specific chromosomal breaks was enhanced. The cells were harvested fairly rapidly after infection to maximize the potential for seeing damage incurred in the first round of division. Previous analyses were performed much later in infection, when there was likely more global damage to the host DNA, as well as loss of a significant fraction of cells because of virally mediated cell death.
A major question for the future is: what genes lie at these breakpoints? The possible targets residing near 1q42 include: the ADPRT locus involved in DNA repair and replication (37); a potential tumor suppressor gene, whose deletion has been connected to the development of gliomas (38); the major 5S rRNA locus (10); and the USH2A gene (39, 40). The possible targets that reside near 1q21 include: a different proposed tumor suppressor gene deleted in several primary breast tumors (41), the PSU1 small nuclear RNA locus (7), and the DFNA7 gene (42). Two of these loci, DFNA7 and USH2A, are of particular interest with respect to HCMV pathogenesis. The DFNA7 gene located within 1q21–23 has been linked to the inheritance of an autosomal dominant, nonsyndromic, progressive hearing loss (42). The deletion of the DFNA7 gene because of HCMV-induced breakage could potentially be linked to the development of the progressive hearing loss observed in infants congenitally infected with HCMV. The USH2A gene, located at 1q41, close to the most prevalent HCMV-induced break, encodes a protein involved in the development of Usher's Syndrome Type II. Usher's Syndrome is an autosomal recessive disorder that affects 3–6% of children born with hearing impairments and is the most frequent cause of combined deafness and blindness in adults (39, 40). These individuals have sensorineural hearing deficiencies at birth and later develop retinal problems. This disease profile is similar to that of children congenitally infected with HCMV. The predicted ORF for the USH2A protein contains both laminin epidermal growth factor and fibronectin type III motifs, and thus the gene may encode a basement membrane-type protein. One could envision a scenario where HCMV-induced breaks at this locus caused deletion of the 1q terminal region (in one or both copies of chromosome 1), which was then passed on to daughter cells. Subsequent deletion of the other USH2A locus, either by loss of heterozygosity or chromosome erosion could abolish all USH2A protein expression, producing an Usher-type syndrome similar to congenital HCMV infection.
In future studies, we will examine cells of neuronal and glial descent. The fetal brain is most susceptible to HCMV-induced damage during the first half of pregnancy, a time when neuronal cell division and differentiation are maximal (12). We know from tissue culture experiments that several cell types within the brain show varying degrees of permissiveness for the HCMV infection, and this susceptibility appears to depend on the origin of the cell and the state of differentiation (43–46). The gamut of permissiveness ranges from the complete susceptibility of human primary retinal glial cells to a total block in viral protein synthesis and DNA replication in undifferentiated glioblastoma cells (43, 45). It is interesting to note that the same cell line can become more fully permissive on differentiation (44, 46).
The observation that chromosomal damage can occur in the absence of de novo viral gene expression becomes very important when taken in the context of the variability of permissiveness and restriction to gene expression observed in different neuronal cell types at various stages of differentiation. These data suggest that in utero, semi- to nonpermissive undifferentiated neural cells might be infected by HCMV and incur chromosomal damage. As these neuronal precursors divide, they could then pass on this damage to daughter cells, which in turn could either divide again or differentiate and subsequently migrate to their positions within the cortex or other regions of the developing brain.
Lack of a requirement for viral protein synthesis also sets HCMV apart from other damage-inducing viruses. Prior studies have shown that the adenovirus type 12 E1B protein is absolutely required for induction of damage in infected cells, and can actually induce damage independently of viral infection (9, 47). In addition, the damage induced by the HCMV-related herpes simplex virus, which appears as the uncoiling of chromosome 1q12–21 and the pericentric regions of chromosomes 9 and 16 rather than specific breakage, also requires immediate early viral protein synthesis (48, 49).
What, then, is the mechanism behind this HCMV-induced chromosome damage? One possibility could be the early physiological changes brought about by viral binding and the rapid yet transient induction of c-fos, c-jun and c-myc mRNAs, which occurs even in the absence of serum and with UV-inactivated virus (19, 23). The underlying mechanism also could be related to the early up-regulation of another class of mRNAs that are a subset of genes normally induced by α interferon in uninfected cells (50, 51). This HCMV-associated induction appears to simply require the exposure of the cell to virions, noninfectious enveloped particles, or dense bodies. Viral binding also triggers the generation of reactive oxygen intermediates in some cells, which could potentially lead to DNA damage (52, 53). Alternatively, a viral or cellular protein component of the incoming virion or the viral DNA itself may be responsible for the induced damage. Whatever the mechanism, it is clear from our experiments that HCMV infection, without the requirement for new viral gene expression, sets the stage for specific damage to chromosome 1. Thus, any or all of these early events could lead to the induction of a DNA damage pathway or to a block in DNA repair processes in which chromosome 1 is particularly susceptible. The challenges are now to decipher the signaling pathway leading to chromosome damage and to assess the importance of these breaks in chromosome 1 for the pathogenesis of HCMV in the human embryo during development.
