Abstract
Saccharomyces cerevisiae PRP17 (CDC40) encodes a second-step pre-mRNA splicing factor with a role in cell division. The functions of Prp17 in specific cell cycle transitions were examined using temperature-sensitive alleles in arrest/release experiments. We find that G1/S and G2/M transitions depend on Prp17. G1-synchronized prp17::LEU2 cells arrest at non-permissive temperatures as unbudded haploid cells with low levels of CLN1, CLB5 and RNR1 transcripts. This indicates a Prp17 execution point at or prior to Start. Reduced levels of α-tubulin protein, a mitotic spindle component, underlie the benomyl sensitivity of prp17 mutants and possibly their G2/M arrest. Splicing of TUB1 and TUB3 transcripts, which encode α-tubulin, was analyzed in prp17 and other second-step factor mutants. TUB1 splicing is inefficient in prp17, prp16 and prp22, and marginally affected in prp18, slu7-1 and psf1-1. TUB3 splicing is similarly affected. In vitro splicing with TUB3 pre-mRNA demonstrates a compromised second step in prp17::LEU2 extracts, implicating a direct role for Prp17 in its efficient splicing. Genomic replacement of an intronless TUB1 gene relieves the benomyl sensitivity of prp17 mutants; however, they remain temperature sensitive, implying multiple limiting factors for mitosis. The data suggest that integration of splicing with the cell cycle is important for G1/S and G2/M transitions.
INTRODUCTION
In eukaryotic cells, an orderly progression through the mitotic cycle is ensured by the highly regulated transition from one phase to the next. In Saccharomyces cerevisiae, as in other eukaryotes, a number of evolutionarily conserved effectors, such as the p34-cyclin-dependent protein kinase Cdc28/Cdc2, play a critical role in driving the division cycle. The movement across major cell cycle transitions (G1/S, G2/M and M/G1) is controlled by timed activation/inactivation of these specific effectors, many of which are very well studied. Cells also use the general cellular/metabolic processes for efficient regulation of cell cycle events. For example, although the ubiquitous 26S proteasome degrades all ubiquitin-tagged proteins, its activity is harnessed by specific ubiquitin ligases (E3 enzymes) to degrade cell cycle phase-dependent substrates. Such regulatory controls exemplify how the general cellular machinery for protein degradation, though not particularly specialized for cell cycle control, is utilized to drive the cell cycle with critical specificity. Although links between the cell cycle and RNA processing events are indicated by genetic studies in budding and fission yeasts and biochemical studies in human cells (1–6), they have not been investigated thoroughly.
In this report, we have investigated the connections between cell division and splicing with a special focus on PRP17/CDC40. PRP17 encodes a protein essential for viability at elevated temperatures (7). Studies on cdc40-1 and a null allele showed a role in both initiation of DNA replication and nuclear division (7,8). Independent genetic screens identified Prp17 as a pre-mRNA splicing factor (9,10). Sequence identity between PRP17 and CDC40 shows them to be the same gene. In vivo analysis of several intron-containing transcripts and in vitro experiments with the ACT1 pre-mRNA implicate a role for Prp17 in the second transesterification reaction during splicing (9,11,12). This function is apparently conserved even in the human Prp17 protein (13,14) that partially complements growth defects of yeast prp17::LEU2. While PRP17 is not essential for viability, it exhibits extensive genetic interactions with PRP18, PRP16, SLU7, PRP8, PRP22, U5 snRNA and U2 snRNA (4,10,11,15,16). With the exception of PRP18, all the others are essential for the second step of splicing.
In addition to Prp17, a few other yeast splicing factors affect the cell cycle. The prp3 allele dbf3-1, the prp8 allele dbf5 and alleles of prp22 and cef1 were uncovered in cell cycle mutant screens (1,17,18). The diverse phenotypes of prp17/cdc40-null cells hint that Prp17 influences more than one stage in the division cycle (7). This prompted us to examine the role of Prp17 closely in specific events of the division cycle. Using different temperature-sensitive missense alleles of prp17 in various arrest/release protocols, we have assessed the execution points for this splicing factor. We show that Prp17 is required for the G1/S and G2/M transitions. The latter requirement is due in part to the highly compromised splicing of α-tubulin transcripts from the TUB1 and TUB3 genes, which contain non-standard introns. This analysis in prp17 and in several second-step splicing factor mutants demonstrates a differential dependence of these cellular RNAs on some splicing factors. The contribution of Prp17 to TUB3 intron splicing was also studied in vitro. We find an inefficient second step even in wild-type extracts, a defect exacerbated in the absence of Prp17. Altogether, our data suggest a role for a non-essential pre-mRNA splicing factor in efficient progression through cell cycle transitions.
MATERIALS AND METHODS
Strains
YPH274Δ17a MATa prp17::LEU2 ade2-101 his3Δ200 ura3-52 lys2 trp1 leu2-3 leu2-112 bar1::hisGURA3hisG was the null allele used. prp17-1 bar::URA3 strain MATa prp17-1 ade2-101 his3Δ200 ura3-52 lys2 bar1::hisGURA3hisG. prp17-2 bar::URA3 strain MATa prp17-2 his3Δ200 ura3-52 trp bar1::hisGURA3hisG. prp17-3 strain MATa ura3-52 his3Δ200 trp1Δ63 lys2-801 ade2-101 leu2Δ1; slu7-1 and psf1-1 strains were obtained from C. Guthrie, UCSF, San Francisco. prp17-4 (AAY80) MATa cdc40 ade2-128 was obtained from A. E. M. Adams, University of Arizona, Tucson. The permissive temperature for all strains was 23°C and the restrictive temperature was 37°C.
Synchronization procedures and flow cytometry analysis
For G1 arrest, cells grown to 1 × 107cells/ml were treated with α-factor (1.0 µg/ml). For BAR+ strains, 5 µg/ml α-factor was added. After >95% G1 arrest, the cells were filtered, washed and inoculated into fresh media kept at 23 or 37°C. For early S phase growth arrest, hydroxyurea (15 mg/ml) was added to the cultures. After 3 h, the synchronized cells were treated as mentioned above. Flow cytometry analysis of DNA content was done as described (19).
Immunofluorescence analysis
Indirect immunofluorescence of the mitotic spindle and 4′,6-diamidino-2-phenylindole staining of the nuclei were performed as described (20). The primary antibody was YOL134 (1:500 in 1% bovine serum albumin–phosphate-buffered saline) and tetramethylrhodamine isothiocyanate TRITC-conjugated goat anti-rat sera (1:30 dilution, Jackson Immunoresearch Company) were the secondary antibodies. Cells were observed on a Zeiss Axioscope microscope and images captured on Axiocam (Zeiss).
