Abstract
ymf39 is a conserved hypothetical protein-coding gene found in mitochondrial genomes of land plants and certain protists. We speculated earlier, based on a weak sequence similarity between Ymf39 from a green alga and the atpF gene product from Bradyrhizobium, that ymf39 might code for subunit b of mitochondrial F0F1-ATP synthase. To test this hypothesis, we have sequenced ymf39 from five protists with minimally derived mitochondrial genomes, the jakobids. In addition, we isolated the mitochondrial ATP synthase complex of the jakobid Seculamonas ecuadoriensis and determined the partial protein sequence of the 19-kDa subunit, the size expected for Ymf39. The obtained peptide sequence matches perfectly with a 3′-proximal region of the ymf39 gene of this organism, confirming that Ymf39 is indeed an ATP synthase subunit. Finally, we employed statistical tests to assess the significance of sequence similarity of Ymf39 proteins with each other, their nucleus-encoded functional counterparts, ATP4/ATP5F, from fungi and animals and α-proteobacterial ATP synthase b-subunits. This analysis provides clear evidence that ymf39 is an atpF homolog, while ATP4/ATP5F appears to be a highly diverged form of ymf39 that has migrated to the nucleus. We propose to designate ymf39 from now on atp4.
INTRODUCTION
Two decades of research in mitochondrial genomics have reinforced the view that mitochondria arose from an α-proteobacterium through an endosymbiotic event that occurred only once in the evolution of eukaryotes. Mitochondria enclose their own DNA (mtDNA), a vestigial bacterial genome containing up to ∼100 genes encoding constituents of this organelle with functions in respiration, oxidative phosphorylation and protein synthesis, but also protein import and maturation, RNA processing, and, in rare instances, transcription (for a review, see 1). However, a few genes have remained resistant to functional identification until now.
The first mitochondrial genome to be sequenced completely was that of Homo sapiens, in 1981 (2,3). At that time, only 6 of the 13 protein-coding genes in animal mtDNAs were identified, i.e. those coding for apocytochrome b, cytochrome oxidase subunits 1 to 3 and ATP synthase subunits 6 and 8. In the following years, all seven predicted, hypothetical protein-coding genes designated urf1-4, 4L, 5 and 6 have been shown to be expressed (4), and to specify subunits of NADH dehydrogenase (5,6) (URF stands for ‘Unassigned Reading Frame’; the current nomenclature is ORF, mnemonic for ‘Open Reading Frame’). Since this first report, numerous additional ORFs have been encountered in mtDNAs of other organisms, in numbers varying extensively across eukaryotes. Mitochondrial ORFs are essentially absent in animals; they are particularly abundant in land plants (e.g. approximately 40 ORFs larger than 100 amino acids in the thale cress, Arabidopsis thaliana) (7,8); whereas fungal and protist mtDNAs harbor a more modest five ORFs on average (for a review, see 9). Many of these new ORFs carry no significant sequence similarity to known or hypothetical proteins, some are conserved but confined to a particular group of organisms (clade-specific ORFs), while a small number are broadly distributed across eukaryotes. It is in the latter two categories of ORFs that one might suspect incognito genes, i.e. genes that belong to the ancestral bacterial set but are difficult to identify due to their high sequence divergence.
A considerable number of mitochondrial ORFs are enclosed in introns. Most of these ORFs are related to endonucleases, reverse transcriptases and RNA maturases and have been shown to play roles in intron splicing and propagation (for reviews, see 10,11). Because such ORFs are apparently absent from extant α-proteobacteria, they are believed to have been introduced secondarily into mitochondria. A further group of conserved mitochondrial ORFs, which probably do not originate from the mitochondrial ancestor, resemble RNA and DNA polymerases otherwise found in mitochondrial plasmids (12). In fact, most if not all rpo and dpo-related ORFs occurring in mtDNAs are likely vestiges of plasmid insertion events, often leaving behind pseudo-genes that are shortened and/or fragmented.
A few mitochondrial ORFs have been shown to be genuine mitochondrion-encoded genes. Among them are var1 of Saccharomyces cerevisiae, the intron-encoded S5 from Neurospora crassa and orf277 (or urfa) from Schizosaccharomyces pombe, all of which have recently been recognized to code for ribosomal protein S3 (13). Another example is the conserved ORF ymf16 found in mtDNAs of plants and several protists, which is likely a homolog of mttB, involved in Sec-independent protein import (14,15). Similarly, ymf19, originally designated ORFb in flowering plants and associated with male sterility, is now known as atp8, specifying a subunit of mitochondrial F0F1-ATP synthase (1). Finally, orf86a, which was initially described in mtDNA of the liverwort Marchantia polymorpha (16), and later also found in protists, has been revealed to be a highly diverged version of the gene coding for succinate dehydrogenase subunit 4 (17,18).
Here, we describe the identification of the mitochondrial ORF ymf39 (originally designated orf25) (19,20), which has remained resistant to identification for more than a decade. This ORF has long been suspected of functional significance as it is present in numerous protist and plant mtDNAs. This notion was reinforced by the finding that ymf39 is transcribed in land plants (20) and specifies an unknown, integral mitochondrial membrane component (21). Therefore, we set out to examine minimally derived Ymf39 proteins, as an attempt to trace back this protein’s line of descent. The organisms of choice for our studies are jakobid-like flagellates (22). In taxonomic terms, jakobid-like flagellates are an informal group, which has been subdivided on morphological and ultrastructural grounds into two families: (i) the Jakobidae [referred to as ‘jakobids’ in the following, but also dubbed ‘core jakobids’ elsewhere (23)], including the genera Jakoba, Reclinomonas, Histiona and Seculamonas; and (ii) the Malawimonadidae or malawimonads (24), currently including only two species, Malawimonas jakobiformis and Malawimonas californiana. Members of the Jakobidae possess the most gene-rich and bacteria-like mitochondrial genomes of all eukaryotes (25; G.Burger and B.F.Lang, unpublished results), while mtDNAs of the phylogenetically distinct Malawimonadidae (22,23) are less gene-rich and resemble rather those of other protists and plants (9).