Acknowledgments
This work was supported in part by National Institutes of Health Training Grant AI 07036, March of Dimes Grant 5-FY98-0727 (to E.A.F.), and National Institutes of Health Grant CA73490 (to D.H.S.).
Abbreviations
- HCMV
human cytomegalovirus
- pi
postinfection
- hpi
hours postinfection
- FFs
foreskin fibroblasts
- moi
multiplicity of infection
References
- 1.Harnden D G. In: Chromosomes and Cancer. German J, editor. New York: Wiley; 1974. pp. 151–190. [Google Scholar]
- 2.Lüleci G, Sakízlí M, Günalp A. Acta Virol. 1980;24:341–345. [PubMed] [Google Scholar]
- 3.AbuBakar S, Au W W, Legator M S, Albrecht T. Environ Mol Mutagen. 1988;12:409–420. doi: 10.1002/em.2860120409. [DOI] [PubMed] [Google Scholar]
- 4.Deng C Z, AbuBakar S, Fons M P, Boldogh I, Hokanson J, Au W W, Albrecht T. Environ Mol Mutagen. 1992;19:304–310. doi: 10.1002/em.2850190407. [DOI] [PubMed] [Google Scholar]
- 5.zur Hausen H. J Virol. 1967;1:1174–1185. doi: 10.1128/jvi.1.6.1174-1185.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steffensen D M, Szabo P, McDougall J K. Exp Cell Res. 1976;100:436–439. doi: 10.1016/0014-4827(76)90176-2. [DOI] [PubMed] [Google Scholar]
- 7.Lindgren V, Bernstein L B, Weiner A M, Francke U. Mol Cell Biol. 1985;5:2172–2180. doi: 10.1128/mcb.5.9.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindgren V, Ares M, Jr, Weiner A M, Francke U. Nature (London) 1985;314:115–116. doi: 10.1038/314115a0. [DOI] [PubMed] [Google Scholar]
- 9.Schramayr S, Caporossi D, Mak I, Jelinek T, Bacchetti S. J Virol. 1990;64:2090–2095. doi: 10.1128/jvi.64.5.2090-2095.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sørensen P D, Lomholt B, Frederiksen S, Tommerup N. Cytogenet Cell Genet. 1991;57:26–29. doi: 10.1159/000133107. [DOI] [PubMed] [Google Scholar]
- 11.Britt W, Alford C. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 2493–2523. [Google Scholar]
- 12.Cinque P, Marenzi R, Ceresa D. Intervirology. 1997;40:85–97. doi: 10.1159/000150536. [DOI] [PubMed] [Google Scholar]
- 13.Fowler K B, McCollister F P, Dahle A J, Boppana S, Britt W J, Pass R F. J Pediatr (Berlin) 1997;130:624–630. doi: 10.1016/s0022-3476(97)70248-8. [DOI] [PubMed] [Google Scholar]
- 14.Boppana S B, Fowler K B, Britt W J, Stagno S, Pass R F. Pediatrics. 1999;104:55–60. doi: 10.1542/peds.104.1.55. [DOI] [PubMed] [Google Scholar]
- 15.Jault F M, Jault J-M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu M, Shenk T. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 18.Dittmer D, Mocarski E S. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albrecht T, Fons M P, Bologh I, AbuBakar S, Deng C Z, Millinoff D. Transplant Proc. 1991;23:48–55. [PubMed] [Google Scholar]
- 20.Hirai K, Watanabe Y. Biochim Biophys Acta. 1976;447:328–339. doi: 10.1016/0005-2787(76)90056-3. [DOI] [PubMed] [Google Scholar]
- 21.Estes J E, Huang E-S. J Virol. 1977;24:13–21. doi: 10.1128/jvi.24.1.13-21.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isom H C. J Gen Virol. 1979;42:265–278. doi: 10.1099/0022-1317-42-2-265. [DOI] [PubMed] [Google Scholar]