Preparation of total RNA and RT–PCR analysis
Cells from early–mid log phase cultures were collected either before or after growth at 37°C for 1 h. Total RNA was extracted (9) and 5–10 µg used to make first-strand cDNA using SuperscriptII (Invitrogen) in a 20 µl reaction. Gene-specific primers for reverse transcription were chosen in exon 2 of TUB1 and TUB3: TUB3 exon 2 primer (5′-CTCTTGCGTAGTTATTGGC-3′); TUB1 exon 2 primer (5′-CCTATCCAGAACATCGCCCAA-3′). A 3 µl aliquot from each reverse transcriptase reaction was used in a 15 µl PCR with 1 µM exon 2 primer and a TUB3-specific exon 1 primer (5′-AGAGAGGTCATTAGTATT-3′) to amplify TUB3 cDNA, or a TUB1-specific exon 1 primer (5′-GAGAGAAGTTATTAGTATT-3′) to amplify TUB1 cDNA. Twenty-five PCR cycles were done for a linear response. The expected RT–PCR product from TUB3 pre-mRNA is 614 bp, and that from TUB3 mRNA is 316 bp. The predicted product from TUB1 pre-mRNA is 481 bp and from its mRNA is 366 bp. As controls for these reactions, U5 snRNA levels were measured using primers U5RT RP (5′-AACGCCCTCCTTACTCATTG-3′) and U5 RT FP (5′-GCAGCTTTACAGATCAAT-3′). All PCR products were resolved on 10% native PAGE gels. Photostimulated luminescence counts were obtained in a phosphorimager.
Construction of a strain for inducible expression of CLN2 in prp17::LEU2 cells
A galactose-inducible version of the G1 cyclin, pGAL-CLN2, was integrated at the URA3 locus. Prior to this integration, the URA3 cassette previously integrated at the bar locus in the strain prp17ΔYPH274 bar:hisg:URA3:hisg was excised by selection on 5-fluoro-orotic acid media. Single bar– and ura– colonies were then transformed with StuI-linearized GAL-CLN2 (pUS152) construct. The integrants were verified by Southern analysis, and northern analysis confirmed galactose-induced expression of CLN2.
In vitro transcription and splicing
The TUB1 minigene construct was generated by PCR amplification of residues +3 to +1028 of the open reading frame (ORF) present in the plasmid RB306 (21). The product, cloned in the PCR blunt vector (Invitrogen), was linearized with AccI and used for in vitro transcription with T7 RNA polymerase. The TUB3 minigene construct was generated by PCR of +3 to +617 bp of the ORF in RB300 (21). The product was cloned in the pBluescript vector. The recombinant was linearized with SnaBI and transcribed with T7 RNA polymerase. ACT1 substrate was prepared from BamHI-linearized p422 (22). In vitro transcription and splicing reactions were done as described (23). The splicing reactions were resolved on 8.5% (19:1) denaturing polyacrylamide gels for reactions with TUB3 transcripts; 6.5% (19:1) gels were used for reactions with ACT1. Quantitations were done by phosphorimager analysis.
Construction and homologous replacement of an intronless TUB1 gene
A 74 nt primer starting 26 bp upstream of the TUB1 ATG, spanning exon 1 and creating a translational fusion to exon 2, was made. In a PCR with a TUB1 exon 2 reverse primer, this 76mer would delete the intron. The PCR product was cloned and sequenced. This intronless DNA fragment was then swapped with wild-type TUB1 present in a yeast centromeric vector pRB326 (21) to create the recombinant: pTUB1Δi RB326. For genomic integration, a SalI–ClaI fragment from pTUB1Δi RB326 was cloned in pRS303. The resultant recombinant was linearized at the BstEII site in exon 2 before integration in the yeast genome.
RESULTS
Prp17 is required for the G1/S transition of the cell cycle
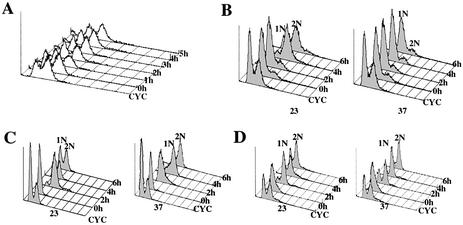
To examine the role of Prp17 in the cell cycle, an asynchronous population of cells with the prp17 null allele were grown at 23°C and then transferred to the restrictive temperature (37°C). By fluorescence-activated cell sorting (FACS) analysis, we note that there are only minimal changes in the DNA content of these cells even after 5 h (Fig. 1A). At 37°C, the culture contained two populations of cells: ∼40% with 1N DNA content and ∼60% with 2N DNA content. This suggests that Prp17 plays a role at two stages of the division cycle.
Figure 1.
Prp17 is required for G1/S and G2/M transitions. FACS analysis of DNA content in prp17::LEU2 cells assayed in various arrest/release protocols. The peaks representing 1N or 2N DNA content are labeled. Indicated to the right of each data plot are the time points shown here. CYC denotes a 23°C cycling culture of prp17::LEU2. (A) Terminal arrest phenotypes in a prp17::LEU2 culture grown at 23°C and shifted to 37°C at time point 0 h. (B) Arrest phenotype in G1-synchronized prp17::LEU2 cells grown at non-permissive temperatures. α-Factor-treated cells were released at time 0 h to either permissive (23°C, left) or non-permissive temperatures (37°C, right). Plots show cellular DNA content in aliquots withdrawn at the indicated time points. (C) prp17::LEU2 cells grown at 23°C were synchronized in early S phase with hydroxyurea (time point 0 h). Cells were then released to 23 and 37°C, and DNA content was monitored. (D) G1-synchronized prp17::LEU2 cells were generated as in (B). These cells were first released for growth at 23°C for 2 h. At this stage, marked time point 0 h, half the culture was shifted to 37°C. DNA content in cells withdrawn subsequently at 2, 4 and 6 h is plotted.
To confirm these observations, and to aid in clearly defining the execution point(s) for PRP17, a number of prp17 alleles were studied in various arrest/release protocols. The alleles chosen were: prp17-1, prp17-2, prp17-3, prp17-4 and prp17::LEU2, the null allele. It is noteworthy that the prp17-1 mutation, an allele analyzed in detail here, lies in the N‐terminal region of the protein that is functionally important (11,16). After G1 synchronization, cells were allowed to re-enter the division cycle at either permissive or non-permissive temperatures. Cellular DNA content was monitored and the status of the mitotic spindle assessed during cell cycle progression. At the permissive temperature, prp17::LEU2 cells grow slowly, enter S phase and subsequently complete mitosis. However, at non-permissive temperatures, these cells arrest uniformly with 1N DNA content (Fig. 1B). The prp17-4 allele also confers a complete G1/S arrest (Table 1). This implicates a requirement for Prp17 during transition to S phase. Interestingly, other alleles of PRP17 behave differently. For instance, prp17-1 cells after release from α-factor synchronization completely arrest with 2N DNA (Table 1). On the other hand, prp17-2 causes predominantly a 1N arrest with only a small proportion of cells progressing slowly through S phase (Table 1). In prp17-2, the mutation lies in the nuclear localization signal, which could retard nuclear trafficking of this protein and cause slow progression through S phase. We conclude, therefore, that at least one execution point for Prp17/Cdc40 is in the late G1 phase. However, the behavior of the prp17-1 mutant raises the possibility of a stringent requirement at G2/M.
