Abstract
RNA editing in chloroplasts alters the RNA sequence by converting C-to-U or U-to-C at a specific site. During the study of the complete nucleotide sequence of the chloroplast genome from the hornwort Anthoceros formosae, RNA editing events have been systematically investigated. A total of 509 C-to-U and 433 U-to-C conversions are identified in the transcripts of 68 genes and eight ORFs. No RNA editing is seen in any of the rRNA but one tRNA suffered a C-to-U conversion at an anticodon. All nonsense codons in 52 protein-coding genes and seven ORFs are removed in the transcripts by U-to-C conversions, and five initiation and three termination codons are created by C-to-U conversions. RNA editing in intron sequence suggests that editing can precede intercistronic processing. The sequence complementary to the edited site is proposed as a distant cis-recognition element.
INTRODUCTION
RNA editing to modify genetic information at the transcript level has been identified in both mitochondria and chloroplasts of land plants (1–4). All the editing involves conversions from a cytidine (C) to uridine (U) residue and in rare cases from U to C by base modification (5). In higher plants, essentially all protein-encoding mRNA is subject to RNA editing in mitochondria but the phenomenon is significantly less frequent in chloroplasts, where it affects only some transcripts (6). Indeed, systematic investigation of Arabidopsis mitochondria has revealed 456 RNA editing sites (7). In contrast, only 25 sites in maize (8), 31 sites in tobacco (9) and 26 sites in black pine (10) have been identified in chloroplasts. In the early land plants, however, numerous editing sites have been observed in chloroplast transcripts (11). In the hornwort Anthoceros formosae, 20 RNA editing sites in the rbcL transcript and 29 sites in the atpB transcript have been identified (12,13). We report here 509 C-to-U and 433 U-to-C conversions in the hornwort chloroplast.
The reaction for RNA editing requires at least three factors; trans-acting factors involved in editing site recognition (14–16), cis-acting elements interacting with the corresponding trans-acting factors (14,17–21) and an editing enzyme catalyzing base modifications (5). In higher plant chloroplast, the cis-acting elements have been identified in the region upstream of the editing site by in vivo (14,17–19,21) and in vitro (16,22) studies. The trans-acting factors and the editing enzyme, forming a hypothetical editing complex (23), are extraplastidic origin (15) and proteins (16,22). Moreover, a RNA segment complementary to edited sequence was shown to have no effect on the RNA editing (24). However, we have found RNA segment complementary to every edited site of the transcripts of atpB and rbcL (13). The complementary sequences, which are longer than 5 nt, are located in the same transcript from Anthoceros chloroplasts. They are not restricted in a specific transcript but are always found in the transcripts examined, except in a group II intron. The role of the complementary sequence is discussed.
MATERIALS AND METHODS
Preparation of RNA and cDNA
Total RNA was isolated and complementary DNA (cDNA) was synthesized as described previously (12). Total cellular nucleic acids were prepared using cetyltrimethylammonium bromide (CTAB) (25) with a slight modification. Frozen thalli (15 g) of A.formosae were disrupted with quartz sand and nucleic acids were extracted in 15 ml of extraction buffer containing 100 mM Tris–HCl (pH 8.0), 20 mM EDTA, 1.4 M NaCl, 1% CTAB and 1% 2-mercaptoethanol at 60°C for 30 min. Total RNA was precipitated by adding LiCl to a final concentration of 2 M after extraction of chloroform–isoamyl alcohol (24/1, v/v). Contaminating DNA was removed from the RNA sample by digestion with RNase-free DNase (Boehringer Mannheim). A cDNA bank was constructed using the total RNA as the template with a commercial kit (cDNA synthesis kit, Takara, Kyoto). Total RNA (0.5 µg) and 20 pmol random hexamer primer were annealed at 70°C for 2 min and cooled to 0°C. They were then mixed with 10 pmol of each nucleotide triphosphate (dNTP), 20 U of recombinant RNase inhibitor and 200 U of MLV reverse transcriptase (Wako, Osaka) in a total volume of 20 µl. The reaction proceeded at 42°C for 60 min and at 94°C for 5 min, then 80 µl of water was added.