MATERIALS AND METHODS
Sequence accession numbers
The here reported novel DNA sequences of ymf39 genes have been deposited in GenBank under the following accession numbers: Jakoba bahamiensis BHBA-1, AY236972; J.libera CB, AY236973; Reclinomonas americana MD, AY236975; R.americana MI, AY236976; M.californiana, AY236974; and Seculamonas ecuadoriensis, AY236977.
Strains
Flagellate strains used in this study, most of which were originally isolated by T.Nerad (American Type Culture Collection), were obtained from the ATCC: J.bahamiensis BHBA-1 (ATCC 50695), J.libera CB (ATCC 50422), R.americana MD (ATCC 50283), R.americana MI (ATCC 50284), M.californiana (ATCC 50740) and S.ecuadoriensis (ATCC 50688).
Culture of S.ecuadoriensis, DNA cloning and sequencing
Seculamonas ecuadoriensis was grown in WCL medium with gentle shaking at 24°C. The protists were fed with live Enterobacter aerogenes (ATCC 13048). One 600-ml culture yielded ∼0.5 g of cells after 8 days. Preparation of mtDNA, cloning and DNA sequencing was performed as described previously (17).
Purification of mitochondrial membranes from S.ecuadoriensis
Approximately 5 g of cells were used for protein-chemical studies. As described in detail elsewhere (26), cells were disrupted by sonication and mitochondrial membranes were purified by centrifugation through sucrose step gradients. Protein complexes were separated by blue-native polyacryl amide gel electrophoresis (27) and subjected to tricine-SDS gel electrophoresis in a second dimension. The resulting bands, which correspond to individual subunits, were cut out of the gel, eluted and digested with trypsin as described in (28). The resulting peptides were analyzed by electrospray ionization tandem mass spectrometry (ESI-MS/MS) on a Q-TOF apparatus (MICROMASS Technology). The sequence obtained from a peptide of the 19-kDa protein was KA{I/L}QEG{I/L}EM{I/L}ER. Other peptides derived from this protein did not yield interpretable results. Note that isoleucine and leucine are not distinguishable by this technique.
Protein sequence and secondary structure analysis
Position-specific iterative BLAST (PSI-BLAST) (29) was performed remotely using the web server at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/BLAST). An initial BLAST search was performed with the S.ecuadoriensis protein in the non-redundant database of GenBank, using the default values for the substitution matrix (Blosum62), gap costs (=11 for opening; =1 for extension), and the statistical significance threshold (E = 10). The matches with E-values better than the threshold were found with two protistan mitochondrial ORFs (E = 7e-29; E = 1e-07) and ATP synthase subunit b from an α-proteobacterium (E = 0.002). With these highest scoring hits, a profile was constructed, with which the same database was searched again (first iteration). To avoid a preponderance of the highly conserved bacterial sequences in the generation of a refined profile, the input to the second iteration consisted of all seven mitochondrion-encoded Ymf39 sequences retrieved in the first iteration, but was restrained to only three bacterial sequences, one each with a low (E = 3e-34), medium (E = 5e-08) and high (E = 0.004) E-value. A third and final iteration was performed including all mitochondrion-encoded (approximately 30) and nucleus-encoded (i.e. two) proteins that were retrieved in the second iteration, and the three bacterial counterparts as before.
Hydrophobicity profiles of proteins were calculated with the PROTSCALE tool at ExPASy (http://ca.expasy.org/tools/) using the amino acid scale of Kyte and Doolittle (30). Membrane-spanning regions were predicted with TMpred (31) at EMBnet (http://www.ch.embnet.org/), using 14 residues as a minimum length of the hydrophobic part of the transmembrane helix. Multiple protein alignments were performed with ClustalW (32), and refined manually.
Statistical tests of the significance of similarity scores were performed with PRSS version 3 (33), available at the EMBnet node server (http://www.ch.embnet.org). This tool first calculates the optimal similarity scores for two given sequences, then repeatedly shuffles the second sequence, recalculates the optimal similarity score using the Smith–Waterman algorithm (34), and compares the new score with the initial score. The characteristic parameters of the extreme-value distribution are then used to estimate the probability that each of the unshuffled sequence scores would be obtained by chance. Only regions in which the sequences can be unambiguously aligned have been used to calculate the significance values. The following parameters were used for PRSS analyses: PAM250 as the scoring matrix (suited for distant proteins) and the default values for the number of shuffles (=200); window size (=10); gap opening penalty (=12) and gap extension penalty (=2).
RESULTS
Ymf39 is an ATP synthase subunit in S.ecuadoriensis
Nearly a decade ago, we discovered a short region of weak sequence similarity between Ymf39 of the green alga Prototheca wickerhamii and the atpF gene product of the α-proteobacterium Bradyrhizobium japonicum. Based on this observation, we considered the possibility that ymf39 codes for subunit b of mitochondrial F0F1-ATP synthase (35). To follow up on this idea, we set out to examine the mitochondrial ATP synthase of the jakobid S.ecuadoriensis. Among Jakobidae, this species is best suited for protein-chemical studies, because it can be cultured in appropriate quantities.