Table 1. Summary of cell cycle phenotypes in prp17 mutants synchronized in G1 or early S phase.
| Allele | Cell cycle phenotype at non-permissive temperature | |||
|---|---|---|---|---|
| α-Factor synchronization | HU synchronization | |||
| DNA content | Spindle morphology | DNA content | Spindle morphology | |
| prp17-1 | 2N | Very short or absent | 2N | Absent |
| prp17-2 | Predominantly 1N | Very short or absent | 2N | Very short or absent |
| prp17-3 | 1N | ND | 2N | ND |
| prp17-4 | 1N | Absent | 2N | Absent |
| prp17::LEU2 | 1N | Absent | 2N | Absent |
| cdc40-1a | 2N | Absent | 2N | Absent |
ND, not defined.
aSee Vaisman et al. (7).
Prp17 is required in the pre-Start phase of the cell cycle
To define better the function of Prp17 during G1/S, we analyzed the window in G1 at which prp17 cells are arrested. More specifically, we asked if in the absence of Prp17 the cells pass through Start and arrest at a post-Start phase. Traditionally, Start is defined as the point in G1 beyond which cells are irreversibly committed to a new division cycle. The emergence of a bud, a burst of transcription of specific genes and initiation of DNA replication are some of the major indicators of this commitment (24,25). We analyzed the Start-dependent expression of CLB5, an S-phase cyclin, in G1-synchronized prp17::LEU2 cells that resume growth at permissive or non-permissive temperatures. CLB5 transcript levels were determined 30 min after release from synchronization. The analysis was repeated in culture aliquots withdrawn at subsequent hourly intervals (Fig. 2A). prp17::LEU2 cells at 23°C accumulate high levels of CLB5 within 2 h of release from α-factor treatment and, by 4 h, the levels decline, indicating passage through G1/S. At 37°C, CLB5 RNA levels do not build up even 5 h after the release from synchronization (Fig. 2A). These data are consistent with the absence in this culture of cells with 2N DNA content. To establish the pre-Start arrest in the prp17::LEU2, a second marker for early S phase was employed. The expression of RNR1, encoding ribonucleotide reductase, is transcriptionally activated in a Start-dependent manner, since high levels of Rnr1 are necessary for S phase. RNR1 transcript levels in the same experimental set-up as described above show no significant increase in prp17 at 37°C (Fig. 2B). Additionally, we assessed the levels of the G1 cyclin CLN1, transcriptional upregulation of which is a prerequisite for Start. While CLN1 levels in prp17 cells at 23°C reach a peak within 30 min of release from synchronization, this increase fails to occur at 37°C (Fig. 2C). Together, the data confirm the pre-Start arrest in prp17 cells and suggest a role for Prp17 in the G1/S window lying between α-factor arrest and Start.
Figure 2.
Transcript levels of a G1 cyclin and two Start-dependent genes in prp17::LEU2 cells. G1-arrested prp17::LEU2 cells were released to fresh medium at 23 or 37°C. Total RNA in samples withdrawn at regular intervals from both cultures was analyzed by northern blots. The lane marked CYC denotes RNA from a cycling culture. (A) Changes in the levels of CLB5 transcripts as prp17::LEU2 cells progress through the cell cycle upon release from G1 synchronization. The bar graph plots the ratio of CLB5/ SNR7 at the various time points tested in cultures at permissive and non-permissive temperatures. (B) Changes in levels of RNR1 transcripts as prp17::LEU2 cells progress through the cell division cycle after release from G1 synchronization. The bar graphs depict the normalized levels of RNR1 at these different time points and conditions. (C) The blot was probed to detect CLN1 transcripts, a G1 cyclin. The levels of CLN1 mRNA were normalized to that of SNR189, a constitutively expressed RNA. The bar graph plots the ratio of CLN1/SNR189 at the various time points tested.
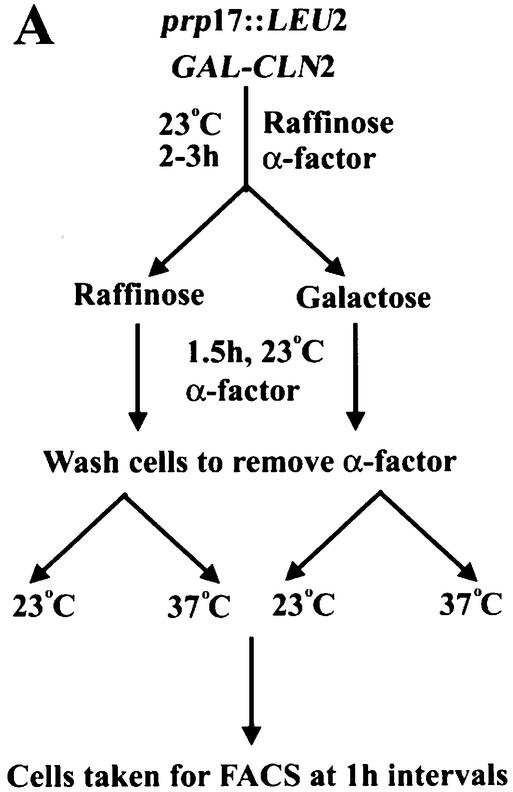
Ectopic induced transcription of CLN2 does not relieve the G1/S block in prp17::LEU2 cells
The failure to accumulate high levels of G1 cyclins could cause the pre-Start arrest in prp17::LEU2 cells. To understand if this is the sole defect in these cells and that there are no other regulatory controls, we have analyzed the consequences of ectopic induction of the functionally redundant CLN2 gene in these arrested cells. A construct where a galactose-inducible transcription control of CLN2 can be obtained was employed. This regulated CLN2 construct was integrated at the URA3 locus of prp17::LEU2 (Materials and Methods) and then used in cell cycle protocols. Such cells grown at 23°C with raffinose as the carbon source keep GAL1-CLN2 repressed and were synchronized with α-factor. At 1.5 h before complete synchronization of cells was achieved, galactose was added to the culture to induce CLN2 transcription, but cells were kept arrested due to the presence of α-factor (Fig. 3A). After complete synchronization, the cells were moved into α-factor-free medium containing either raffinose in the control sample or galactose in the experimental set-up. Both control and experimental cultures were grown at 23 and 37°C (Fig. 3A). Cells maintained in a CLN2-repressed state (i.e. raffinose medium, Fig. 3B, top left) at 23°C grow slowly yet pass through Start and complete DNA synthesis by 4 h. As expected, at 37°C these cells without CLN2 overexpression remain arrested with 1N DNA content (Fig. 3B, top right), reflecting the prp17::LEU2 phenotype. Notably, in cultures where CLN2 is induced (galactose), the cells do not pass through Start and fail to complete DNA synthesis at 37°C (Fig. 3B, bottom right). Thus, we find that overexpression of CLN2, a G1 cyclin whose levels are a determinant for passage through Start, cannot compensate for the G1/S arrest in prp17::LEU2 cells at non-permissive temperature. It is highly unlikely that the CLN2 transcript is a direct target for Prp17 as it lacks introns. These data point to multiple targets for Prp17 and indicate that the pathway leading to G1 cyclin expression is not the only limiting factor in prp17::LEU2 cells.
Figure 3.