Sequencing of cDNA
The cDNA was amplified using a Taq polymerase (Ex Taq, Takara) and appropriate primers with a Mini Cycler (PTC-150, MJ Research), as described previously (12). The primers (accession no. AB086179) for the 5′ and 3′ of each transcript were designed from the genomic sequence 20–50 nt upstream and downstream of the coding region, respectively, so as to be able to sequence the entire coding region. A typical reaction (50 µl) contained 25 pmol of each primer pair, 10 mM of each dNTP and 1.25 U of Taq polymerase. One thermo cycle was 96°C for 1 min, annealing for 2 min and 72°C for 2 min. The annealing temperature was adjusted to the Tm value of the primer pair. The reaction proceeded through 30 cycles, followed by 72°C for 5 min. Amplified cDNA was directly analyzed using a BigDye Terminator Cycle Sequencing Ready Reaction (ver. 3.0, Applied Biosystems) with a 310 Genetic Analyzer (Perkin Elmer). Additional primers within the cDNA were also designed from the genomic sequence and used to analyze the sequence of long cDNA. Amplified cDNA, which was not homogeneous, was separated by agarose gel electrophoresis and the homogeneous cDNA was extracted from the gel using a QIAquick Gel Extraction Kit (Qiagen, Tokyo). It was used for direct sequencing, or ligated into Bluescript vector (Stratagene) using a Blunting Kination Ligation Kit (Takara) and introduced into Escherichia coli DH5α. The plasmid was prepared by standard alkaline lysis (26) and used for sequencing.
Analysis of RNA editing
RNA editing was analyzed by comparing each sequence of cDNA with that of genomic DNA. When the sequences did not fit, the genomic DNA and cDNA were re-examined after the re-amplification of genomic DNA and the cDNA library, respectively, using the same primer pair.
RESULTS
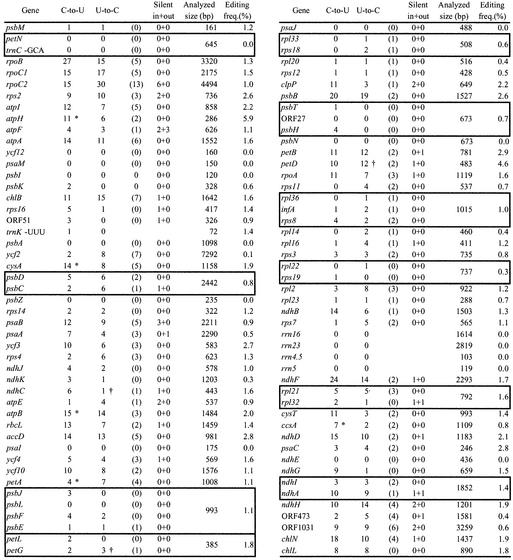
cDNA for every transcript expected from the complete chloroplast genome of A.formosae (27) has been synthesized and its nucleotides sequenced (accession nos AB087416– AB087494 and AB097086), except for some tRNAs. A comparison of the sequences of genomic DNA and cDNA has revealed RNA editing at 509 sites in the form of C-to-U conversions and at 433 sites in the form of U-to-C conversions. The efficiency of RNA editing at individual sites is virtually complete. The overall conversion of nucleotides examined is 1.2%. The results are summarized in Figure 1. RNA editing has been found in the transcripts of 68 protein-coding genes and eight ORFs, and one tRNA. No editing has been found in any rRNA or the rest of the tRNA examined. The editing frequency of 942 sites in a genome of 161 kb is even higher than the Arabidopsis mitochondrial editing frequency of 456 sites in 367 kb (7).
Figure 1.
Number of editing sites in the chloroplast of A.formosae. The number of RNA editing sites converting C-to-U and U-to-C is shown. The number of those converting nonsense codons into sense codons is shown in parentheses. Editing sites, which do not alter the coding amino acid are shown as silent (in) and those found outside of the coding region as (out). Editing frequency shows percentage of editing sites per analyzed size (bp). Editing sites on co-transcripts are shown in boxes. *Containing the creation of an initiation codon. (†) Containing the creation of a termination codon.
The most notable feature of Anthoceros RNA editing is the numerous U-to-C conversions. The number of U-to-C conversions is comparable with that of C-to-U conversions, though this type of RNA editing had been described as rare and called reverse type editing (28,29). It is not restricted to the transcripts of rbcL and atpB (12,13), but widely observed in the transcripts of many genes. Indeed, nonsense codons in 52 protein-coding genes and seven ORFs become sense codons through U-to-C conversions.