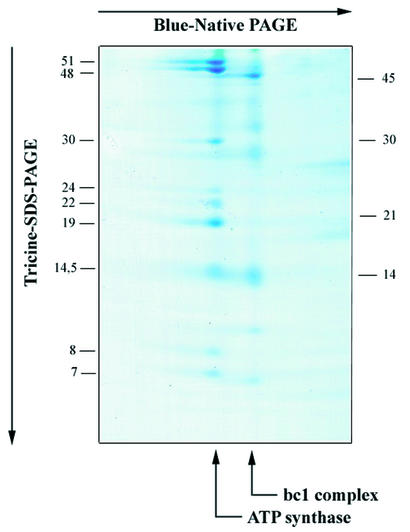
First, we determined the DNA sequence of the S.ecuadoriensis ymf39, which is 573 nt in length. In a second step, we characterized the ATP synthase of this protist, a complex with apparent weight of 550 kDa, as reported recently (26). Separation of this enzyme complex by two-dimensional-gel electrophoresis revealed at least nine subunits of sizes 51, 48, 30, 24, 22, 19, 14, 8 and 7 kDa (Fig. 1). As the gene sequence of the S.ecuadoriensis ymf39 predicts a protein of 190 residues, we extracted a subunit of the corresponding size (∼19 kDa) from the gel, and analyzed several of its fragments by electrospray ionization tandem mass spectrometry. One of the peptides yielded a spectrum with a total of 12 consecutive, unambiguous positions. Sequence comparison revealed that this peptide perfectly matches a stretch in the C-terminal region of the deduced protein sequence of the S.ecuadoriensis ymf39, and thus confirms that Ymf39 is indeed a mitochondrion-encoded ATP synthase subunit.
Figure 1.
Separation of ATP synthase subunits of S.ecuadoriensis. Membrane complexes were separated by blue-native polyacryl amide gel electrophoresis in the first dimension, and ATP synthase subunits were separated by Tricine–SDS polyacryl amide gel electrophoresis in the second dimension. Sizes of standard proteins are indicated in the right margin, and inferred sizes of ATP synthase subunits are given in the left margin. The protein complex from S.ecuadoriensis was identified by the characteristic subunit pattern, and sequence analysis of selected subunits using mass spectrometry (26).
Identification of Ymf39 homologs
The only published gene sequences from jakobid-like flagellates are from R.americana NZ (25) and M.jakobiformis (36). To better define the degree of sequence conservation among putatively minimally derived Ymf39 proteins, we sequenced the corresponding gene from four additional jakobids, J.bahamiensis, J.libera, R.americana MD and R.americana MI, as well as one malawimonad, M.californiana. In the three R.americana strains, Ymf39 is 191–197 residues long, with 86.4–86.9% identity over the entire length of the protein. Sequence identities between Ymf39 of R.americana strains, S.ecuadoriensis, J.bahamiensis and J.libera range from 38.8 to 48.4% and extend over essentially the entire protein except the C-terminal 10–20 residues. In contrast, sequence similarities among malawimonads, as well as between malawimonads and jakobids, are confined to an approximate 70 residue N‐terminal stretch only.
To find out which of the bacterial and eukaryotic/nuclear-encoded F0F1-ATP synthase subunits may correspond to Ymf39, we employed position-specific iterative BLAST analysis (29). An initial BLAST search with the S.ecuadoriensis protein identified three sequences with an E-value higher than the threshold, namely Ymf39 of R.americana and the cryptophyte alga Rhodomonas salina, and ATP synthase subunit b (AtpF) from the α-proteobacterium B.japonicum. A profile was constructed using the three highest scoring hits, with which the database was searched again in three subsequent iterations. These searches returned more mitochondrion-encoded, putative Ymf39 proteins, AtpF from other α-proteobacteria including Rickettsia prowazekii and conorii, which are close relatives of mitochondria (37,38), and finally also nucleus-encoded proteins. These nucleus-encoded proteins are described in GenBank as ATP synthase subunit 4 and ATP synthase chain b, encoded by the genes ATP4 (fungi) and ATP5F (animals), whereas one is unassigned (Anopheles gambiae gene; NCBI record EAA01034). This latter protein revealed strong similarity with ATP synthase subunit b from Drosophila (not shown), and is therefore considered an ATP5F homolog. The nucleus-encoded genes and their products will be referred to in the following as ATP4 and ATP synthase chain 4 (ATP4), respectively.
To summarize the above, PSI-BLAST analyses including the newly generated jakobid Ymf39 sequences have permitted us to identify a total of nearly 50 potential homologs, which are encoded in mitochondrial, bacterial and nuclear genomes. Interestingly, these three protein classes differ in their average length by about 20 residues: ∼170 ± 10 for α-proteobacterial AtpF, ∼190 ± 60 for mitochondrion-encoded Ymf39 and 210 ± 5 for nucleus-encoded ATP4 (mature proteins not including the pre-protein that is required for import into mitochondria). Possible implications of this observation will be discussed in a later section.
Sequence similarities between mitochondrion-encoded Ymf39, nucleus-encoded ATP4 and bacterial AtpF
As our working hypothesis posited, Ymf39 sequences of jakobids are remarkably similar to α-proteobacterial AtpF. Figure 2 shows multiple protein alignments of Ymf39 and AtpF. The resemblance between jakobid and bacterial proteins is mostly found within a stretch of more than 90 residues in the N‐terminal region.