- 23.Boldogh I, AbuBakar S, Deng C Z, Albrecht T. J Virol. 1991;65:1568–1571. doi: 10.1128/jvi.65.3.1568-1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speir E, Modali R, Huang E-S, Leon M B, Sahwl F, Finkel T, Epstein S E. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 26.Muganda P, Mendoza O, Hernandez J, Qian Q. J Virol. 1994;68:8028–8034. doi: 10.1128/jvi.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvant B S, Fortunato E A, Spector D H. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muralidhar S J, Doniger J, Medelson E, Araujo J C, Kashanchi F, Azumi N, Brady J N, Rosenthal L J. J Virol. 1996;70:8691–8700. doi: 10.1128/jvi.70.12.8691-8700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonin L R, McDougall J K. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortunato E A, Spector D H. J Virol. 1998;72:2033–2039. doi: 10.1128/jvi.72.3.2033-2039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamashiro J C, Hock L J, Spector D H. J Virol. 1982;42:547–557. doi: 10.1128/jvi.42.2.547-557.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morello C S, Cranmer L D, Spector D H. J Virol. 1999;73:7678–7693. doi: 10.1128/jvi.73.9.7678-7693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker D, Hudson J. Arch Virol. 1987;92:103–119. doi: 10.1007/BF01310066. [DOI] [PubMed] [Google Scholar]
- 35.Navarro D, Paz P, Tugizov S, Topp K, La Vail J, Pereira L. Virology. 1993;197:143–158. doi: 10.1006/viro.1993.1575. [DOI] [PubMed] [Google Scholar]
- 36.Nichols W W. Annu Rev Microbiol. 1970;24:479–500. doi: 10.1146/annurev.mi.24.100170.002403. [DOI] [PubMed] [Google Scholar]
- 37.Baumgartner M, Schneider R, Auer B, Herzog H, Schweiger M, Hirsch-Kauffmann M. Cytogenet Cell Genet. 1992;61:172–174. doi: 10.1159/000133400. [DOI] [PubMed] [Google Scholar]
- 38.Li Y S, Ramsay D A, Fan Y S, Armstrong R F, Del Maestro R F. Cancer Genet Cytogenet. 1995;84:46–50. doi: 10.1016/0165-4608(95)00065-8. [DOI] [PubMed] [Google Scholar]
- 39.Sumegi J, Wang J Y, Zhen D K, Eudy J D, Talmadge C B, Li B F, Berglund P, Weston M D, Yao S F, Ma-Edmonds M, et al. Genomics. 1996;35:79–86. doi: 10.1006/geno.1996.0325. [DOI] [PubMed] [Google Scholar]
- 40.Eudy J D, Weston M D, Yao S, Hoover D M, Rehm H L, Ma-Edmonds M, Yan D, Ahmad I, Cheng J J, Ayuso C, et al. Science. 1998;280:1753–1757. doi: 10.1126/science.280.5370.1753. [DOI] [PubMed] [Google Scholar]
- 41.Bièche I, Champème M H, Lidereau R. Clin Cancer Res. 1995;1:123–127. [PubMed] [Google Scholar]
- 42.Fagerheim T, Nilssen O, Raeymaekers P, Brox V, Moum T, Elverland H H, Teig E, Omland H H, Fostad G K, Tranebjaerg L. Hum Mol Genet. 1996;5:1187–1191. doi: 10.1093/hmg/5.8.1187. [DOI] [PubMed] [Google Scholar]
- 43.Jault F M, Spector S A, Spector D H. J Virol. 1994;68:959–973. doi: 10.1128/jvi.68.2.959-973.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poland S D, Bambrick L L, Dekaban G A, Rice G P. J Infect Dis. 1994;170:1267–1271. doi: 10.1093/infdis/170.5.1267. [DOI] [PubMed] [Google Scholar]
- 45.Burd E M, Pulido J S, Puro D G, O'Brien W J. Invest Ophthalmol Visual Sci. 1996;37:1957–1966. [PubMed] [Google Scholar]
- 46.Spiller O B, Borysiewicz L K, Morgan B P. J Gen Virol. 1997;78:3349–3356. doi: 10.1099/0022-1317-78-12-3349. [DOI] [PubMed] [Google Scholar]
- 47.Liao D, Yu A, Weiner A M. Virology. 1999;254:11–23. doi: 10.1006/viro.1998.9512. [DOI] [PubMed] [Google Scholar]
- 48.Chenet-Monte C, Mohammad F, Celluzzi C M, Schaffer P A, Farber F E. Virol Res. 1986;6:245–260. doi: 10.1016/0168-1702(86)90073-0. [DOI] [PubMed] [Google Scholar]
- 49.Peat D S, Stanley M A. J Gen Virol. 1986;67:2273–2277. doi: 10.1099/0022-1317-67-10-2273. [DOI] [PubMed] [Google Scholar]
- 50.Zhu H, Cong J P, Shenk T. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shibutani T, Johnson T M, Yu Z X, Ferrans V J, Moss J, Epstein S E. J Clin Invest. 1997;100:2054–2061. doi: 10.1172/JCI119738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Speir E, Yu Z X, Ferrans V J, Huang E S, Epstein S E. Circ Res. 1998;83:210–216. doi: 10.1161/01.res.83.2.210. [DOI] [PubMed] [Google Scholar]