The RNA editing replaced 868 out of 24275 amino acids corresponding to a 3.6% change in sequence identity. The general tendency is for RNA editing to increase the proportion of hydrophobic amino acid codons (7). This is also the case in Anthoceros chloroplasts. As shown in Table 1, the three most frequent amino acid transitions are Ser to Leu at 136 sites, Ser to Phe at 104 sites and Pro to Leu at 87 sites, which are modifications of hydrophilic to hydrophobic residues. The reverse modifications reducing hydrophobicity are rather rare and include transitions of Leu to Pro at 60 sites, Cys to Arg at 40 sites and Leu to Ser at 26 sites. The RNA editing is required to form a functional protein structure, because the amino acid sequence deduced from the genomic sequence of rbcL will not allow the known three-dimensional structure of the Rubisco large subunit (K.Yura, K.Kobayashi and M.Go, manuscript in preparation).
Table 1. Alteration of amino acid identity by RNA editing in Anthoceros chloroplast.
| Alteration | ||
|---|---|---|
| From | To | No. of site |
| Ser | Leua | 136 |
| Non | Gln | 110 |
| Ser | Phea | 104 |
| Pro | Leua | 87 |
| Leua | Pro | 60 |
| Non | Arg | 54 |
| Thr | Ilea | 41 |
| Cysa | Arg | 40 |
| Pro | Ser | 31 |
| Leua | Ser | 26 |
| Phea | Ser | 25 |
| Ser | Pro | 22 |
| Vala | Alaa | 18 |
| His | Tyr | 18 |
| Alaa | Vala | 17 |
| Tyr | His | 14 |
| Leua | Phea | 13 |
| Phea | Pro | 9 |
| Ilea | Thr | 9 |
| Arg | Trp | 8 |
| Thr | Meta | 8 |
| Phea | Leua | 5 |
| Pro | Phea | 4 |
| Arg | Cysa | 3 |
| Gln | Non | 3 |
| Meta | Thr | 2 |
| Trp | Arg | 1 |
Non represents nonsense codon. aHydrophobic amino acid based on the criteria of Kyte and Doolittle (45).
One of the most notable modifications is the conversion from nonsense to sense codons; 110 nonsense codons are converted into Gln and 54 into Arg. They are found in 52 protein-coding genes and seven ORFs, showing that RNA editing is essential to the translation of functional proteins. Numbers of editing sites at different codon positions are shown in Table 2. The most frequent editing of C-to-U is at the second position and then at the first, and that of U-to-C conversion is at the first position and then at the second. In total, 58.6% of RNA editing is found at the second position, 38.4% at the first position and only 3.0% at the third position. Editing sites are restricted at the first and second codon positions in the case of Arabidopsis mitochondria (7). The biased distribution of editing sites within the codons is not explained by the general distribution of pyrimidines within the mRNA of the Anthoceros chloroplast genome, because 41.4, 53.0 and 52.5% of pyrimidines are estimated to occur at the first, second and third position of the codon, respectively, from the overall codon usage of the Anthoceros chloroplast genome.
Table 2. RNA editing frequency at different codon positions in Anthoceros chloroplast.
| Codon | Number of editing sites | Distribution | Pyrimidine | ||
|---|---|---|---|---|---|
| C-to-U | U-to-C | Sum | (%) | (%) | |
| First | 94 | 264 | 358 | 38.4 | 41.4 |
| Second | 397 | 149 | 546 | 58.6 | 53.0 |
| Third | 12 | 16 | 28 | 3.0 | 52.5 |
| Sum | 503 | 429 | 932 | ||
The total number of RNA editing sites for C-to-U and U-to-C conversions is shown at different codon positions. The distribution (%) of the total number of editing sites in each codon position and the percentage (%) of pyrimidines in the overall codon usage are shown.
Silent conversion
More than 95% of RNA editing alters amino acid identity or converts a nonsense codon into a sense codon. However, several silent conversions, which do not alter the amino acid, are also observed in the transcripts of seventeen genes and one ORF. These occur mainly at the third position of the codon as observed at 28 sites. However, two peculiar silent conversions have been observed. One is a conversion at the first codon position where one CUG codon and four CUA codons for Leu are changed into UUG and UUA, respectively, by C-to-U conversion, and two UUA codons for Leu are changed into CUA by U-to-C conversion. The other involves two sites of RNA editing within one codon whereby three CCG, one CCA and two CCC codons for Pro are changed into UUG, UUA and CUU for Leu, respectively, by two C-to-U conversions, in which only the change at the second position affects the amino acid identity. RNA editing outside of coding regions has been detected at nine sites in the Anthoceros, though only one case is known in higher plant chloroplasts (30). One of them locates in a putative Shine–Dalgarno (SD) sequence of ndhA, where the base conversion by RNA editing can increase a base pairing with the 3′ end of the 16S rRNA and form the conserved SD sequence of the gene, suggesting the requirement of RNA editing for effective translation. RNA editing in the SD sequence is not known in chloroplasts but is seen in plant mitochondria (31).