Figure 2.
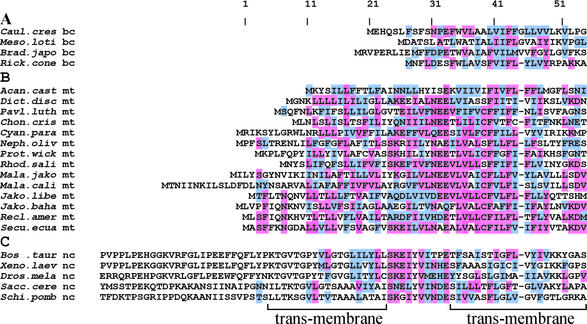
Multiple protein alignments of jakobid and cryptomonad mitochondrial Ymf39, and α-proteobacterial AtpF. Three or more identical amino acids within a column are highlighted in magenta. Amino acids with a positive PAM250 value relative to the magenta-labeled residue in a given column are highlighted in light blue. Sequences used are (taxonomy and GenBank accession no. in parentheses): Caulo.cres., Caulobacter crescentus (α-proteobacterium; NP_419184); Meso.loti., Mesorhizobium loti (α-proteobacterium; NP_107736); Brad.japo., B.japonicum (α-proteobacterium; AAF78803); Rick.cono., Rickettsia conorii (α-proteobacterium; NP_359661); Secu.ecua., S.ecuadoriensis (jakobid; reported here); Recl.amer., R.americana NZ (jakobid; NC_001823); Jako.baha., J.bahamiensis (jakobid; reported here); Jako.libe., J.libera (jakobid; reported here); Rhod.sali., R.salina (cryptophyte; NC_002572). bc, bacterial gene; mt, mitochondrion-encoded gene; nc, nucleus-encoded gene.
Sequence similarities between Ymf39 proteins from jakobids and non-jakobids, as well as among non-jakobid eukaryotes, are less pronounced, and restricted to a region corresponding to residues 11–54 in the S.ecuadoriensis protein (Fig. 3B). Finally, although the nucleus-encoded proteins resemble one another moderately throughout the complete length (not shown), their similarity with Ymf39 proteins is quite weak, and is confined to the same, short region between positions 11 and 54 (Fig. 3B and C).
Figure 3.
Multiple protein alignments of bacterial AtpF, mitochondrion-encoded Ymf39 and nucleus-encoded ATP4. (A) Bacterial AtpF; (B) mitochondrion-encoded Ymf39; (C) nucleus-encoded ATP4 (also designated ATP5F or PVP). Highlighting of amino acids followed similar rules as in Figure 2. In a given column, an amino acid is highlighted in magenta when four or more residues are identical in alignment B; amino acids with a positive PAM250 value relative to the magenta-labeled residue in a given column are shown in light blue. Sequences used, taxonomy and GenBank accession nos as in legend to Figure 2. Acan.cast., A.castellanii (mycetozoa; NC_001637); Dict.disc., D.discoideum (mycetozoa; NC_000895); Pav.luth., P.lutheri (haptophyte alga; reported here); Chon.cris., C.crispus (rhodophyte alga; NC_1677); Cyan.para., C.paradoxa (glaucocystophyte alga; reported here); Neph.oliv., Nephroselmis oliva (green alga; NC_000927); Prot.wick., P.wickerhamii (green alga; NC_001613); Mala.jako. M.jakobiformis (malawimonad; NC_002553); Mala.cali., M.californiana (malawimonad; reported here). Bos.taur., Bos taurus (chordata; P13619); Xeno.laev., Xenopus laevis (chordata; AAF31360); Sacc.cere., Saccharomyces cerevisiae (fungi; NP_015247); Schi.pomb., Schizosaccharomyces pombe (fungi; CAA22340).
Significance of similarities between Ymf39, ATP4 and AtpF
To assess the significance of the sequence alignments shown in Figures 2 and 3, we performed the statistical test PRSS (33), using the best-aligned regions. The obtained support values are listed in Table 1. First we compared the significance of pair-wise alignments of proteins within each of the three classes. At a confidence level of E-value <0.005, Ymf39 of S.ecuadoriensis is significantly similar to the (mitochondrion-encoded) counterparts of green algae, plants and the mycetozoon Dictyostelium discoideum. Exceptions are Pavlova lutheri (haptophyte), Chondrus crispus (rhodophyte), Cyanophora paradoxa (glaucocystophyte) and Acanthamoeba castellanii (mycetozoon), whose Ymf39 alignments with the S.ecuadoriensis protein yielded E-values two to three times higher than the confidence limit. However, pair-wise alignments of the protein from the haptophyte, rhodophyte and glaucocystophyte with that of the cryptophyte R.salina are significant (Table 1). Only the alignment of the A.castellanii Ymf39 with any of its counterparts does not withstand the significance test, i.e. it is highly divergent as are other mitochondrial genes of this organism (39).