(A) A schematic representation of the experimental strategy used to assess the effects of ectopic CLN2 expression on G1-arrested prp17::LEU2 cells. A galactose-inducible version of G1 cyclin CLN2 was integrated at the URA3 locus in prp17::LEU2 cells. The cells were grown in non-inducing conditions and treated with α-factor for 2–3 h, following which an aliquot of the culture received galactose for induction of CLN2. After complete synchronization, both cultures were released from α-factor arrest and tested for cell cycle progression at 23 and 37°C. (B) The DNA content as determined by FACS analysis in the four cultures is plotted. The growth conditions and time point of analysis are shown on the right.
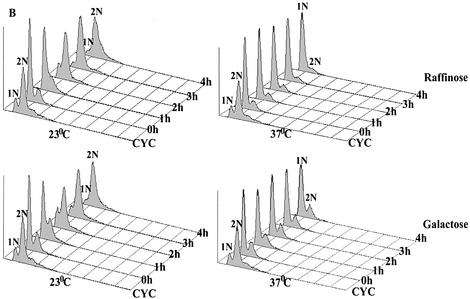
G2/M transition defines the second point of Prp17 requirement
Prompted by the complete G2/M arrest of prp17-1, we next examined if the alleles causing a strict G1/S arrest are also defective for the temporally later G2/M transition. prp17:: LEU2 cells were synchronized in early S phase by hydroxyurea (HU) treatment so that they were already past their G1/S execution point. Subsequently, cell cycle progression was monitored at 23 and 37°C. In prp17::LEU2 cells released at permissive temperature, DNA was replicated by 2 h and mitosis completed by 4 h (Fig. 1C). However, at the non-permissive temperature, these cells arrested with 2N DNA (Fig. 1C), a large bud and an undivided nucleus. Similar results were obtained with other missense alleles of prp17 (Table 1). Thus, all alleles of prp17 in this experimental regime arrest with 2N DNA and fail to undergo nuclear division. Further, to prove conclusively the requirement for Prp17 beyond S phase initiation, yet another set of experiments was done. Here, prp17::LEU2 cells were synchronized in G1 by α-factor treatment and then allowed to resume growth at permissive conditions for 2 h, thereby allowing them to pass Start and initiate S phase. After this time interval, half of the culture was shifted to non-permissive temperature while the other half was maintained at the permissive temperature. In parallel, wild-type cells were subjected to identical experimental conditions. Consistent with results described in the previous sections, prp17::LEU2 cells shifted to non- permissive temperature arrest with 2N DNA (Fig. 1D). Collectively, these experiments clearly demonstrate that Prp17/Cdc40 is required for both the G1/S and G2/M transitions.
Spindle maintenance defects in G2/M-arrested prp17 cells—tubulin as a target for regulation by splicing
Earlier reports have implicated CDC40/PRP17 in spindle assembly/maintenance (7). Since we find allele-specific phenotypes for onset of S phase, we examined the status of the mitotic spindle in various prp17 mutants, with special attention paid to alleles that progress through S phase after Prp17 inactivation. The mitotic spindle in cells that were allowed to resume the division cycle after synchronization with α-factor or HU treatment was monitored by indirect immunoflorescence of α-tubulin. We find a spindle maintenance defect within 2 h of exposure to 37°C (data not shown), and by 3 h cells do not have any spindles or have very short spindles (Table 1). The prp17 alleles differ only in the severity of spindle defects.
We have investigated possible reasons underlying microtubule instability by examining post-transcriptional regulation of tubulin. The spindle consists of microtubules that are polymers of the α–β tubulin heterodimer. Saccharomyces cerevisiae has a single essential β-tubulin gene, TUB2, which does not contain any introns. There are two functional α-tubulin genes, TUB1 and TUB3, which share 90% homology entirely within their coding sequences. In crude extracts from prp17-1 cells passing through S and G2, there is a significant reduction (∼80%) in α-tubulin protein levels when this splicing factor is inactivated (data not shown). Similar effects are known in cef1-13, and in isy1Δ syf2Δ double mutants. These mutations in splicing factors also cause a G2/M arrest (6,26).
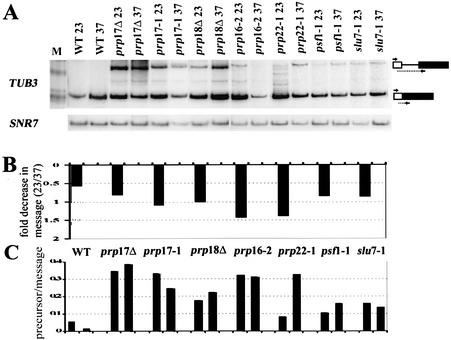
Consequently, we investigated regulation of α-tubulin by mRNA splicing. Both TUB1 and TUB3 genes contain introns that have been speculated targets for regulation by splicing. While the TUB1 intron has consensus cis-recognition elements, it is a small intron (115 nt). The TUB3 intron (298 nt) is unusual in that the distance between the branch point and the 3′ splice site is 135 nt, as opposed to 36–43 nt in most other yeast introns. We and others (4,15,27) have hypothesized that Prp17 is required for improving the efficiency of splicing for transcripts with non-canonical intron features. Therefore, the splicing efficiency of both TUB1 and TUB3 pre-mRNAs was examined. Since these genes share extensive nucleotide homology, we devised a semi-quantitative RT–PCR strategy to measure pre-mRNA and mRNA levels for each transcript. As a measure of splicing efficiency, we have determined the decrease in mRNA and also the change in precursor mRNA to mRNA ratio after inactivation of temperature-sensitive mutations in splicing factors. The effects caused by prp17 and prp18 were compared since both are non-essential auxiliary factors for the second step. We have also examined TUB1 and TUB3 splicing in other second-step factor mutants: prp16, slu7 and prp22. Prp16 is an essential helicase required to bring about a critical conformational change at the 3′ splice site. It is suggested to act at or about the step of action of Prp17. The contribution of Slu7 and Prp22 to splicing of artificial substrates with varying distances between branch point and 3′ splice site has implicated them in the splicing of introns with increased distances between these specific intron elements (28,29). Furthermore, new alleles of prp22 have been identified in a screen for mutants that arrest in mitosis with no microtubules (17). The psf1-1 mutation in PRP8 was also included in this study, since it identifies a Prp8 domain involved in 3′ splice site definition during the second step (30).
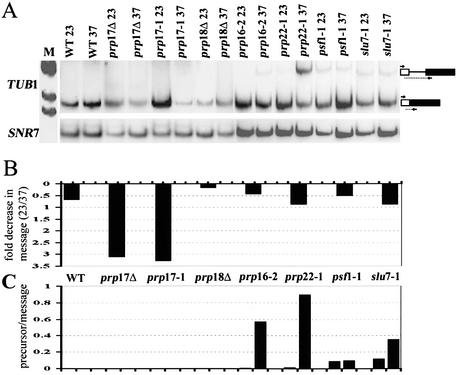
The splicing of TUB1 was compromised in prp17::LEU2, as revealed by the drastic decrease in mRNA; this contrasts with the lack of any significant change in TUB1 mRNA levels in similarly treated wild-type cells (Fig. 4A and B). This effect is also observed upon inactivation of the missense mutant prp17-1, confirming that the decreased TUB1 mRNA levels are seen in the absence of active Prp17 at higher temperatures. None of the other mutations in second-step splicing factors prp18::HIS3, prp16-2, slu7-1, prp22-1 and psf1-1 greatly alter TUB1 mRNA levels (Fig. 4A and B). Despite the depleted mRNA levels in prp17 mutants, an increase in TUB1 pre-mRNA is not detectable (Fig. 4A and C). Failure to build up precursors or intermediates in prp17 mutants has been reported for other intron-containing transcripts (14,31,32). We observe prp18 mutations to behave similarly; there is no increase in TUB1 pre-mRNA levels (Fig. 4A and C). Notably there is no reduction in TUB1 mRNA levels in the absence of Prp18 (Fig. 4B). In fact, an increase in mRNA levels is observed at 37°C. Strikingly, of the essential second-step factors, prp16 and prp22, but not slu7-1 and psf1-1, cause increased levels of TUB1 pre-mRNAs (Fig. 4A and C). Thus, taking the combined effects of a decrease in mRNA levels and an increase in the pre-mRNA/mRNA ratio, we infer that TUB1 intron splicing requires Prp17, Prp16 and Prp22.
Figure 4.
(A) RT–PCR analysis to measure changes in TUB1 intron splicing after temperature-dependent inactivation of mutant second-step splicing factors. cDNA was generated using RNA from each strain, grown at permissive temperatures or after shift to non-permissive temperatures for 60 min. The RT–PCR products corresponding to the spliced mRNA and the unspliced precursor are indicated to the right. The lane M indicates the pBR322 HinfI markers used. The U5 snRNA (SNR7) levels were measured concurrently to provide a normalization control since it does not contain an intron. These cDNAs were quantitated by phosphorimager analysis. (B) The bar graphs plotted depict the fold decrease in normalized mRNA levels that occur in each strain after shift to 37°C. (C) The bar graphs show the normalized TUB1 pre-mRNA/mRNA ratio at 23°C or after shift to 37°C in each of the strains.
The consequence of mutations in second-step splicing factors for TUB3 intron splicing was also examined. The TUB3 transcripts are poorly spliced in the absence of Prp17, as evidenced by an increase in the precursor/message ratio (Fig. 5A–C). The poor splicing of TUB3 in prp17::LEU2 is replicated in the temperature-sensitive prp17-1 strain (Fig. 5A). Similar to the effects on TUB1 splicing, TUB3 splicing is inefficient in prp16 and prp22 mutants. The effects of prp18::HIS3, slu7-1 and psf1-1 are comparatively lower. Collectively, these data indicate a greater dependence of TUB1 and TUB3 introns on PRP17, PRP16 and PRP22.
Figure 5.
(A) RT–PCR analysis of TUB3 splicing efficiency in mutants of second-step splicing factors. Reverse transcription with primers specific to TUB3 was performed on the same RNA as that used in Figure 4. These RNAs were quantitated by phosphorimager analysis, following which the mRNA and the pre-mRNA values were normalized to that of U5 snRNA. (B) The fold decrease in normalized TUB3 mRNA levels occurring in each mutant after temperature inactivation is shown. (C) The ratio of unspliced TUB3 pre-mRNA to mRNA in each strain grown at permissive temperature or after shift to non-permissive conditions is plotted.
A second-step defect for the splicing of the TUB3 intron in cell-free extracts from prp17::LEU2
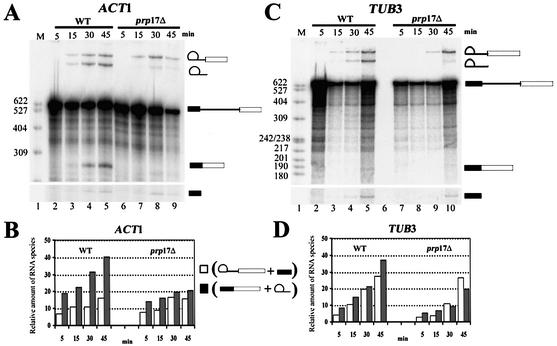
The reduction in TUB1 and TUB3 spliced mRNA in prp17 cells could arise as an indirect consequence of reduced transcription or altered decay kinetics for these mRNAs. To probe the direct relevance of the in vivo data, the splicing of the TUB1 and TUB3 transcripts was examined in vitro. Segments of the TUB1 and TUB3 genes were cloned as minigenes to generate splicing substrates through in vitro transcription. As a control, ACT1 pre-mRNA was also generated. The latter is spliced efficiently both in vitro and in vivo, and is the prototype in nearly all such analyses. The TUB3, TUB1 and ACT1 substrates were examined in vitro particularly for the kinetics of the second splicing reaction. This was studied in two different cell-free extracts, one from a wild-type strain and the other from an isogenic strain bearing a null allele of PRP17. The ACT1 intron is spliced efficiently in wild-type extracts; products of step 1 appear rapidly at 23°C (Fig. 6A, lanes 2–5) and the second step occurs without any delay (Fig. 6B). At any time point, the products from the second step are in larger amounts than those from the first step. In contrast, the splicing of the ACT1 intron is inefficient for step 2 in extracts from prp17::LEU2; products from the second step are at significantly lower levels. This defect persists even at later time points (Fig. 6A, lanes 6–9, and B). These observations reinforce the general requirement for Prp17 for improving the rate of the second step for ACT1 and concur with data obtained through immunodepletion of Prp17 from wild-type extracts (12). The complete absence of Prp17 does not fully abolish the second step, since some mRNA is formed. Similar consequences are noted in in vitro reactions that lack active Prp18 (33,34). The splicing of the TUB3 pre-mRNA in vitro is marginally slow, as compared with ACT1 (∼10 min delay) for step 1 even in wild-type extracts (Fig. 6C and D). Even more striking is a delay in the kinetics of splicing step 2 (Fig. 6C and D). In prp17::LEU2, the products from the TUB3 substrate are at low levels and a build up of the intermediates from reaction 1 occurs (compare Fig. 6A, lanes 8 and 9 with C, lanes 9 and 10; Fig. 6B and D). This inefficiency of TUB3 splicing is reproducible even in different batches of extracts (data not shown). The TUB1 intron was not spliced from in vitro transcripts even in wild-type extracts, perhaps due to the small intron size and extremely poor splicing efficiency, and therefore could not be investigated further.
Figure 6.
In vitro splicing of ACT1 and TUB3 in wild-type and prp17::LEU2 extracts. (A) 32P-labeled ACT1 pre-mRNA was reacted with a wild-type extract at 23°C. Aliquots were withdrawn at the time points given above each lane (lanes 2–5). The RNA products were analyzed by denaturing PAGE. The RNA species lariat–exon 2, lariat intron, precursor, mRNA and exon 1 are indicated to the right. The splicing reaction performed with ACT1 substrate and extracts from prp17::LEU2 were run on lanes 6–9. (C) 32P-labeled TUB3 pre-mRNA substrate reacted with wild-type (lanes 2–5) or prp17::LEU2 extract (lanes 7–10). The reaction aliquots were withdrawn at the time points given above each lane. The RNA species were resolved on denaturing PAGE and are represented schematically on the right. (B and D) Quantitation of the splicing reactions 1 and 2 for the substrate ACT1 (B) or TUB3 (D). In each case, quantitations were done for reactions with wild-type extract and also prp17::LEU2 extracts. Open bars indicate the amounts of lariat–exon 2 plus exon 1 resulting from reaction 1 at the given time point. Filled-in bars indicate the amounts of lariat intron plus mRNA resulting from reactions 1 and 2 at the given time point. The quantitations of pre-mRNA, lariat–exon 2, lariat intron, mRNA and exon 1 were obtained through phosphorimager analysis of the gels. The levels of the RNA intermediates and product RNAs were normalized to the levels of precursor RNA in each lane.
While the splicing reactions have not been modeled kinetically, we have estimated the likely rate of the second step in these reactions, adopting the approximations of Horowitz and Abelson (34). Quantitative analyses of the time course of splicing shown in Figure 6A and B were done to estimate the rate of the second step. The rate of change in mRNA occurring within a 15 min time frame over the concentration of lariat intermediate was taken as an approximation of k2, i.e. the rate of the second splicing reaction. ACT1 is a somewhat better substrate than TUB3 for the second step in wild-type extracts (Table 2). This small difference in the estimated rate of the second step is accentuated in the absence of Prp17 (Table 2), indicating a direct role for this non-essential factor in splicing of TUB3, with its non-standard intron.
Table 2. Estimated in vitro rate constants for second step with ACT1 and TUB3 substrates.
| In vitro splicing substrate | k2 valuea | Fold decrease in k2 | |
|---|---|---|---|
| Wild-type extract | prp17Δ extract | ||
| ACT1 | 0.030 | 0.023 | 1.3 |
| TUB3 | 0.025 | 0.013 | 1.92 |
aΔ[mRNA]/Δt ∼ k2 [lariat intermediate] (34).
TUB1Δi gene does not rescue the growth defects of prp17-1; multiple splicing targets for Prp17 in G2/M
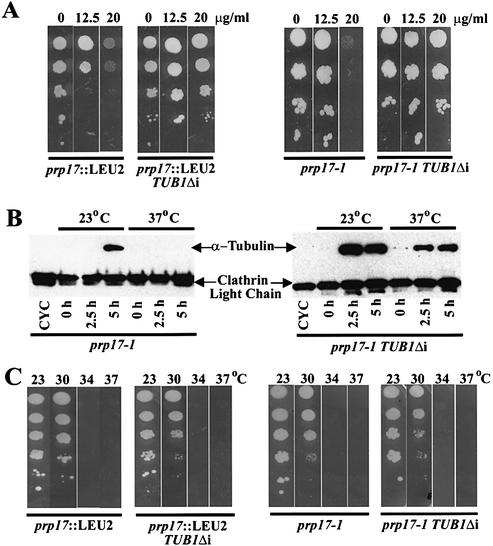
A precise deletion of the TUB1 intron was created by a loopout PCR on a plasmid-borne TUB1 gene (Materials and Methods). The resulting TUB1Δi plasmid was linearized and integrated at its genomic locus so as to generate a single functional copy of the TUB1 gene without the intron. The consequences of this intronless gene were examined in wild-type, cold-sensitive tub1-1, prp17-1 and prp17::LEU2 strains. We resorted to the expression of TUB1Δi from the endogenous promoter at its chromosomal location since the expression levels and the stoichiometry of α-tubulin to β-tubulin are critical for efficient assembly and maintenance of mitotic spindles. Overexpres sion of TUB2, encoding β-tubulin, causes a growth arrest at G2/M (35). The TUB1Δi gene fully rescued the cold sensitivity of tub1-1, demonstrating the TUB1Δi construct to be functional (data not shown). The instability of mitotic spindles in prp17 mutant strains makes them mildly sensitive to benomyl, a microtubule-depolymerizing drug. The TUB1Δi construct rescues this phenotype in both prp17-1 and prp17::LEU2 strains (Fig. 7A). We have determined α-tubulin protein levels in prp17-1 cells and compared them with the levels in prp17-1 TUB1Δi. These experiments were performed in synchronized cultures obtained through HU treatment. α-Tubulin in the parent prp17-1 cells is observed only at permissive temperatures (Fig. 7B). The inability to generate α-tubulin at non-permissive temperatures is alleviated by the TUB1Δi gene (Fig. 7B). Nevertheless, the temperature-sensitive growth phenotype of prp17-1 is not rescued (Fig. 7C). Notably, prp17-1 displays only a G2/M arrest (Table 1). Taken together, these data indicate that in prp17 mutants, multiple limiting factors affect progression through G2/M.
Figure 7.
Consequences of an intronless TUB1 gene (TUB1Δi) on phenotypes of prp17-1 and prp17::LEU2. (A) Benomyl sensitivity of prp17:: LEU2 and prp17-1; their rescue by TUB1Δi. The panels on the left show the growth at 23°C of serial dilutions of prp17::LEU2 or prp17::LEU2 TUB1Δi cultures. The growth of the cells in the absence or presence of benomyl at varying concentrations (indicated on the top) is shown. The panels on the right are from similar experiments with prp17-1 and prp17-1 TUB1Δi cells. (B) Western blot of α-tubulin protein levels. HU-synchronized prp17-1 cells (left panel) or prp17-1 TUB1Δi cells (right panel) were released at time 0 h to either permissive (23°C) or non-permissive temperatures (37°C). α-Tubulin protein levels in aliquots withdrawn subsequently at the indicated time points are shown here. Clathrin light chain protein levels served as a loading control. (C) Growth at various temperatures of prp17 mutants with or without TUB1Δi. Serial dilutions of cells of the genotype indicated at the bottom were grown at the temperatures given at the top of each plate. The temperature sensitivity of prp17-1 or prp17::LEU2 is not rescued.
DISCUSSION
Earlier experiments have shown Prp17 to be a second-step splicing factor, suggesting a role as a general spliceosome component (14,11,12). However, since cells lacking Prp17 are viable at 23°C, it is thought to encode a function auxiliary to an essential, second-step splicing factor, possibly Prp8 (4,15,27). Interestingly, the behavior of a null allele of prp17 implied a role for Prp17/Cdc40 in efficient progression through S phase and mitosis (7,8). It remained unclear if discrete transitions in the cell division cycle require Prp17. In this report, using a null mutant and a number of alleles with missense mutations in different regions of the protein, we demonstrate a requirement for Prp17 particularly during G1/S and G2/M cell cycle transitions. Several other lines of evidence gathered here strongly support this conclusion. Strikingly, even the weakest allele, prp17-1 (with a G127A alteration), fails to complete mitosis; cells arrest with 2N DNA but without mitotic spindles (Table 1). No defect (kinetic or otherwise) for entry into and completion of S phase is detected in this mutant (data not shown). Our data from other prp17 alleles confirm that Prp17 is required for entry into S phase (Table 1). These results strongly suggest a role for this factor at these two distinct cell cycle transitions. One possible reason underlying the G1/S arrest in prp17 mutants may be the inefficient or lack of splicing of several intron-containing transcripts whose products encode factors needed for the metabolic state of G1 cells. The hypothesis is supported by our finding that ectopic overexpression of a G1 cyclin, whose levels are one of the critical elements for transition to S phase, cannot force an S-phase entry in these G1-arrested prp17 cells. The alternative possibility for the G1 arrest in prp17 is a direct role for Prp17 in DNA replication. This is a plausible situation since PRP17 exhibits genetic interactions with CLF1 (SYF3), a splicing factor that recently was shown to interact with replication origin protein, Orc2 (36). Prp17 thus joins a host of factors that influence specific early and late events in the regulated cascade of the cell division cycle. Mechanistically how this might be achieved has been speculated upon and tested to some extent here.
PRP17 is not the only second-step splicing factor that affects the cell division cycle. Prp8, Slu7, Prp16, Prp18 and Prp22 are other splicing factors whose functions are required for the second step of splicing. Amongst them, alleles of prp22 and prp16 have been uncovered recently in genetic screens for mitosis defects (17,37), and alleles of prp8 (dbf5) have been reported previously (1). The lack of mitotic spindles in the new alleles of prp16 and prp22 lead to the hypothesis that α-tubulin transcripts are targets for regulation by splicing. While the contributions of these factors to TUB1 or TUB3 splicing was not investigated, the hypothesis was supported by the rescue of spindle defects in these new strains by an intronless TUB1 gene (37). Our data from the analysis of TUB1 and TUB3 implicate these transcripts to be in vivo substrates dependent on Prp17. By examining the splicing of these transcripts in mutants for several second-step factors, we show that Prp17, Prp16 and Prp22 contribute to the efficient splicing of TUB1 and TUB3. Genetic interactions between PRP17 and PRP16 are known; high copy expression of PRP16 partially rescues the growth defect of prp17::LEU2 (12). Further, in vitro studies implicate that the second step of splicing can be divided into two events where Prp16 and Prp17 act at an early ATP-dependent step (12). Therefore, the similar effects of Prp16 and Prp17 on TUB1 and TUB3 intron splicing that we observe are not surprising. Prp18 and Slu7 are predicted to act in concert in a step subsequent to Prp16 and Prp17. Inactivation of either protein has marginal consequences on TUB1 and TUB3 splicing efficiency, with Prp18, the non-essential factor, perhaps having a somewhat greater role. Slu7 is predicted to be required for splicing of introns where the distance between the branch point and the 3′ splice site is >7 nt (28). We find here that despite TUB3 having a 135 nt distance between these cis elements and TUB1 having a 37 nt spacing feature, both of these precursors are spliced in the absence of active Slu7. The lack of dependence on Slu7 is consistent with the marginal role of Prp18, its partner in this step. Our results and predictions for the contribution of Prp18 to TUB3 splicing agree with the data obtained from genome-wide analysis of yeast mRNA processing (31). Any minor role for Prp18 in TUB3 splicing may arise from a recently described new domain in Prp18. The latter contributes to second-step splicing independent of Prp18–Slu7 association (38).
Through in vitro splicing reactions, we validate the role of Prp17 in increasing the rate of the second splicing step without being absolutely required for the reaction. Previous analyses have deduced the same through immunodepletion of Prp17 from wild-type extracts (12). It is as yet unknown if human Prp17 behaves similarly. Attempts to deplete hPrp17 from HeLa cell extracts were not successful (13). Nevertheless, hPrp17 associates with spliceosomes prior to the second step, in wild-type substrates and in artificial substrates with greater distances between the BP and 3′ss (13). The in vitro data reported here performed with extracts from the prp17 null strain irrevocably point to its auxilliary role in the second step of splicing. This is the case for at least two different transcripts, ACT1 and TUB3. By studying TUB3 intron splicing in vitro, we show that this substrate is more sensitive to the absence of Prp17. Our analysis of the TUBΔi gene in prp17-1 and prp17::LEU2 cells suggests other critical limiting components in addition to α-tubulin for progression through mitosis.
We observe a partial reduction in the first step of splicing in prp17::LEU2 extracts, particularly with the TUB3 substrate. The genetic interaction between Prp17 and factors in the Prp19-associated complex may explain this observation (4). Prp19 and associated proteins act early during spliceosome assembly, at or around the time of loading U4/U6 onto the spliceosome (39). In the absence of Prp17, this step may be inefficient. Perhaps events leading from the association of Prp19 with the spliceosome to the second-step function of Prp22 are compromised in TUB1 and TUB3 in the absence of Prp17. In fact, depletion of CEF1, a component of Prp19-associated complex (40,41), blocked both splicing reactions in vitro, and its genetic depletion causes a G2 arrest (6,42). The precise role of CEF1 or PRP17 is not clear. Collectively, our results, together with the other recent studies discussed here, suggest that multiple interactions of Prp17 with other essential splicing factors such as Prp16 or Prp22 improve the second step of the splicing reaction. Our data provide clues to the nature of the involvement of the pre-mRNA splicing apparatus in cell cycle progression.
REFERENCES
- 1.Shea J.E., Toyn,J.H. and Johnston,L.H. (1994) The budding yeast U5 snRNP Prp8 is a highly conserved protein which links RNA splicing with cell cycle progression. Nucleic Acids Res., 22, 5555–5564. [DOI] [PMC free article] [PubMed]
- 2.Lundgren K., Allan,S., Urushiyama,S., Tani,T., Oshima,Y., Frendewey,D. and Beach,D. (1996) A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell, 7, 1083–1094. [DOI] [PMC free article] [PubMed]
- 3.Seghezzi W., Chua,K., Shanahan,F., Gozani,O., Reed,R. and Lees,E. (1998) Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol. Cell. Biol., 18, 4526–4536. [DOI] [PMC free article] [PubMed]
- 4.Ben Yehuda S., Dix,I., Russell,C.S., McGarvey,M., Beggs,J.D. and Kupiec,M. (2000) Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics, 156, 1503–1517 [DOI] [PMC free article] [PubMed]
- 5.Russell C.S., Ben Yehuda,S., Dix,I., Kupiec,M. and Beggs,J.D. (2000) Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA, 6, 1565–1572. [DOI] [PMC free article] [PubMed]
- 6.Burns C.G., Ohi,R., Mehta,S., O’Toole,E.T., Winey,M., Clark,T.A., Sugnet,C.W., Ares,M.,Jr and Gould,K.L. (2002) Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol., 22, 801–815. [DOI] [PMC free article] [PubMed]
- 7.Vaisman N., Tsouladze,A., Robzyk,K., Ben Yehuda,S., Kupiec,M. and Kassir,Y. (1995) The role of Saccharomyces cerevisiae Cdc40p in DNA replication and mitotic spindle formation and/or maintenance. Mol. Gen. Genet., 247, 123–136. [DOI] [PubMed]
- 8.Boger-Nadjar E., Vaisman,N., Ben-Yehuda,S., Kassir,Y. and Kupiec,M. (1998) Efficient initiation of S-phase in yeast requires Cdc40p, a protein involved in pre-mRNA splicing. Mol. Gen. Genet., 260, 232–241. [DOI] [PubMed]
- 9.Vijayraghavan U., Company,M. and Abelson,J. (1989) Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev., 3, 1206–1216. [DOI] [PubMed]
- 10.Frank D., Patterson,B. and Guthrie,C. (1992) Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol. Cell. Biol., 12, 5197–5205. [DOI] [PMC free article] [PubMed]
- 11.Lindsey L.A., Chawla,G., Srinivasan,N., Vijayraghavan,U. and Garcia-Blanco,M.A. (2000) The carboxy terminal WD domain of the pre-mRNA splicing factor Prp17p is critical for function. RNA, 6, 1289–1305. [DOI] [PMC free article] [PubMed]
- 12.Jones M.H., Frank,D.N. and Guthrie,C. (1995) Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl Acad. Sci. USA, 92, 9687–9691. [DOI] [PMC free article] [PubMed]
- 13.Zhou Z. and Reed,R. (1998) Human homologs of yeast Prp16 and Prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J., 17, 2095–2106. [DOI] [PMC free article] [PubMed]
- 14.Lindsey L.A. and Garcia-Blanco,M.A. (1998) Functional conservation of the human homolog of the yeast pre-mRNA splicing factor Prp17p. J. Biol. Chem., 273, 32771–32775. [DOI] [PubMed]
- 15.Umen J.G. and Guthrie,C. (1995) The second catalytic step of pre-mRNA splicing. RNA, 1, 869–885. [PMC free article] [PubMed]
- 16.Seshadri V., Vaidya,V.C. and Vijayraghavan,U. (1996) Genetic studies of the PRP17 gene of Saccharomyces cerevisiae: a domain essential for function maps to a nonconserved region of the protein. Genetics, 143, 45–55. [DOI] [PMC free article] [PubMed]
- 17.Hwang L.H. and Murray,A.W. (1997) A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol. Biol. Cell, 10, 1877–1887. [DOI] [PMC free article] [PubMed]
- 18.McDonald W.H., Ohi,R., Smelkova,N., Frendewey,D. and Gould,K.L. (1999) Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol., 19, 5352–5362. [DOI] [PMC free article] [PubMed]
- 19.Lim H.H., Loy,C.J., Zaman,S. and Surana,U. (1996) Dephosphorylation of threonine 169 of Cdc28 is not required for exit from mitosis but may be necessary for start in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 4573–4583. [DOI] [PMC free article] [PubMed]
- 20.Kilmartin J.V. and Adams,A.E.M. (1984) Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol., 98, 922–933. [DOI] [PMC free article] [PubMed]
- 21.Schatz P.J., Solomon,F. and Botstein,D. (1986) Genetically essential and nonessential alpha-tubulin genes specify functionally interchangeable proteins. Mol. Cell. Biol., 6, 3722–3733. [DOI] [PMC free article] [PubMed]
- 22.O’Keefe R.T., Norman,C. and Newman,A.N. (1996) The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell, 86, 679–689. [DOI] [PubMed]
- 23.Newman A.J., Lin,R.-J., Cheng,S.C. and Abelson,J. (1985) Molecular consequences of specific intron mutations in yeast mRNA splicing in vivo and in vitro. Cell, 68, 743–754. [DOI] [PubMed]
- 24.Levine K., Tinkelenberg,A.H. and Cross,F. (1995) The CLN gene family central regulators of cell cycle START in budding yeast. Prog. Cell Cycle Res., 1, 101–104. [DOI] [PubMed]
- 25.Pringle J.R. and Hartwell,L.H. (1981) The Saccharomyces cerevisiae cell cycle. In Strathern,J.D., Jones,E.W. and Broach,J.R. (eds), The Molecular Biology of the Yeast Saccharomyces cerevisiae: Life Cycle and Inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 97–142
- 26.Dahan O. and Kupiec,M. (2002) Mutations in genes of Saccharomyces cerevisiae encoding pre-mRNA splicing factors cause cell cycle arrest through activation of the spindle checkpoint. Nucleic Acids Res., 30, 4361–4370. [DOI] [PMC free article] [PubMed]
- 27.Dagher S.F. and Fu,X.D. (2001) Evidence for a role of Sky1p-mediated phosphorylation in 3′ splice site recognition involving both Prp8 and Prp17/Slu4. RNA, 7, 1284–1297. [DOI] [PMC free article] [PubMed]
- 28.Brys A. and Schwer,B. (1996) Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA, 2, 707–717. [PMC free article] [PubMed]
- 29.Schwer B. and Gross,C.H. (1998) Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J., 17, 2086–2094. [DOI] [PMC free article] [PubMed]
- 30.Umen J.G. and Guthrie,C. (1995) A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev., 9, 855–868. [DOI] [PubMed]
- 31.Clark T.A., Sugnet,C.W. and Ares,M. (2002) Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science, 296, 2146–2152. [DOI] [PubMed]
- 32.Xu D., Field,D.J., Tang,S.J., Moris,A., Bobechko,B.P. and Friesen,J.D. (1998) Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol. Cell. Biol., 18, 2055–2066. [DOI] [PMC free article] [PubMed]
- 33.Vijayraghavan U. and Abelson,J. (1990) PRP18, a protein required for the second reaction in pre-mRNA spicing. Mol. Cell. Biol., 10, 324–232. [DOI] [PMC free article] [PubMed]
- 34.Horowitz D.S. and Abelson,J. (1993) A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of pre-mRNA splicing in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 2959–2970. [DOI] [PMC free article] [PubMed]
- 35.Stevenson L.F., Kennedy,B.K. and Harlow,E. (2001) A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl Acad. Sci. USA, 98, 3946–3951. [DOI] [PMC free article] [PubMed]
- 36.Zhu W., Rainville,I.R., Ding,M., Bolus,M., Heintz,N.H. and Pederson,D.S. (2002) Evidence that the pre-mRNA splicing factor Clf1p plays a role in DNA replication in Saccharomyces cerevisiae. Genetics, 160, 1319–1333. [DOI] [PMC free article] [PubMed]
- 37.Biggins S., Bhalla,N., Chang,A., Smith,D.L. and Murray,A.W. (2001) Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics, 159, 453–470. [DOI] [PMC free article] [PubMed]
- 38.Bacikova D. and Horowitz,D.S. (2002) Mutational analysis identifies two separable roles of the Saccharomyces cerevisiae splicing factor Prp18. RNA, 8, 1280–1293. [DOI] [PMC free article] [PubMed]
- 39.Chen C.-H., Yu,W.-C., Tsao,T.Y., Wang,L-Y., Chen,H.-R., Lin,J.-Y., Tsai,W.-Y. and Chen,S.-C. (2002) Functional and physical interactions between components of the Prp19-associated complex. Nucleic Acids Res., 30, 1029–1037. [DOI] [PMC free article] [PubMed]
- 40.Tsai W.Y., Chow,Y.T., Chen,H.R., Huang,K.T., Hong,R.I., Jan,S.P., Kuo,N.Y., Tsao,T.Y., Chen,C.H. and Cheng,S.C. (1999) Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem., 274, 9455–9462. [DOI] [PubMed]
- 41.Chen H.R., Tsao,T.Y., Chen,C.H., Her,L.S., Hsu,M.M. and Cheng,S.C. (1999) Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 5406–5411. [DOI] [PMC free article] [PubMed]
- 42.Burns C.G., Ohi,R., Krainer,A.R. and Gould,K.L. (1999) Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 13789–13794. [DOI] [PMC free article] [PubMed]