Another unique RNA editing is found in an overlapped gene. The 3′ portion of psbD and the 5′ portion of psbC overlap each other, where CCUGU sequence is changed into CCCGU by a U-to-C editing. The UGU coding Cys is altered into CGU coding Arg in the psbD transcript, but the CCU coding Pro into CCC does not alter amino acid in the psbC transcript.
RNA editing in immature transcripts
Nucleotide sequences of the cDNAs to several co-transcripts have been analyzed to determine editing sites. In the co-transcript of petG-petL, seven editing sites have been observed including three of U-to-C conversions in the petG region. Numerous editing sites have also been observed in the other co-transcripts; 11 in psbJ-psbL-psbF-psbE, 5 in psbH-psbN-psbT, 25 in ndhA-ndhI, 3 in rps18-rpl33, 19 in psbD-psbC, 10 in rpl36-infA-rps8, 2 in rpl22-rps19 and 13 in rpl21-rpl32 (Fig. 1). Some of them will be the mature poly-cistronic mRNA, however at least psbH-psbT and rpl36-rps8 will be cleaved into the mono-cistronic mRNAs because they contain putative terminator or stabilizer sequence (32) in front of the following gene. These suggest that the RNA editing can precede intercistronic processing.
RNA editing has also been detected in an intron-containing immature transcript of ndhA, namely at the 3′ end of the 5′ exon corresponding to the intron-binding site (IBS). The IBS serves in tertiary interaction with the exon-binding site (EBS) and then in trans-esterification to the 5′ exon (33). The IBS sequence 5′-CUUUAC of ndhA from Anthoceros is edited to 5′-CUCUAC by U-to-C conversion, which is complementary to the EBS sequence. This edited IBS sequence is well conserved within several land plants, suggesting that the RNA editing is required for the tertiary interaction between IBS and EBS, and allows precise splicing. These findings of RNA editing in immature transcripts support the idea that the editing reaction takes place prior to splicing (34,35).
No editing has been detected in the immature transcript of trnK-UUU, however cDNA synthesized within the tRNA has revealed a C-to-U conversion at the 3′ position of the anticodon. This is the first finding of RNA editing in the anticodon of a plant tRNA, though RNA editing in tRNA has been detected in several plant mitochondria (36–39) and in an anticodon of the mitochondrial tRNA of protists (40).
RNA editing in introns
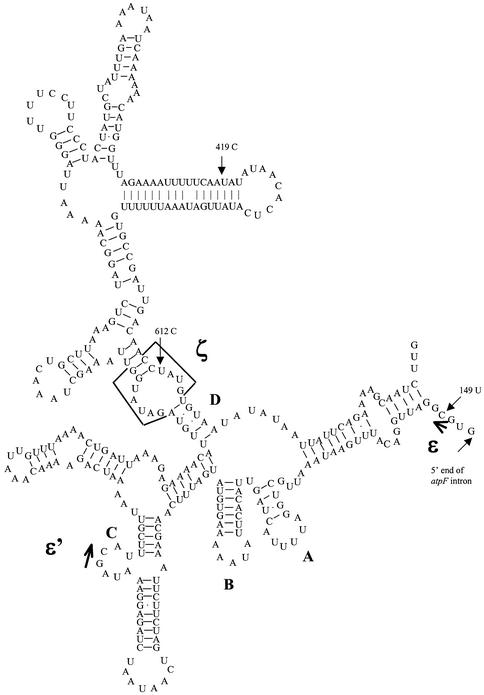
Three editing sites have been found in the intron sequence of the unspliced atpF transcript in Anthoceros chloroplasts (Fig. 2). The first at position 149 relative to the translation start site is located 4 nt downstream of the 5′ end of the intron, which corresponds to the ε site of the conserved group II intron (33,41). The 2 nt GC, which is known to involve ε-ε’ interaction and to be required for self-splicing activity (42), have been created from GU by a U-to-C conversion. The second at position 419 is found in the predicted helix of domain I of the group II intron, which could strengthen the helix by the conversion of an A-C mispair to an A-U pair. The same type of RNA editing in group II introns has been reported in plant mitochondria (43). The third is a C-to-U conversion at 612 corresponding to the ζ locus of the group II intron, which is called U381 by Boudvillain and Pyle (42), and has an important role in the catalysis of self-splicing through its tertiary contact with domain 5. These findings of RNA editing in the intron also support that the process is a prerequisite for splicing.