Table 1. Statistical significance of similaritiesa among Ymf39, AtpF and ATP4 proteins.
| Taxab | S.ecua.mt | R.amer.mt | M.jako.mt | R.sali.mt | P.wick.mt | D.disc.mt | C.cris.mt | C.para.mt | P.luth.mt | A.cast.mt | S.cere.nc | R.coni.bc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S.ecua.mt 11–54 | 1.3e-06 | 3.6e-05 | 0.0010 | 0.0027 | 0.0030 | 0.0022 | 0.0058 | 0.013 | 0.020 | 0.011 | 0.035 | – |
| S.ecua.mt 19–91 | 2.6e-09 | 3.5e-07 | – | 2.3e-04 | – | – | – | – | – | – | – | – |
| R.sali.mt 11–54 | 2.3e-4 | 0.0012 | 0.0011 | 1.2e-07 | 4.4e-04 | 0.0052 | 5.7e-04 | 0.0014 | 2.5e-04 | 0.022 | – | – |
| B.japo.bc 27–91 | 7.5e-06 | 5.3e-05 | – | 0.033 | 0.013 | – | – | – | – | – | – | 4.9e-04 |
The nuclear-encoded ATP4 sequences from animals share significant sequence similarity with those from fungi over the entire length of the protein, confirming earlier reports from others (40). Surprisingly, within the region where Ymf39 proteins are most conserved (residue 11–54 in the S.ecuadoriensis protein), the similarity between ATP4 proteins from animals and fungi is only marginal. As expected, pair-wise alignments between AtpF genes are highly significant (E-values between 1e-04 and 1e-07).
Secondly, we compared the significance of alignments of proteins from different classes. No support was obtained for a similarity between nucleus-encoded ATP4 and bacterial AtpF, while the E-values for the alignment of Ymf39 with ATP4 are inconsistent: the similarity of the S.ecuadoriensis protein with that from S.pombe is significant (although weak), but not significant with that of S.cerevisiae. However, strong support was obtained for pair-wise alignments between Ymf39 of jakobids and AtpF from the α-proteobacteria included in this analysis, testifying to the evolutionary descent of Ymf39 via the bacterial ancestor of mitochondria.
Secondary structure similarities between Ymf39, ATP4 and AtpF
Despite limited sequence similarities, mitochondrion-encoded Ymf39 and nucleus-encoded ATP synthase chain 4 resemble one another remarkably at the secondary structure level (see also 41). The inferred hydrophobic profiles and folding patterns display a pronounced bi-partite structure with an N‐terminal hydrophobic portion (residues 1 to approximately 54) and the uniformly hydrophilic remainder of the protein (residues approximately 55 to end). In addition, the N‐terminal region can be folded into two α-helical barrels of more than 13 residues, each of which being able to span a lipid bilayer (Fig. 3C). Such a bi-partite structure is typical for proteins that are membrane-anchored and extend into the aqueous space, such as the stalk-forming components of the F0 moiety of ATP synthase.
Remarkably, bacterial AtpF proteins have a single potential trans-membrane domain, whereas two such domains are present in the mitochondrial proteins. It has been speculated that the additional membrane-spanning segment of ATP4, and this view would also apply to Ymf39, has evolved for interacting with the g-subunit that is only present in the mitochondrial, but not in the bacterial (and chloroplast) enzyme (42).
DISCUSSION
The structure of jakobid F0F1-ATP synthase is of intermediate complexity
F0F1-ATP synthase is an enzyme complex found in mitochondria, chloroplasts and bacteria that catalyzes the formation of ATP by utilizing a trans-membrane proton gradient generated during electron transport through the respiratory chain. These three enzymes are clearly evolutionarily related in that the organellar ATP synthases are of bacterial descent: the mitochondrial enzyme originates from an α-proteobacterial and the chloroplast enzyme from a cyanobacterial ancestor, respectively.
Bacterial ATP synthases consist of only eight subunits in total. Three subunits (chain a, b and c) are organized in the proton-translocating, membrane-traversing F0 sector, while the other five (subunits α to ε) form the peripheral catalytic F1 moiety (for a review see 41). Mitochondrial F0F1-ATP synthases are considerably more complex. In plants, this enzyme complex consists of 12 or 13 different proteins, 10 of which (but not including subunit b) have been investigated by N‐terminal sequencing and immunological methods (43,44). Animal (beef) and fungal (yeast) mitochondrial ATP synthases appear to contain as many as 16 different proteins: nine (instead of three) subunits in the F0 moiety (A–G, F6 and A6L), three subunits in F1 as in the bacterial system, plus two additional components, the oligomycin-sensitivity conferring protein (OSCP) and the inhibitor protein (IF1). Recent reports even describe a total of 19 or 20 different subunits in yeast mitochondrial ATP synthase, four of which are non-essential to structural integrity or in vitro activity (45,46).
Here, we report that ATP synthase in S.ecuadoriensis consists of only nine subunits indicating that the jakobid enzyme is of lower complexity than that of animals, fungi and even plants. However, it should be noted that the number of subunits in the jakobid might have been underestimated, because apolar subunits of mitochondrial membrane complexes tend to stain poorly, and because some subunits may be present in sub-stoichiometrical proportion within the complex. Nevertheless, the molecular mass of the S.eculamonas ATP synthase complex is ∼5% smaller than that of plants, which corroborates the view that the jakobid enzyme has indeed the lowest number of subunits among all eukaryotes.
The specific function of Ymf39
In the present report we provide evidence that Ymf39 in S.ecuadoriensis, and by implication, the mitochondrion-encoded Ymf39 of all protists and plants, is a homolog (i.e. an evolutionary descendant) of the bacterial F0F1-ATP synthase subunit b. This raises the question about the role that this subunit plays in the multi-component enzyme complex.
The structure of F0F1-ATP synthases consists of three parts, a headpiece, i.e. sector F1 that carries out ATP synthesis/hydrolysis, a base piece embedded in the membrane that conducts the protons through a pore across the membrane (F0), and two connecting stalks. It is believed that the activities of F0 and F1 are coupled through a rotary motion, in which subunit γ functions as the rotating stalk, whereas subunit b, together with subunit δ in the bacterial system (and subunits d, F6 and OSCP in mitochondria), forms the static link (for recent reviews, see 45–48). This implies that the role of subunit b is strictly architectural, which would be consistent with the low sequence conservation of the mitochondrion-encoded homologs as discussed above.
The evolutionary relatedness of mitochondrion-encoded Ymf39 and nucleus-encoded ATP4
The ATP synthase subunits Ymf39 and ATP4 are obviously functional counterparts, because all eukaryotes, in which a nuclear ATP4 gene has been reported, lack mtDNA-encoded ymf39. On this basis we can predict that, vice versa, all eukaryotes whose mtDNA lack ymf39, such as apicomplexan, chlamydomonad, choanoflagellate, ciliate, stramenopile and kinetoplastid protists (1), possess a nucleus-encoded ATP4 gene. Moreover, organisms whose mtDNA does contain ymf39, such as plants, jakobids, mycetozoa and cryptophyte, glaucocystophyte, haptophyte, red and most green algae (1), should lack nucleus-encoded ATP4. Ongoing nuclear genome and EST sequencing programs will provide the data to test this hypothesis.
Given that ATP4 and Ymf39 are functionally equivalent but share very little sequence similarity, the question arises whether they share a common ancestor, as do Ymf39 and bacterial AtpF. ATP4 could indeed originate from mtDNA-encoded ymf39 via transfer from the mitochondrial to the nuclear genome, with the accelerated evolutionary rate of the nuclear gene being a result of the adaptations to the new genetic context (see e.g. 49). Such a migration of mitochondrial genes to the nucleus has taken place numerous times in the evolutionary history of eukaryotes. A well-documented example of a mitochondrial protein whose gene is located in mtDNA in some eukaryotes but in the nucleus in others is succinate dehydrogenase (SDH), a complex of the mitochondrial respiratory chain. SDH subunits 2–4 are encoded by mtDNAs in jakobids, the cryptophyte R.salina, red algae and some plants, but are otherwise absent from mtDNAs of the majority of eukaryotes, including malawimonads (1). In turn, nucleus-encoded genes specifying Sdh2 have been identified in a number of animals and fungi. Phylogenetic analysis, including bacterial, mtDNA-encoded and nucleus-encoded Sdh2 protein sequences, strongly suggests that the nuclear Sdh2 gene originated by transfer from a mitochondrial genome in which it originally resided (17). Note that phylogenetic analyses including ATP4, Ymf39 and AtpF have not been attempted, because of the insufficient number of sites that can be aligned unambiguously.
Another possibility is that ATP4 is a genuinely nuclear gene that has been recruited to functionally substitute the ancestral atpF in proto-mitochondrial mtDNA. An example of such a functional replacement is mitochondrial RNA polymerase. In jakobids, this enzyme belongs to the bacterial α2ββ′ type with all subunits encoded by mtDNA (25), whereas most other eukaryotes possess a nucleus-encoded RNA polymerase which resembles that of T3/T7-phages at the level of sequence, secondary structure and subunit composition (50,51). It appears unlikely that in such a highly integrated protein complex as ATP synthase some subunits would have been substituted, while the majority (Atp1, 3, 6, 8 and 9) remain of ancestral type (52). Therefore, we favor the notion that the nuclear ATP4 gene is a highly derived homolog of ymf39/atpF.
Variable contributions of the nuclear and mitochondrial genome to ATP synthase across eukaryotes
With a total of six, the largest number of mitochondrion-encoded ATP synthase subunits is found in jakobids. The corresponding genes and proteins are atp1 (F1-subunit α), atp3 (F1-subunit γ), atp4 (F0-subunit b), atp6 (F0-subunit g), atp8 (F0-subunit F6) and atp9 (F0-subunit A6L). Mitochondrial DNAs of plants contain four atp genes (atp1, 6, 8 and 9); those of most fungi contain three (atp6, 8 and 9); and animals only two (atp6 and atp8) or one (nematode mtDNAs lack atp8) (53). Finally, some protists, such as chlamydomonads and apicomplexans, do not even possess a single mitochondrion-encoded atp gene (for a review, see 1). In these latter organismal groups, the entire complex must be encoded by nuclear genes, translated on cytoplasmatic ribosomes and imported into mitochondria. As mentioned earlier, the total number of ATP synthase subunits is considerably higher in animals and fungi than in plants and jakobids. This suggests a correlation between the number of subunit genes that have emigrated to the nucleus and the total number of subunits of a given complex, a correlation also observed in the cytochrome oxidase, cytochrome bc1 and NADH dehydrogenase complexes. Therefore, we expect that in chlamydomonads and apicomplexans, which have lost all atp genes from mtDNA, the total number of ATP synthase subunits will be even larger than in animals and fungi. Genomics and proteomics studies of protistan eukaryotes are well underway, and are bound to reveal the evolutionary trends of mitochondrial enzyme complexes and the organelle as a whole.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank T. Nerad (ATCC), who has isolated these important jakobid and malawimonad protists studied here. We also thank I. Plante for library construction and DNA sequencing, D. Dvzorno, I. Plante and Y. Zhu for DNA sequencing, M. Baumgaertner for peptide sequencing, S. Kannan for assistance in data analysis and E. O’Brien for critical reading of the manuscript. This work was supported by the ‘Deutsche Forschungsgemeinschaft’, the ‘Fonds der Chemischen Industrie’, and the ‘Canadian Institutes of Health Research’ (CIHR, MSP-14226). G.B. is Canadian National Associate and B.F.L. is Imasco Fellow in the program of Evolutionary Biology of the Canadian Institute for Advanced Research (CIAR), whom we thank for salary and interaction support.
DDBJ/EMBL/GenBank accession nos+AY236972–AY236977
REFERENCES
- 1.Gray M.W., Lang,B.F., Cedergren,R., Golding,G.B., Lemieux,C., Sankoff,D., Turmel,M., Brossard,N., Delage,E., Littlejohn,T.G., Plante,I., Rioux,P., Saint-Louis,D., Zhu,Y. and Burger,G. (1998) Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res., 26, 865–878. [DOI] [PMC free article] [PubMed]
- 2.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H., Smith,A.J., Staden,R. and Young,I.G. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed]
- 3.Anderson S., de Bruijn,M.H., Coulson,A.R., Eperon,I.C., Sanger,F. and Young,I.G. (1982) Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J. Mol. Biol., 156, 683–717. [DOI] [PubMed]
- 4.Michael N.L., Rothbard,J.B., Shiurba,R.A., Linke,H.K., Schoolnik,G.K. and Clayton,D.A. (1984) All eight unassigned reading frames of mouse mitochondrial DNA are expressed. EMBO J., 3, 3165–3175. [DOI] [PMC free article] [PubMed]
- 5.Chomyn A., Mariottini,P., Cleeter,M.W., Ragan,C.I., Matsuno-Yagi,A., Hatefi,Y., Doolittle,R.F. and Attardi,G. (1985) Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature, 314, 592–597. [DOI] [PubMed]
- 6.Chomyn A., Cleeter,M.W., Ragan,C.I., Riley,M., Doolittle,R.F. and Attardi,G. (1986) URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit. Science, 234, 614–618. [DOI] [PubMed]
- 7.Unseld M., Marienfeld,J.R., Brandt,P. and Brennicke,A. (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nature Genet., 15, 57–61. [DOI] [PubMed]
- 8.Marienfeld J., Unseld,M. and Brennicke,A. (1999) The mitochondrial genome of Arabidopsis is composed of both native and immigrant information. Trends Plant Sci., 4, 495–502. [DOI] [PubMed]
- 9.Lang B.F., Gray,M.W. and Burger,G. (1999) Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet., 33, 351–497. [DOI] [PubMed]
- 10.Belfort M. and Roberts,R.J. (1997) Homing endonucleases: keeping the house in order. Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed]
- 11.Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed]
- 12.Griffith A.J. (1995) Natural plasmids of filamentous fungi. Microbiol. Rev., 59, 673–685. [DOI] [PMC free article] [PubMed]
- 13.Bullerwell C.E., Burger,G. and Lang,B.F. (2000) A novel motif for identifying rps3 homologs in fungal mitochondrial genomes. Trends Biochem. Sci., 25, 363–365. [DOI] [PubMed]
- 14.Bogsch E.G., Sargent,F., Stanley,N.R., Berks,B.C., Robinson,C. and Palmer T. (1998) An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem., 273, 18003–18006. [DOI] [PubMed]
- 15.Weiner J.H., Bilous,P.T., Shaw,G.M., Lubitz,S.P., Frost,L., Thomas,G.H., Cole,J.A. and Turner,R.J. (1998) A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell, 93, 93–101. [DOI] [PubMed]
- 16.Oda K., Yamato,K., Ohta,E., Nakamura,Y., Takemura,M., Nozato,N., Ajashi,K., Kanegae,T., Ogura,Y., Kohchi,T. et al. (1992) Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J. Mol. Biol., 223, 1–7. [DOI] [PubMed]
- 17.Burger G., Lang,B.F., Reith,M. and Gray,M.W. (1996) Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc. Natl Acad. Sci. USA, 93, 2328–2332. [DOI] [PMC free article] [PubMed]
- 18.Viehmann S., Richard,O., Boyen,C. and Zetsche K. (1996) Genes for two subunits of succinate dehydrogenase form a cluster on the mitochondrial genome of Rhodophyta. Curr. Genet., 29, 199–201. [DOI] [PubMed]
- 19.Dewey R.E., Levings,C.S.,3rd and Timothy,D.H. (1986) Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell, 44, 439–449. [DOI] [PubMed]
- 20.Stamper S.E., Dewey,R.E., Bland,M.M. and Levings,C.S.,3rd (1987) Characterization of the gene urf13-T and an unidentified reading frame, ORF 25, in maize and tobacco mitochondria. Curr. Genet., 12, 457–463. [DOI] [PubMed]
- 21.Prioli L.M., Huang,J. and Levings,C.S.,3rd (1993) The plant mitochondrial open reading frame orf221 encodes a membrane-bound protein. Plant Mol. Biol., 23, 287–295. [DOI] [PubMed]
- 22.O’Kelly C.J. (1993) The jakobid flagellates: structural features of Jakoba, Reclinomonas and Histiona and implications for the early diversification of eukaryotes. J. Euk. Microbiol., 40, 627–636.
- 23.Edgcomb V.P., Roger,A.J., Simpson,A.G., Kysela,D.T. and Sogin,M.L. (2001) Evolutionary relationships among ‘jakobid’ flagellates as indicated by alpha- and beta-tubulin phylogenies. Mol. Biol. Evol., 18, 514–22. [DOI] [PubMed]
- 24.O’Kelly C.J., Farmer,M.A. and Nerad,T.A. (1999) Ultrastructure of Trimastix pyriformis (Klebs) Bernard et al.: similarities of Trimastix species with retortamonad and jakobid flagellates. Protist, 150, 149–162. [DOI] [PubMed]
- 25.Lang B.F., Burger,G., O’Kelly,C.J., Cedergren,R., Golding,G.B., Lemieux,C., Sankoff,D., Turmel,M. and Gray,M.W. (1997) An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature, 387, 493–497. [DOI] [PubMed]
- 26.Marx S., Baumgärtner,M., Kannan,S., Braun,H.P., Lang,B.F. and Burger,G. (2003) Structure of the bc1 complex from Seculamonas ecuadoriensis, a jakobid flagellate with an ancestral mitochondrial genome. Mol. Biol. Evol., 20, 145–153. [DOI] [PubMed]
- 27.Schägger H., Cramer,W.A. and von Jagow,G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem., 217, 220–230. [DOI] [PubMed]
- 28.Kruft V., Eubel,H., Jansch,L., Werhahn,W. and Braun,H.P. (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol., 127, 1694–1710. [PMC free article] [PubMed]
- 29.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- 30.Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed]
- 31.Hoffmann K. and Stoffel,W. (1993) TMpred. Biol. Chem. Hoppe-Seyler, 374, 166.
- 32.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed]
- 33.Pearson W.R. (1996) Effective protein sequence comparison. Methods Enzymol., 266, 227–258. [DOI] [PubMed]
- 34.Smith T.F. and Waterman,M.S. (1983) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed]
- 35.Wolff G., Plante,I., Lang,B.F., Kück,U. and Burger,G. (1994) Complete sequence of the mitochondrial DNA of the chlorophyte alga Prototheca wickerhamii. Gene content and genome organization. J. Mol. Biol., 237, 75–86. [DOI] [PubMed]
- 36.Burger G. (2000) GenBank accession no. NC_002553.
- 37.Andersson S.G., Zomorodipour,A., Andersson,J.O., Sicheritz-Ponten,T., Alsmark,U.C., Podowski,R.M., Naslund,A.K., Eriksson,A.S., Winkler,H.H. and Kurland,C.G. (1998) The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature, 396, 133–140. [DOI] [PubMed]
- 38.Gray M.W. (1998) Rickettsia, typhus and the mitochondrial connection. Nature, 396, 109–110. [DOI] [PubMed]
- 39.Burger G., Plante,I., Lonergan,K.M. and Gray,M.W. (1995) The mitochondrial DNA of the amoeboid protozoon, Acanthamoeba castellanii: complete sequence, gene content and genome organization. J. Mol. Biol., 245, 522–537. [DOI] [PubMed]
- 40.Velours J., Arselin,G., Paul,M.F., Galante,M., Durrens,P., Aigle,M. and Guerlin,B. (1989) The yeast ATP synthase subunit 4: structure and function. Biochimie, 71, 903–915. [DOI] [PubMed]
- 41.Boyer P.D. (1997) The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem., 66, 717–749. [DOI] [PubMed]
- 42.Soubannier V., Vaillier,J., Paumard,P., Coulary,B., Scheffer,J. and Velours,J. (2002) In the absence of the first membrane-spanning segment of subunit 4(b), the yeast ATP synthase is functional but does not dimerize or oligomerize. J. Biol. Chem., 277, 10739–10745. [DOI] [PubMed]
- 43.Hamasur B. and Glaser,E. (1992) Plant mitochondrial F0F1 ATP synthase. Identification of the individual subunits and properties of the purified spinach leaf mitochondrial ATP synthase. Eur. J. Biochem., 205, 409–416. [DOI] [PubMed]
- 44.Jänsch L., Kruft,V., Schmitz,U.K. and Braun,H.P. (1996) New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J., 9, 357–368. [DOI] [PubMed]
- 45.Mueller D.M. (2000) Partial assembly of the yeast mitochondrial ATP synthase. J. Bioenerg. Biomembr., 32, 391–400. [DOI] [PubMed]
- 46.Velours J. and Arselin,G. (2000) The Saccharomyces cerevisiae ATP synthase. J. Bioenerg. Biomembr., 32, 383–390. [DOI] [PubMed]
- 47.Noji H. and Yoshida,M. (2001) The rotary machine in the cell, ATP synthase. J. Biol. Chem., 276, 1665–1668. [DOI] [PubMed]
- 48.Dunn S.D., McLachlin,D.T. and Revington,M. (2000) The second stalk of Escherichia coli ATP synthase. Biochim. Biophys. Acta, 1458, 356–363. [DOI] [PubMed]
- 49.Adams K.L., Daley,D.O., Qiu,Y.L., Whelan,J. and Palmer,J.D. (2000) Repeated recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature, 408, 354–357. [DOI] [PubMed]
- 50.Cermakian N., Ikeda,T.M., Miramontes,P., Lang,B.F., Gray,M.W. and Cedergren,R. (1997) On the evolution of the single-subunit RNA polymerases. J. Mol. Evol., 45, 671–681. [DOI] [PubMed]
- 51.Cermakian N., Ikeda,T.M., Cedergren,R. and Gray,M.W. (1996) Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res., 24, 648–654. [DOI] [PMC free article] [PubMed]
- 52.Gray M.W., Burger,G. and Lang,B.F. (2001) The origin and early evolution of mitochondria. Genome Biol., 2, 1018.1–1018.5. [DOI] [PMC free article] [PubMed]
- 53.Wolstenholme D.R., Macfarlane,J.L., Okimoto,R., Clary,D.O. and Wahleithner,J.A. (1987) Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc. Natl Acad. Sci. USA, 84, 1324–1328. [DOI] [PMC free article] [PubMed]