Figure 2.
Schematic representation of the predicted secondary structure of domain I in the group II intron of Anthoceros atpF. Sub-domains A, B, C and D together with ε, ε’ and ζ sites are shown. Edited sites 149, 419 and 612 positions from the translation start, and the 5′ end of the intron are shown with arrows.
Guide-type RNA
As shown in our previous paper (12,13), atpB and rbcL transcripts contain a sequence complementary to every edited site. The sequence is longer than 5 nt and located in the same transcript. The existence of the complementary sequence is not restricted in the transcripts of atpB and rbcL, but they are always detected in the editing site examined, except in a group II intron. Most complementary sequences locate in the coding region of the transcript but some locate in the introns as shown in Table 3. The short complementary sequences such as pentamer or hexamer can easily be found, however the longer one is the more difficult. For example, the probability to find the dodecamer 5′-CAUUGGAUCAUA, complementary to edited site 473 in the ndhB transcript, from an AU biased sequence containing 67% A + U is 1.2 × 10–7, where the AU content of 67% is conventionally assumed from the overall AT content of Anthoceros chloroplast genome. Indeed the identical sequence is not found in the rest of the chloroplast genome. Another unique complementary sequence is found in the intron of trnK-UUU, where a complementary sequence 3′-CGAAAAUU-5′ in the intron could form double-stranded structure with the 3′ end of the first exon 5′-GCUUUCAA containing anticodon of the pre-tRNA, and mispair A-C can be corrected into A-U by C-to-U editing. This might be an IBS–EBS-like interaction for splicing, though the splicing mechanism of chloroplast pre-tRNA is not known. Thus, RNA sequence containing the nucleotide to be edited might form a double stranded structure with the complementary strand during a temporal secondary or tertiary interaction, and the complementary sequence might act as a distant cis-recognition element determining the editing site, though we have had no experimental evidence of this in chloroplasts until now.
Table 3. Possible double-stranded structure determining editing sites in the transcript of ndhB.
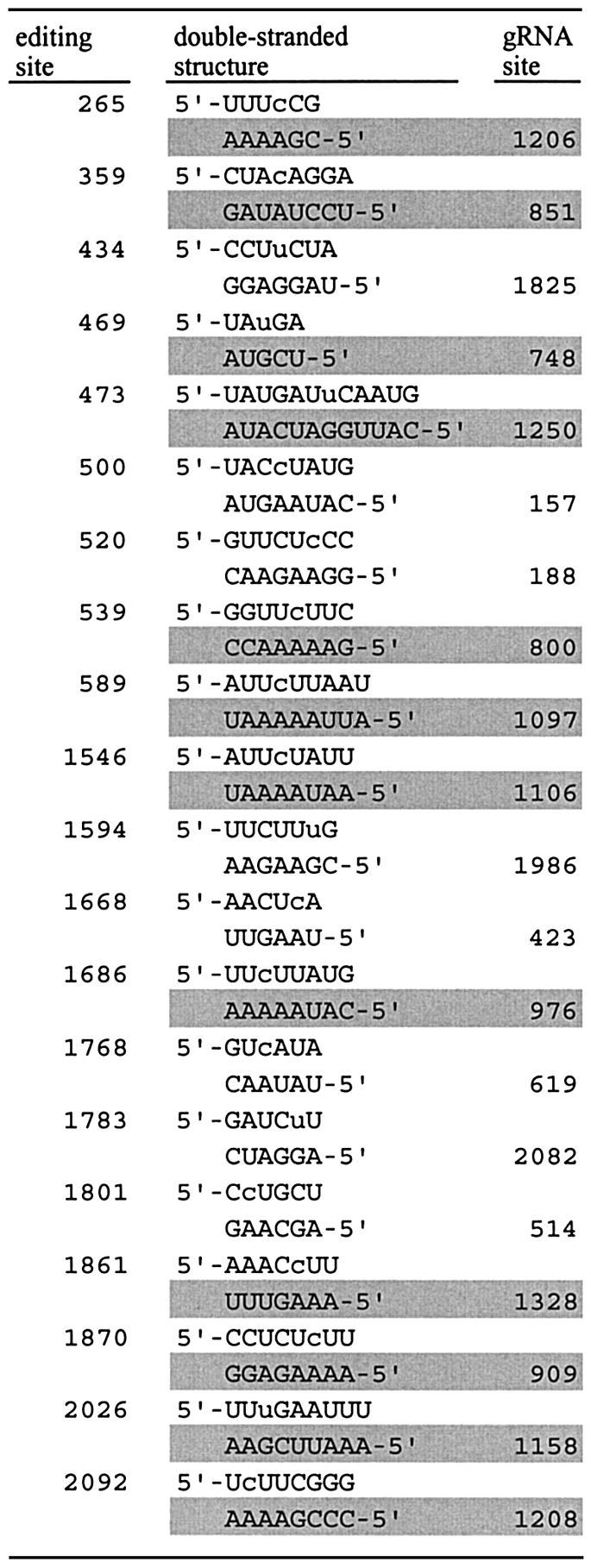
Nucleotide to be edited is shown with lower case letter and the nucleotide position is shown from the translation start. The guide-type RNA (gRNA) is shown in reverse orientation together with the nucleotide position corresponding to the editing site. The complementary strand located in the intron is shaded.
DISCUSSION
Although we have predicted that non-Watson–Crick G-U base pairs will be converted to G-C by U-to-C editing, we do not know why the G-U pairs that naturally occur in the tRNA and rRNA secondary structure are not edited. However, it could be that the recognition of editing site requires some additional factors such as the upstream element reported by Hermann and Bock (21).
It is theoretically difficult to find such a long guide-type RNA sequence from a limited length of random sequence as described in our previous paper (13). However, it is not certain whether the complementary sequence does concern as a site-determining factor in RNA editing or not. Therefore, we tried to detect the complementary sequences similar to those of Anthoceros in the atpB and rbcL of Marchantia and Nitella sp. in which RNA editing has not been found, but failed to detect any homologous double-stranded structures. However, possible guide-type RNA sequences have been found in the chloroplasts of lycopsids, such as Selaginella moellendorffii (K.Ueda et al. unpublished), and in ndhB transcript from tobacco (data not shown). Studies in vitro might elucidate the meaning of the complementary sequence and the mechanism of RNA editing in chloroplasts.
A characteristic of RNA editing in early land plants is the presence of U-to-C conversions and numerous editing sites. In the lycopsid Selaginella uncinata, 112 editing sites corresponding to 7.7% of nucleotides have been found in the atpB transcript, which includes four U-to-C conversions. U-to-C conversions are also found in the rbcL transcript from chloroplasts of Lycopodium clavatum and Isoetes asiatica (K.Ueda, unpublished). In the fern Pteridium, 94 editing sites have been detected in the transcripts of 18 genes including two sites of U-to-C conversion (K.Yamada and T.Wakasugi, unpublished). These characters of RNA editing are not restricted to chloroplasts but found in mitochondria of early land plants. In the cox1 transcripts from A.formosae, 13 editing sites including four U-to-C conversions have been found (K.Kadowaki, unpublished). Numerous editing sites of U-to-C conversion together with the removal of non-sense codons are reported in the mitochondrial transcripts of other Anthoceros species (44), possibly suggesting that the editing processes in the two plant cell organelles have common roots.
In conclusion, RNA editing in the hornwort chloroplast shows that almost all editing serves to reconstruct functional RNA. (i) Nonsense codons in almost half the genes are repaired as sense codons by RNA editing, (ii) >95% of codons containing editing sites are converted to conserved forms to translate functional proteins, (iii) the anticodon of tRNA-Lys is corrected and made functional, (iv) a SD sequence is created for efficient translation, (v) an intron sequence is corrected for precise splicing, and (vi) possible guide-type RNA sequence, a distant cis-recognition element, is detected for every editing site except in a group II intron.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Yamada and Wakasugi of Toyama University, Ueda of Kanazawa University, Kadowaki of Agrobiological Resource, and Go of Nagoya University for providing un-published data. This work was supported in part by a Grant-in-Aid from the Ministry of Education (Japan).
DDBJ/EMBL/GenBank accession nos:
REFERENCES
- 1.Covello P.S. and Gray,M.W. (1989) RNA editing in plant mitochondria. Nature, 341, 662–666. [DOI] [PubMed]
- 2.Gualberto J.M., Lamattina,L., Bonnard,G., Weil,J.H. and Grienenberger,J.M. (1989) RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature, 341, 660–662. [DOI] [PubMed]
- 3.Hiesel R., Wissinger,B., Schuster,W. and Brennicke,A. (1989) RNA editing in plant mitochondria. Science, 246, 1632–1634. [DOI] [PubMed]
- 4.Hoch B., Maier,R.M., Appel,K., Igloi,G.L. and Koessel,H. (1991) Editing of a chloroplast mRNA by creation of an initiation codon. Nature, 353, 178–180. [DOI] [PubMed]
- 5.Yu W. and Schuster,W. (1995) Evidence for a site-specific cytidine deamination reaction involved in C to U RNA editing of plant mitochondria. J. Biol. Chem., 270, 18227–18233. [DOI] [PubMed]
- 6.Hirose T., Fan,H., Suzuki,J.Y., Wakasugi,T., Tsudzuki,T., Koessel,H. and Sugiura,M. (1996) Occurrence of silent RNA editing in chloroplasts: its species specificity and the influence of environmental and developmental conditions. Plant Mol. Biol., 30, 667–672. [DOI] [PubMed]
- 7.Giege P. and Brennicke,A. (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA, 96, 15324–15329. [DOI] [PMC free article] [PubMed]
- 8.Maier R.M., Neckermann,K., Igloi,G.L. and Koessel,H. (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol., 251, 614–628. [DOI] [PubMed]
- 9.Hirose T., Kusumegi,T., Tsudzuki,T. and Sugiura,M. (1999) RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol. Gen. Genet., 262, 462–467. [DOI] [PubMed]
- 10.Wakasugi T., Hirose,T., Horihata,M., Tsudzuki,T., Koessel,H. and Sugiura,M. (1996) Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl Acad. Sci. USA, 93, 8766–8770. [DOI] [PMC free article] [PubMed]
- 11.Yoshinaga K. (1997) RNA editing in land plants. In Iwatsuki,K. and Raven,P.H. (eds), Evolution and Diversification of Land Plants. Springer, Tokyo, pp. 45–60.
- 12.Yoshinaga K., Iinuma,H., Masuzawa,T. and Ueda,K. (1996) Extensive RNA editing of U to C in addition to C to U substitution in the rbcL transcripts of hornwort chloroplasts and the origin of RNA editing in green plants. Nucleic Acids Res., 24, 1008–1014. [DOI] [PMC free article] [PubMed]
- 13.Yoshinaga K., Kakehi,T., Shima,Y., Iinuma,H., Masuzawa,T. and Ueno,M. (1997) Extensive RNA editing and possible double-stranded structures determining editing sites in the atpB transcripts of hornwort chloroplasts. Nucleic Acids Res., 25, 4830–4834. [DOI] [PMC free article] [PubMed]
- 14.Chaudhuri S., Carrer,H. and Maliga,P. (1995) Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J., 14, 2951–2957. [DOI] [PMC free article] [PubMed]
- 15.Bock R. and Koop,H.U. (1997) Extraplastidic site-specific factors mediate RNA editing in chloroplasts. EMBO J., 16, 3282–3288. [DOI] [PMC free article] [PubMed]
- 16.Hirose T. and Sugiura,M. (2001) Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J., 20, 1144–1152. [DOI] [PMC free article] [PubMed]
- 17.Bock R., Hermann,M. and Koessel,H. (1996) In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J., 15, 5052–5059. [PMC free article] [PubMed]
- 18.Bock R., Hermann,M. and Fuchs,M. (1997) Identification of critical nucleotide positions for plastid RNA editing site recognition. RNA, 3, 1194–1200. [PMC free article] [PubMed]
- 19.Chaudhuri S. and Maliga,P. (1996) Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J., 15, 5958–5964. [PMC free article] [PubMed]
- 20.Chaudhuri S. and Maliga,P. (1997) New insights into plastid RNA editing. Trends Plant Sci., 2, 5–6.
- 21.Hermann M. and Bock,R. (1999) Transfer of plastid RNA-editing activity to novel sites suggests a critical role for spacing in editing-site recognition. Proc. Natl Acad. Sci. USA, 96, 4856–4861. [DOI] [PMC free article] [PubMed]
- 22.Miyamoto T., Obokata,J. and Sugiura,M. (2002) Recognition of RNA editing sites is directed by unique proteins in chloroplasts: biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol. Cell. Biol., 22, 6726–6734. [DOI] [PMC free article] [PubMed]
- 23.Bock R. (2000) Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie, 82, 549–557. [DOI] [PubMed]
- 24.Bock R. and Maliga,P. (1995) In vivo testing of a tobacco plastid DNA segment for guide RNA function in psbL editing. Mol. Gen. Genet., 247, 439–443. [DOI] [PubMed]
- 25.Gawel N.J. and Jarret,R.L. (1991) A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol. Biol. Rep., 9, 262–266.
- 26.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual (Second edition). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Kugita M., Kaneko,A., Yamamoto,Y., Takeya,Y., Matsumoto,T. and Yoshinaga,K. (2003) The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucleic Acids Res., 31, 716–721. [DOI] [PMC free article] [PubMed]
- 28.Schuster W., Hiesel,R., Wissinger,B. and Brennicke,A. (1990) RNA editing in the cytochrome b locus of the higher plant Oenothera berteriana includes a U-to-C transition. Mol. Cell. Biol., 10, 2428–2431. [DOI] [PMC free article] [PubMed]
- 29.Gualberto J.M., Weil,J.H. and Grienenberger,J.M. (1990) Editing of the wheat coxIII transcript: evidence for twelve C to U and one U to C conversions and for sequence similarities around editing sites. Nucleic Acids Res., 18, 3771–3776. [DOI] [PMC free article] [PubMed]
- 30.Kudla J. and Bock,R. (1999) RNA editing in an untranslated region of the Ginkgo chloroplast genome. Gene, 234, 81–86. [DOI] [PubMed]
- 31.Schuster W., Unseld,M., Wissinger,B. and Brennicke,A. (1990) Ribosomal protein S14 transcripts are edited in Oenothera mitochondria. Nucleic Acids Res., 18, 229–233. [DOI] [PMC free article] [PubMed]
- 32.Stern D.B. and Gruissem,W. (1987) Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell, 51, 1145–1157. [DOI] [PubMed]
- 33.Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed]
- 34.Sutton C.A., Conklin,P.L., Pruitt,K.D. and Hanson,M.R. (1991) Editing of pre-mRNAs can occur before cis- and trans-splicing in Petunia mitochondria. Mol. Cell. Biol., 11, 4274–4277. [DOI] [PMC free article] [PubMed]
- 35.Ruf S., Zeltz,P. and Koessel,H. (1994) Complete RNA editing of unspliced and dicistronic transcripts of the intron-containing reading frame IRF170 from maize chloroplasts. Proc. Natl Acad. Sci. USA, 91, 2295–2299. [DOI] [PMC free article] [PubMed]
- 36.Marechal-Drouard L., Ramamonjisoa,D., Cosset,A., Weil,J.H. and Dietrich,A. (1993) Editing corrects mispairing in the acceptor stem of bean and potato mitochondrial phenylalanine transfer RNAs. Nucleic Acids Res., 21, 4909–4914. [DOI] [PMC free article] [PubMed]
- 37.Binder S., Marchfelder,A. and Brennicke,A. (1994) RNA editing of tRNA(Phe) and tRNA(Cys) in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet., 244, 67–74. [DOI] [PubMed]
- 38.Marechal-Drouard L., Kumar,R., Remacle,C. and Small,I. (1996) RNA editing of larch mitochondrial tRNA(His) precursors is a prerequisite for processing. Nucleic Acids Res., 24, 3229–3234. [DOI] [PMC free article] [PubMed]
- 39.Fey J., Tomita,K., Bergdoll,M. and Marechal-Drouard,L. (2000) Evolutionary and functional aspects of C-to-U editing at position 28 of tRNA(Cys)(GCA) in plant mitochondria. RNA, 6, 470–474. [DOI] [PMC free article] [PubMed]
- 40.Alfonzo J.D., Blanc,V., Estevez,A.M., Rubio,M.A. and Simpson,L. (1999) C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J., 18, 7056–7062. [DOI] [PMC free article] [PubMed]
- 41.Qin P.Z. and Pyle,A.M. (1998) The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol., 8, 301–308. [DOI] [PubMed]
- 42.Boudvillain M. and Pyle,A.M. (1998) Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J., 17, 7091–7104. [DOI] [PMC free article] [PubMed]
- 43.Carrillo C., Chapdelaine,Y. and Bonen,L. (2001) Variation in sequence and RNA editing within core domains of mitochondrial group II introns among plants. Mol. Gen. Genet., 264, 595–603. [DOI] [PubMed]
- 44.Steinhauser S., Beckert,S., Capesius,I., Malek,O. and Knoop,V. (1999) Plant mitochondrial RNA editing. J. Mol. Evol., 48, 303–312. [DOI] [PubMed]
- 45.Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed]