Abstract
Thus far, no transcription factor IIIA (TFIIIA) from higher plants has been cloned and characterized. We have cloned and characterized TFIIIA and ribosomal protein L5 from Arabidopsis thaliana. Primary sequence comparison revealed a high divergence of AtTFIIIA and a relatively high conservation of AtL5 when compared with other organisms. The AtTFIIIA cDNA encodes a protein with nine Cys2-His2-type zinc fingers, a 23 amino acid spacer between fingers 1 and 2, a 66 amino acid spacer between fingers 4 and 5, and a 50 amino acid non-finger C-terminal tail. Aside from the amino acids required for proper zinc finger folding, AtTFIIIA is highly divergent from other known TFIIIAs. AtTFIIIA can bind 5S rDNA, as well as 5S rRNA, and efficiently stimulates the transcription of an Arabidopsis 5S rRNA gene in vitro. AtL5 identity was confirmed by demonstrating that this protein binds to 5S rRNA but not to 5S rDNA. Protoplast transient expression assays with green fluorescent protein fusion proteins revealed that AtTFIIIA is absent from the cytoplasm and concentrated at several nuclear foci including the nucleolus. AtL5 protein accumulates in the nucleus, especially in the nucleolus, and is also present in the cytoplasm.
INTRODUCTION
Ribosome biogenesis in eukaryotic cells requires the synthesis of RNAs by all three nuclear RNA polymerases. RNA polymerase II (pol II) produces mRNAs that encode ribosomal proteins, while the 5.8S, 18S and 28S rRNAs are co-transcribed in the nucleolus by RNA polymerase I. Tran scription factor IIIA (TFIIIA) is pol III transcription factor specifically required for transcription of 5S rRNA genes. It binds to the internal control region of the 5S rRNA genes as the first step in the assembly of a transcription complex, allowing the recruitment of TFIIIC, TFIIIB and pol III (1,2).
TFIIIA has been studied extensively in Xenopus laevis where it was first isolated from oocytes (3). Analysis of TFIIIA sequences from several species including X.laevis (3) and other frog species (4,5), human (6), catfish (7), mouse, rat (8), Saccharomyces cerevisiae (9) and Schizosaccharomyces pombe (10) has revealed remarkably poor conservation of primary sequence. All known TFIIIAs, except in S.cerevisiae and S.pombe, have a similar organization: nine consecutive zinc fingers of the Cys2-His2 type, followed by a C-terminal domain of unknown structure required for the support of transcription of 5S rRNA genes in X.laevis (11). The S.cerevisiae TFIIIA protein bears an 81 amino acid spacer insertion between zinc fingers 8 and 9. In S.pombe, TFIIIA contains an unprecedent tenth zinc finger. TFIIIA has been shown to bind to the 5S rRNA in Acanthamoeba castellanii (12) and X.laevis, where it is involved in a network of interactions that couple 5S rRNA synthesis to accumulation of ribosomal proteins (13). Purified proteins containing TFIIIA activity, isolated from tulip (14) and maize (15), have also been shown to bind 5S rRNA.
Ribosomal protein L5 is also known to bind specifically to 5S rRNA and is involved in its nucleocytoplasmic transport (16–19). After transcription, 5S rRNA binds either to its own transcription factor IIIA or to ribosomal protein L5, forming 7S or 5S ribonucleoprotein particles (RNPs), respectively [reviewed in Pieler and Rudt (20)]. It has been suggested that the 5S RNP acts as a precursor to ribosome assembly by delivering 5S rRNA from the nucleoplasm to the nucleolar assembly site of 60S pre-ribosomal subunits (21).
Studies in X.laevis oocytes have shown that 5S rRNA can be exported from the nucleus to the cytoplasm, for storage (16,22). Pre-vitellogenic oocytes of amphibians and fish accumulate two major RNP particles, the 7S RNP and the 42S RNP. The 42S RNP is composed of various tRNAs and oocyte-specific 5S rRNA, along with two proteins, p50 and p43, a 5S rRNA-binding protein (23,24). In fully grown oocytes, 7S and 5S RNPs migrate out of the nucleus and accumulate in the cytoplasm (16,22). 5S rRNA must then re-enter the nucleus to ensure that the ribosome can be fully assembled (21,25). The nuclear re-entry of 5S rRNA is mediated exclusively by the ribosomal protein L5 (26–28). The most plausible interpretation for cytosolic export of 5S rRNA in X.laevis may lie in control of 5S rRNA synthesis, as suggested by the current view of the feedback regulation mechanism (13). Cytoplasmic storage sites for 5S rRNA have not been observed in mammalian somatic cells.
To date, no TFIIIA from higher plants has been cloned and characterized. We have identified, cloned and characterized TFIIIA and L5 from Arabidopsis thaliana. Primary sequence comparison revealed a high divergence of AtTFIIIA and a relatively high conservation of AtL5 compared with other organisms. As previously shown in Xenopus, we have demonstrated that AtTFIIIA can bind to 5S rDNA, as well as to 5S rRNA, the gene product. AtTFIIIA is able to stimulate the transcription of an Arabidopsis 5S rRNA gene in vitro. AtL5 identity was confirmed by showing that this protein binds to 5S rRNA but not to 5S rDNA. Protoplast transient expression assays with green fluorescent protein (GFP) fusion proteins indicate that AtL5 protein accumulates in the nucleus and in the nucleolus. AtL5 is also present in the cytoplasm, probably incorporated in the large ribosomal subunit in association with 5S rRNA. AtTFIIIA is only detected in the nucleus, with a strong accumulation in the nucleolus and at additional foci, suggesting that AtTFIIIA can be imported efficiently from the nucleoplasm into the nucleolus. We assume that the additional foci found in the nucleoplasm represent accumulation of AtTFIIIA on transcribed 5S rDNA loci or into Cajal bodies.
MATERIALS AND METHODS
Isolation of the Arabidopsis TFIIIA cDNA and purification of recombinant protein
A cDNA encoding the putative Arabidopsis TFIIIA homolog was amplified by PCR from an Arabidopsis cDNA library (29) using primers designed according to the Arabidopsis sequence database. The direct primer (5′-ATCATAGGATCCTGGCGGAAGAAGCTAAAG-3′) and reverse primer (5′-ATTACAGGATCCCTAGCAAGTTTCGTG-3′) included an BamHI restriction site (underlined). After PCR amplification and BamHI digestion, the coding sequence of TFIIIA was cloned into the pGEX-5X-1 expression vector (Amersham Biosciences). In the resulting construct named pGEX-AtTFIIIA, AtTFIIIA is fused to the C-terminal end of GST. Prior to expression in bacteria, sequencing was performed to verify the sequence of the cDNA and the translational fusion.
To express the AtTFIIIA recombinant protein, pGEX-AtTFIIIA was transformed into Escherichia coli BLR (DE3) cells. A fresh 2 ml starter culture of BLR (DE3)/pGEX-AtTFIIIA was used to inoculate 200 ml of LB containing 100 µg/ml ampicillin and 12.5 µg/ml tetracycline. The culture was grown at 37°C to an OD600 of 0.3–0.4. Recombinant protein expression was then induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) followed by an incubation of 2–3 h at 30°C. Induction was verified by SDS–PAGE analysis followed by Coomassie blue staining. Induced cells were harvested by centrifugation at 4000 g for 15 min at 4°C, and resuspended in 1 ml of ice-cold buffer A (20 mM Tris–HCl pH 8.0, 150 mM NaCl). Cell lysis was performed by addition of 100 µg/ml lysozyme, followed by a 15 min incubation at 30°C and sonication. Soluble cell extract containing AtTFIIIA was recovered by centrifugation at 12 000 g at 4°C for 30 min, and saved at 4°C for purification.
The extracts containing the GST–AtTFIIIA recombinant protein or GST were mixed with glutathione–agarose (Sigma) for 30 min at room temperature, and unbound protein was removed by three washes in 1 M NaCl and three washes in 1× phosphate-buffered saline (PBS). Subsequently, bound protein was removed by elution with 10 mM reduced glutathione pH 7.6 (Sigma). Eluates were analyzed by SDS–PAGE.
Isolation of the Arabidopsis L5 cDNA and purification of recombinant protein
As for AtTFIIIA, the cDNA encoding the Arabidopsis L5 homolog was amplified by PCR from the Arabidopsis cDNA library, using primers designed according to the L5 cDNA sequence present in the database (accession no. AY081701). The direct primer (5′-ATTCTATGAATTCTTGGTGTTTGTG-3′) and reverse primer (5′-ATTCTATGAATTCTTACTCTTCATCG-3′) included an EcoRI restriction site. After PCR amplification and EcoRI digestion, the L5 cDNA was cloned into the pGEX-5X-1 expression vector. In the resulting construct named pGEX-AtL5, AtL5 is fused to the C-terminal end of GST. Prior to expression in bacteria, sequencing was performed to check the sequence of the cDNA (accession no. AY186611) and the translational fusion.
Expression and purification of the GST–AtL5 recombinant fusion protein were as described above for AtTFIIIA, except that cells were grown at 37°C after induction with 1 mM IPTG.
DNA gel retardation assays
The 238 bp NotI fragment of pGEMT-5S containing the transcribed region and a part of the spacer sequence of an A.thaliana 5S rRNA gene was labeled by a fill-in reaction performed with 25 µCi of [α-32P]dCTP (3000 Ci/mmol), 100 µM each dATP, dGTP and dTTP, 1× Klenow buffer [10 mM Tris–HCl pH 7.9, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol (DTT)], 100 ng of the 5S DNA fragment and 25 U of Klenow fragment (Amersham). After a 30 min incubation at room temperature, the labeled fragment was purified with a PCR purification kit (Qiagen).
Recombinant proteins (see legends to figures for concentrations) were incubated with 1 µl (20 000 c.p.m., 1–3 ng, ∼1 nM) of the labeled 5S rDNA fragment and variable concentrations of unlabeled DNA in 20 µl reactions containing buffer EMSA (20 mM Tris–HCl pH 7.5, 7 mM MgCl2, 10 µM ZnCl2, 1 mM DTT, 10% glycerol, 70 mM KCl) supplemented with 10 µg/ml poly(dI–dC) and 100 µg/ml bovine serum albumin (BSA). Unlabeled DNA referred to as ‘specific’ is the 238 bp NotI fragment of pGEMT-5S, while ‘non-specific’ refers to a 490 bp EcoRI fragment from the plasmid pGEMT-APT1 that contains the cDNA of the adenine phosphoribosyltransferase (APT1) gene (30). The reaction mixtures were incubated at 25°C for 30 min and then quickly cooled on ice before addition of 3 µl of loading buffer [50% (v/v) glycerol, 1 mg/ml bromophenol blue]. The samples were loaded onto an 8% polyacrylamide gel containing 5% glycerol in 25 mM Tris–HCl pH 8, 200 mM glycine. Prior to loading, the gels were pre-run at 50 V for 30 min. After loading, electrophoresis was continued for an additional 2 h at 140 V at room temperature. The gels were dried, and visualized on a PhosphorImager.
RNA gel retardation assays
The transcribed sequence of an A.thaliana 5S rRNA gene was fused to a T7 promoter by PCR. Labeled 5S rRNA was then synthesized in vitro using the SP6/T7 transcription kit (Roche), following the manufacturer’s instructions. Non-specific RNA was generated similarly using the first 120 bp of the APT1 cDNA fused to a T7 promoter. Transcripts were separated on an 8% acrylamide gel containing 7 M urea, and RNAs were eluted from the gel by an overnight incubation at 37°C in elution buffer (0.5 M ammonium acetate, 1 mM EDTA, 0.2% SDS). After ethanol precipitation, RNAs were resuspended in diethylpyrocarbonate (DEPC)-treated water. Before use in the gel retardation assays, the RNA probes were incubated for 10 min at 65°C in renaturation buffer (50 mM Tris–HCl pH 8.0, 50 mM KCl, 5 mM MgCl2) and allowed to cool slowly to room temperature. RNA gel retardation assays were then performed according to the procedure described above for DNA gel retardation.
DNase I footprinting assay
For footprinting of the template strand, 5′-labeled 5S rDNA was generated by PCR using primers OLIGO5 (5′-TATATACGATGGCATTGCATATAC-3′) and 32P-labeled 1037PE-rev ([32P]5′-GGAAACAGCTATGACCATGGAGGGATGCAACACGAGGAC-3′) from a 5S rDNA transcribed unit (1037). For footprinting of the RNA-like strand, 5′-labeled OLIGO5 primer was used instead of labeled 1037PE-rev. Binding was performed in a 50 µl volume containing 30 mM HEPES-KOH pH 7.9, 3 mM MgSO4, 80 mM KOAc, 0.1 mM EDTA, 2 mM DTT, 10% glycerol, 0.8 µg of poly(dI–dC)·poly(dI–dC), 0.15 pmol of labeled 168 bp double-stranded DNA including the –28 to +120 region of 5S rDNA, and ∼400 ng of recombinant protein (GST or GST–AtTFIIIA). After incubation for 15 min at room temperature, 5 µl of Ca2+/Mg2+ solution (5 mM CaCl2 and 10 mM MgCl2) was added, and then 0.33 U of DNase I was added to the mixture and incubated for just 1 min at room temperature. After phenol/chloroform extraction and ethanol precipitation, the 32P-labeled fragments were separated on an 8% polyacrylamide gel containing 7 M urea and TBE. Radioactivity was detected using a Bio-Imaging Analyzer BAS-2000 II (Fuji Photo Film, Japan).
In vitro transcription assays
In vitro transcription reactions from Arabidopsis 5S rDNA in tobacco nuclear extracts were done as previously described (31) with minor modifications. Briefly, the reaction was performed in a 20 µl volume containing 30 mM HEPES-KOH pH 7.9, 3 mM MgSO4, 80 mM KOAc, 0.1 mM EGTA, 2 mM DTT, 10% glycerol, 0.5 mM each of ATP, CTP, UTP, 25 mM GTP, 120 kBq of [α-32P]GTP, 0.2 pmol of circular plasmid containing 5S rDNA, 0.5 µg/ml α-amanitin and ∼50 µg of tobacco nuclear extract. After adding ∼50, 100 and 200 ng each of recombinant protein (GST–AtTFIIIA or GST), the mixture was kept on ice for 10 min, then tobacco nuclear extract was added and incubated on ice for 10 min. The reaction was initiated by addition of NTPs and then incubated at 28°C for 90 min. The 32P-labeled RNA was extracted with phenol/chloroform and chloroform treatment, and precipitated with ethanol. The extracted RNA was separated on an 8% polyacrylamide gel containing 7 M urea and TBE. Radioactivity was detected and measured using BAS-2000 II.
Protoplast transient expression assay with GFP fusion proteins
The NcoI-AtTFIIIA cDNA was PCR amplified using the primers (5′-ATTCTATGAATCTTGGTGTTTGTG-3′) and (5′-ATTCTATGAATTCTTACTCTTCATCG-3′) and inserted at the NcoI restriction site of the GFP fusion vector pAVA393 (32) containing a cytosolic derivative of the GFP5 cDNA (33) driven by the constitutive 35S cauliflower mosaic virus (CaMV) promoter. In the resulting construct, AtTFIIIA is fused to the N-terminal region of the GFP5. Similarly, the AtL5 cDNA was amplified by PCR using the primers (5′-ATTCTATCCATGGTGTTTGTGAAG-3′) and (5′-AATCTATCCATGGACTCTTCATCG-3′) including NcoI restriction sites, and cloned into pAVA393. The resulting constructs, named pAVA-AtTFIIIA and pAVA-AtL5, respectively, were verified by sequencing.
Protoplasts were prepared from Arabidopsis cultured cells and transformed as described (34) with minor modifications (C.I.White, unpublished). Transformations were performed with 50 µg of plasmids (pAVA393, pAVA-AtTFIIIA or pAVA-AtL5) purified using the Plasmid midiprep Kit (Qiagen). Protoplasts were observed 30 h after transformation using a ZEISS Axioplan 2 microscope.
Results
Amplification and cloning of the putative Arabidopsis TFIIIA and L5 cDNAs
Searching Cys2-His2-type multi-zinc finger proteins in the Arabidopsis database resulted in the identification of only one predicted protein containing nine zinc fingers (accession no. AAG51140). As the presence of nine zinc fingers is the only common feature between TFIIIA proteins characterized to date, we considered this protein to be a likely candidate to be Arabidopsis TFIIIA (named AtTFIIIA). The open reading frame encoding this protein (accession no. AC069273) is carried by the BAC clone F28P5 mapped on chromosome 1. A cDNA encoding the putative AtTFIIIA protein was amplified by PCR from a cDNA library using primers designed according to the sequence of the predicted cDNA. The sequence of the obtained cDNA (accession no. AY186610) revealed that the predicted second exon was 66 bp too long in its 3′ end. Furthermore, predicted exons 5 and 6 were 20 and 43 bp too short in their 3′ and 5′ end, respectively. The remainder of the coding sequence was identical to the database prediction.
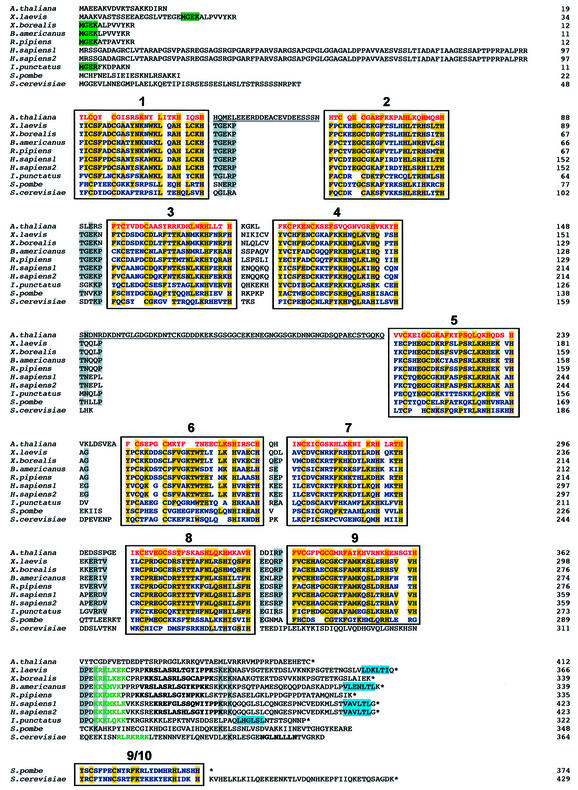
The 412 amino acid encoded protein contains nine Cys2-His2-type zinc fingers as well as 19 amino acid N-terminal and 50 amino acid C-terminal non-zinc finger tails. The nine zinc finger sequences comply with the F/I-X-C-X(2–4)-C-X3-F-X1-K-X(2–3)-L-X2-H-X(3–5)-H consensus. Sequence similarity between AtTFIIIA and any other known TFIIIA is dominated by the seven residues found in each zinc finger that are required for proper folding of the protein. These include three hydrophobic residues: F or I in Arabidopsis (but F or Y in other species), F, L and two Zn2+-coordinating cysteines and histidines. Excluding these conserved residues, the overall identity of AtTFIIIA to known TFIIIAs is low. For instance, there is only 26% sequence identity between A.thaliana and X.laevis, and 17% sequence identity between A.thaliana and S.cerevisiae. Alignment of TFIIIA sequences reveals that AtTFIIIA is organized differently from its homologs (Fig. 1). AtTFIIIA contains two unique 23 and 66 amino acid spacers located between zinc fingers 1 and 2, and 4 and 5, respectively. Amongst known TFIIIAs, such a long spacer sequence (81 amino acids) exists between fingers 8 and 9 in S.cerevisiae TFIIIA. The S.pombe TFIIIA carries a unique tenth zinc finger which has no equivalent in other organisms. Several sequence motifs have been defined in the different TFIIIA sequences, but none of them could be found either in the N- and C-terminus tails, or in the spacers of the AtTFIIIA protein sequence. We searched for the N-terminal MGEK motif characteristic of the smaller oocyte form of X.laevis TFIIIA (3–5,35), the nuclear localization signal (NLS) (7,8), the transcription-activating signal (TAS) (7,11,36) and the nuclear export signal (NES) (7,37) found in some of the reported TFIIIA sequences (Fig. 1), but could not identify any of them in the terminal tails nor in the spacer sequences of AtTFIIIA.
Figure 1.
Primary sequence alignment of known TFIIIAs with Arabidopsis TFIIIA. Sequences were aligned manually to match each of the nine zinc fingers of AtTFIIIA with the corresponding finger of the other TFIIIAs. Zinc fingers are framed and numbered. Note that the ninth finger from S.cerevisiae was aligned with finger 10 from S.pombe only for illustration. Non-aligned regions between fingers 1 and 2, and 4 and 5 in the AtTFIIIA sequence are underlined. AtTFIIIA zinc fingers are in red, and zinc fingers of other organisms are shown in blue. Conserved residues (in >50% of the sequences) in the zinc fingers or in non-finger regions are highlighted in yellow or gray, respectively. The oocyte motif MGEK/R and the NES motifs are highlighted in green and blue, respectively. NLS motifs are written in green characters, and TAS motifs are indicated in black bold type. Asterisk marks the end of the sequence. The residue number is shown to the right of each line of sequence.
The cDNA encoding the putative Arabidopsis 5S rRNA-binding ribosomal protein L5 (AtL5) was PCR amplified from the cDNA library, using primers designed according to the sequence of the cDNA present in the database. Unlike AtTFIIIA, the primary sequence of the putative AtL5 protein revealed a high degree of conservation (∼54% identity between Arabidopsis and human, see Fig. 2).
Figure 2.
Primary sequence alignment of AtL5 with some known L5s. Sequences were aligned using the CLUSTALW program. Identical residues (in 100% of the sequences) are highlighted in black, and conserved residues (in >50% of the sequences) are highlighted in gray. The residue number is shown to the right of each line of sequence.
Sequence-specific DNA-binding activity
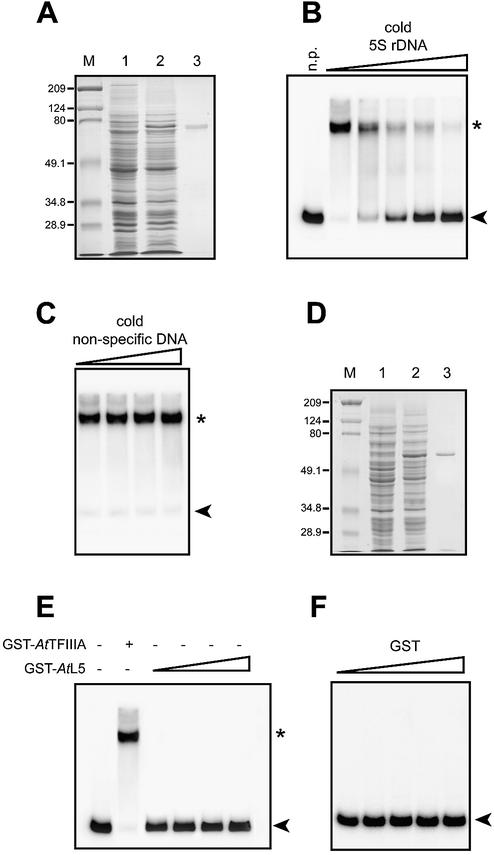
To investigate the biochemical properties of the putative AtTFIIIA and AtL5 proteins, the cloned cDNAs were used for expression of the proteins in the E.coli BLR(DE3) strain. Two recombinant GST fusion proteins (GST–AtTFIIIA and GST–AtL5) were purified (Fig. 3A and D).
Figure 3.
DNA binding assays of GST–AtTFIIIA and GST–AtL5 recombinant proteins. (A and D) SDS–PAGE of GST–AtTFIIIA and GST–AtL5 recombinant proteins, respectively. Shown are results for uninduced E.coli BLR (DE3) (lane 1), IPTG-induced cells (lane 2) and purified recombinant proteins (lane 3). Proteins were visualized by staining with Coomassie brilliant blue. The sizes (in kDa) of molecular mass markers run in lane M are indicated on the left. Increasing concentrations (1–170 nM) of unlabeled 5S rDNA (B) or non-specific competitor DNA (C) were added to binding reactions including labeled 5S rDNA and GST–AtTFIIIA (2.5 ng/µl). Binding reactions were performed with increasing concentrations (2.5– 10 ng/µl) of either AtL5 (E) or GST alone (F). Arrowheads indicate free (unbound) probe, and protein–DNA complexes are indicated by an asterisk. n.p., no protein.
One property of TFIIIA is to bind specifically to the 5S rRNA gene. Gel retardation assays demonstrate that the recombinant protein indeed possesses 5S rDNA-binding activity, although the GST protein alone does not (Fig. 3B, C and F). Competition experiments revealed that the DNA-binding activity of the AtTFIIIA recombinant protein is 5S sequence specific (Fig. 3B and C). As expected, GST–AtL5 protein does not bind to the 5S rRNA gene (Fig. 3E). The equilibrium binding constant (Kd) for the interaction between AtTFIIIA and 5S rDNA, reflecting the affinity of AtTFIIIA for the 5S rRNA gene, was determined using a gel mobility shift assay as described previously (10). We measured a Kd of 0.33 nM (SE ±0.05) for the interaction between AtTFIIIA and 5S rDNA. This value is comparable with those reported previously for other known TFIIIAs (10,38,39).
DNase I footprinting
To analyze further the binding of AtTFIIIA to the 5S rRNA gene, DNase I footprinting was performed on both the template and the RNA-like strands (Fig. 4), using recombinant GST–AtTFIIIA and GST as a control. The protection pattern of the AtTFIIIA protein extends from position +43 to +97 on the template strand and from +45 to +103 on the RNA-like strand. Unprotected regions are found from +63 to +76 on the template strand and from +71 to +76 on the RNA-like strand. This protection pattern is similar to those observed for X.laevis [+47 to +96; (40)], and recently reported for A.castellanii [+44 to +97; (12)] and S.pombe [+45 to +95; (10)]. However, we did not observe any DNase I-hypersensitive site around position +63 as described for these known TFIIIA proteins (10,12,40). As expected, GST alone does not produce any protection along the 5S gene.
Figure 4.
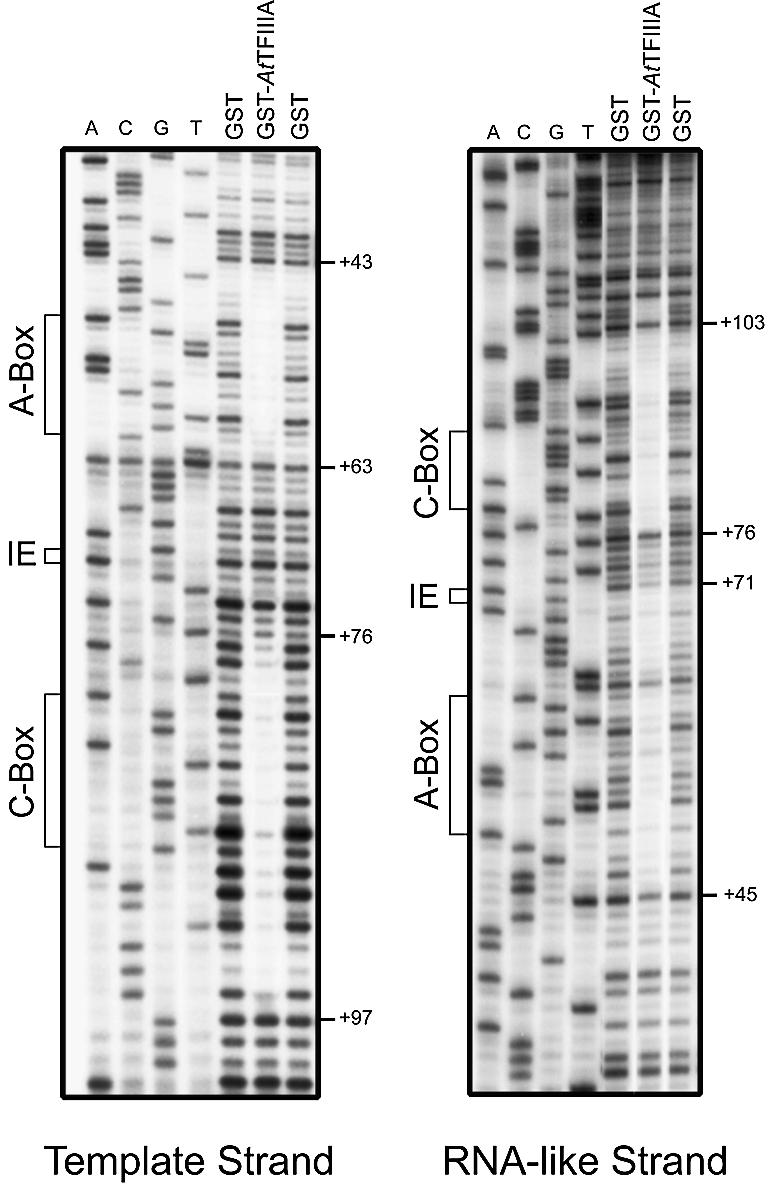
DNase I footprint of AtTFIIIA. DNase I cleavage of each strand is presented. Numbers to the right of each gel indicate the position of DNase I cleavage relative to the start site of transcription (+1). The positions of the A-box, the intermediate element (IE) and the C-box are shown to the left of each gel.
5S rRNA-binding activity of putative AtTFIIIA and AtL5
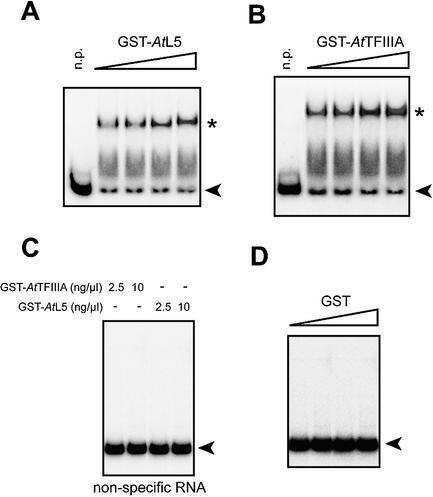
Another characteristic of TFIIIA is to bind not only to the 5S rRNA gene, but also to 5S rRNA, the gene product. Similarly, the L5 ribosomal protein can bind to 5S rRNA, forming the 5S RNP. To investigate this common property between TFIIIA and L5 on the putative AtTFIIIA and AtL5 proteins, we performed gel retardation assays using an in vitro transcribed 5S rRNA. Incubation of increasing concentrations of GST–AtTFIIIA or GST–AtL5 protein with 5S rRNA resulted in the appearance of a 5S rRNA–protein complex with slower mobility in non-denaturing gel electrophoresis (Fig. 5A and B). Control experiments showed that GST alone does not bind to 5S rRNA (Fig. 5D) and that neither GST–AtL5 nor GST–AtTFIIIA bind to a non-specific RNA (Fig. 5C).
Figure 5.
5S rRNA binding analysis of GST–AtTFIIIA and GST–AtL5 proteins. A constant amount of labeled 5S rRNA probe synthesized in vitro was incubated with increasing concentrations (2.5–10 ng/µl) of GST–AtL5 (A), GST–AtTFIIIA (B) or GST (D) proteins and then subjected to gel mobility shift analysis. As a control, GST–AtTFIIIA and GST–AtL5 proteins were incubated with non-specific RNA (C). Arrowheads indicate free (unbound) probe, and protein–RNA complexes are indicated by an asterisk. n.p., no protein.
Taken together, these results demonstrate that the putative AtTFIIIA protein indeed possesses a 5S rRNA-binding activity and that AtL5 is the actual Arabidopsis homolog of the ribosomal L5 protein.
Transcriptional activity of the putative AtTFIIIA protein
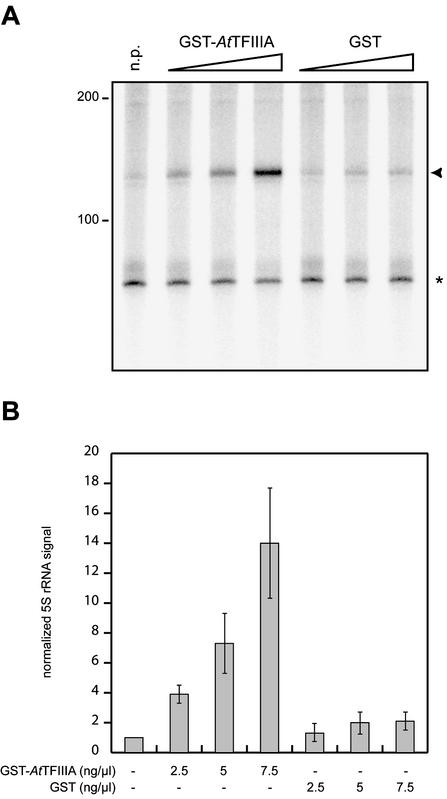
To test the ability of putative AtTFIIIA to support transcription of the Arabidopsis 5S rRNA gene, we used an in vitro transcription system from tobacco cells (41,42). This system can transcribe an Arabidopsis 5S rRNA gene without added recombinant AtTFIIIA (Fig. 6A, first lane) (31). Transcription of the 5S rRNA gene was dramatically stimulated by the addition of recombinant GST–AtTFIIIA, while not significantly modified by the addition of GST alone (Fig. 6A). Addition of GST–AtTFIIIA did not stimulate the transcription of a tRNA gene (data not shown). The 5S rRNA signal normalized to the endogenous tRNA signal was quantified and shows a 14-fold increase upon addition of 7.5 ng/µl of GST–AtTFIIIA (Fig. 6B).
Figure 6.
In vitro transcription assays. (A) Reactions were performed without added recombinant protein (n.p.) or with increasing concentrations (2.5–7.5 ng/µl) of GST–AtTFIIIA or GST. (B) Quantification and graphical representation of the data from several independent experiments similar to that shown in (A). The asterisk and arrowhead indicate endogenous tRNA and 5S rRNA, respectively. Molecular sizes are indicated (in nucleotides) next to the gel.
Taken together with 5S rDNA- and 5S rRNA-binding activities, the ability of putative AtTFIIIA to stimulate 5S rRNA gene transcription efficiently in vitro clearly demonstrates that the protein we have characterized is indeed A.thaliana TFIIIA.
Cellular localization of AtTFIIIA and AtL5 proteins
To investigate the cellular localization of the AtTFIIIA and AtL5 proteins, translational fusions of AtTFIIIA and AtL5 with an enhanced version of GFP (mGFP5) were made. Each construct was then transiently transfected to Arabidopsis cells protoplasts and monitored for GFP expression 30 h post-transfection.
As previously reported (43,44), and expected due to its small size, expression of GFP alone resulted in cytoplasmic and nuclear signals, the nucleolus being clearly devoid of signal (Fig. 7A and B). AtL5–GFP protein accumulates in the nucleus with significant nucleolar enrichment. A faint signal was observed in the cytoplasm, indicating that the protein is also present in this cellular compartment (Fig. 7C and D). This result is in good agreement with previous observations in somatic mammalian cells (21).
Figure 7.
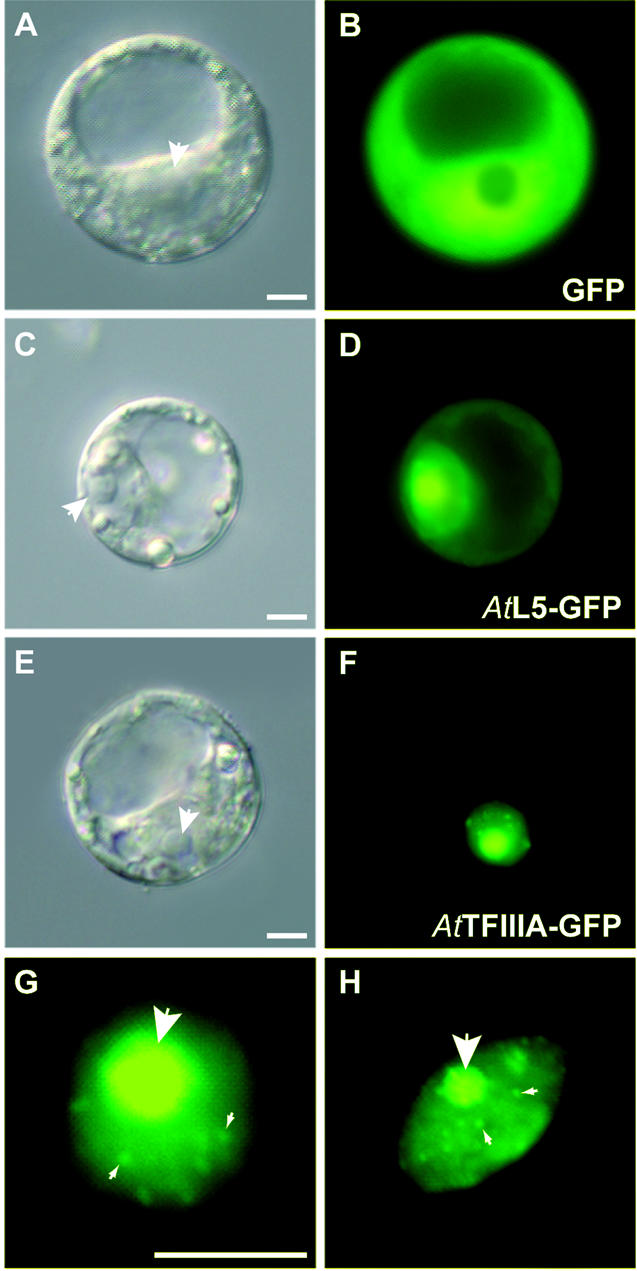
Cellular localization of AtTFIIIA and AtL5 proteins. Protoplasts were transformed with constructs expressing the proteins indicated and were observed 30 h post-transfection (B, D and F). Corresponding differential interference contrast (DIC) images are shown in (A), (C) and (E), respectively. White arrowheads indicate the nucleolus. (G and H) Close-up views of two protoplasts nuclei expressing the GFP–AtTFIIIA fusion protein. Small arrowheads indicate some of the bright nuclear foci present in addition to the nucleolus (large arrowhead). Scale bar (A–H) 10 µm.
In contrast, expression of the AtTFIIIA–GFP protein resulted in complete absence of fluorescence in the cytoplasm (Fig. 7E and F), suggesting the presence of a functional NLS in AtTFIIIA although not identified by sequence comparison (see Fig. 1). The nuclear localization of AtTFIIIA was heterogeneous, with a higher accumulation of the fusion protein in the nucleolus together with additional foci (Fig. 7F–H). As for AtL5, we conclude that AtTFIIIA is imported efficiently from the nucleoplasm to the nucleolus.
DISCUSSION
We report here the first cloning and characterization of TFIIIA from a higher plant, A.thaliana. We have shown that the protein has a specific 5S rDNA-binding activity together with a 5S rRNA-binding property as previously reported for X.laevis and A.castellanii TFIIIAs (12,45). Moreover, AtTFIIIA can stimulate the transcription of an Arabidopsis 5S rRNA gene efficiently in vitro.
In the Arabidopsis database, only predicted TFIIIA cDNA was present. AtTFIIIA cDNA escaped sequencing programs probably because of the low content of AtTFIIIA in somatic cells. Indeed, TFIIIA was first purified from X.laevis oocytes which were shown to contain up to 1012 molecules per cell (3). For comparison, somatic A.castellanii and HeLa cells were estimated to contain only 170 and 400 molecules of TFIIIA, respectively (12,46). Rat TFIIIA was purified from breast tumor, and tumoral cells are known to contain enhanced pol III transcription [for a review see Brown et al. (47)]. The reason for our success in the amplification of AtTFIIIA cDNA could lie in the fact that we used a cDNA library from metabolically active Arabidopsis cells.
AtTFIIIA bears nine Cys2-His2-type zinc fingers including the conserved residues required for proper folding and specific 5S rDNA binding. Irregular spacing between these residues in the zinc fingers, C-X(2–4)-C-X(11–12)-H-X(3–5)-H, may play a role in the alignment of AtTFIIIA zinc fingers along the 5S rRNA gene. The overall sequence identity between AtTFIIIA and yeast or vertebrate homologs ranges between 17 and 26%. Thus the divergence is too important to detect any convincing pattern of higher similarity between Arabidopsis and mammals, amphibians or yeasts. Apart from zinc fingers, known TFIIIAs contain several sequence motifs. In the non-finger N-terminal region, the mammalian, yeast and Arabidopsis TFIIIAs lack the conserved MGEK/R motif characteristic of the smaller oocyte form of TFIIIA from amphibians, also found in catfish TFIIIA purified from immature ovarian tissue (7). We assume that Arabidopsis does not synthesize an oocyte form of TFIIIA as already proposed for mammals (8). Vertebrate TFIIIAs contain a variable length (49–68 amino acids) non-zinc finger region at the C-terminus, which contains a TAS, approximately 25 amino acids long, in amphibians (5,11) and mammals (8). In S.cerevisiae, the 81 amino acid spacer located between fingers 8 and 9 exhibits a leucine-rich oligopeptide required for transcription (48), which differs in sequence from the amphibian or mammalian TAS motifs. In Arabidopsis, the TAS domain could lie in the 66 amino acid spacer between fingers 4 and 5 or, alternatively, in the 50 amino acid long C-terminal tail. We could not find the NLS consensus sequence (KKKM/LKXK) present in the C-terminal region of vertebrates TFIIIAs (8) nor the RLRKRRK NLS found in S.cerevisiae (48). Nonetheless, AtTFIIIA–GFP fusion experiments have clearly revealed a nuclear accumulation and an absence of the protein in the cytoplasm of Arabidopsis protoplasts, indicating the presence of a functional NLS in AtTFIIIA which remains to be identified. Finally, a NES (LXXLTI) has been identified and functionally tested in amphibians (37). A similar motif, based on sequence comparison, has been described in catfish (LXXLSL) (7) and humans (VAVLTL) (8). No similar sequence motif could be found in AtTFIIIA primary sequence and, accordingly, AtTFIIIA–GFP fusion protein does not localize in the cytoplasm of transformed protoplasts.
The nuclear export of 5S rRNA by TFIIIA for subsequent accumulation at distinct cytoplasmic storage sites has only been reported in Xenopus oocytes (16,22). Pre-vitellogenic oocytes store 5S rRNA in the cytoplasm as either 7S (5S rRNA–TFIIIA complex) or 42S RNPs. In the 42S RNP particle, the p43 protein binds to the 5S rRNA (23,24). Mature oocytes represent a particular cell type where TFIIIA accumulates to 1012 molecules per cell. TFIIIA or L5 protein binds to 5S rRNA, and each of these two RNPs migrates out of the nucleus and accumulates in the cytoplasm, prior to development. Cytoplasmic storage sites for 5S rRNA have not been observed in somatic mammalian cells (19). This cytoplasmic phase of the 5S RNA biosynthetic pathway is probably unique to oocytes and does not occur in somatic cells (18).
We show here that the Arabidopsis ribosomal protein L5 binds to the 5S rRNA and accumulates in the nucleolus. Immediately after transcription, 5S RNA is transiently associated with the La protein which, amongst other things, functions in transcription termination of pol III transcripts (49,50). After association with La, 5S RNA is bound by ribosomal protein L5 to form a 5S rRNP particle. Then the 5S RNP migrates to the nucleolus to participate in large ribosomal subunit assembly. L5 accumulates in the nucleolus at a concentration which greatly exceeds that of assembling ribosomal subunits (18). Rosorius et al. (19) have shown that the binding of L5 protein to 5S rRNA correlates with its ability to accumulate in the nucleolus, as previously demonstrated for the nucleolin protein (51). After nucleolar localization, the 5S RNP becomes incorporated into large ribosomal subunits and is then exported from the nucleus to the cytoplasm. In Arabidopsis protoplasts, the AtL5–GFP fusion protein localizes predominantly to the nucleolus and to a lesser extent to the nucleoplasm, and exhibits a faint homogenous staining pattern in the cytoplasm which probably reveals the protein incorporated into ribosomes. This pattern is in good agreement with previous observations made in monkey and human cells using an anti-5S RNP antibody (21).
In Arabidopsis protoplasts, the AtTFIIIA–GFP fusion protein was concentrated in the nucleolus and at several nuclear foci. Since 5S rDNA transcription occurs in the nucleoplasm and AtTFIIIA specifically binds to 5S rDNA, these nuclear foci could correspond to the transcribed 5S rDNA loci. In the germinal vesicle of Xenopus oocytes, it was reported previously, using polyclonal antibodies, that TFIIIA localizes in nuclear organelles, called Cajal bodies, which were assumed to be the primary site for assembly of the transcription machinery of the nucleus (52,53). Hence, we hypothesize that the fluorescent foci observed along with the nucleolus could also correspond to Cajal bodies where AtTFIIIA would accumulate and be incorporated in the pol III transcription machinery before delivery to the chromosomal sites of 5S rDNA transcription. In germinal vesicle of Xenopus oocytes, TFIIIA was only detected in Cajal bodies but not in nucleoli (52,53). To our knowledge, our results describe for the first time the localization of TFIIIA in somatic cells and its presence in the nucleolus. The reason for the nucleolar accumulation of AtTFIIIA remains unclear because nucleoplasm–nucleolus shuttling of 5S rRNA is known to be mediated by L5. However, as AtTFIIIA binds 5S rRNA, this nucleoplasm–nucleolus trafficking could occur as 7S RNP.
A model in which a network of nucleic acid–protein interactions, involving TFIIIA, L5, 5S rRNA and 5S rDNA, regulates 5S rRNA synthesis has been proposed (13). In this model, formation of 7S RNP regulates 5S rRNA synthesis because it lowers the amount of free TFIIIA available for 5S rDNA transcription. In Xenopus oocytes, 7S RNPs are sequestered in the cytoplasmic compartment. In the light of our results, we formulate the hypothesis that in somatic cells, 7S RNPs are stored in the nucleolus.
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. G. von Arnim for providing GFP fusion plasmids, J. Y. Bleuyard for helpful advice with protoplasts transformation, C. Cuvillier for technical assistance, Dr J. Obokata for helpful discussions, and C. Bousquet-Antonelli for critical reading of the manuscript. This work was supported by the CNRS and by the Université Blaise Pascal. O.M. is the recipient of a fellowship from the Ministère de l’enseignement supérieur et de la recherche.
DDBJ/EMBL/GenBank accession nos+AY186610, AC069273, AAG51140, AY081701, AY186611
REFERENCES
- 1.Bieker J.J., Martin,P.L. and Roeder,R.G. (1985) Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell, 40, 119–127. [DOI] [PubMed] [Google Scholar]
- 2.Setzer D.R. and Brown,D.D. (1985) Formation and stability of the 5 S RNA transcription complex. J. Biol. Chem., 260, 2483–2492. [PubMed] [Google Scholar]
- 3.Ginsberg A.M., King,B.O. and Roeder,R.G. (1984) Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell, 39, 479–489. [DOI] [PubMed] [Google Scholar]
- 4.Gaskins C.J. and Hanas,J.S. (1990) Sequence variation in transcription factor IIIA. Nucleic Acids Res., 18, 2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaskins C.J., Smith,J.F., Ogilvie,M.K. and Hanas,J.S. (1992) Comparison of the sequence and structure of transcription factor IIIA from Bufo americanus and Rana pipiens. Gene, 120, 197–206. [DOI] [PubMed] [Google Scholar]
- 6.Drew P.D., Nagle,J.W., Canning,R.D., Ozato,K., Biddison,W.E. and Becker,K.G. (1995) Cloning and expression analysis of a human cDNA homologous to Xenopus TFIIIA. Gene, 159, 215–218. [DOI] [PubMed] [Google Scholar]
- 7.Ogilvie M.K. and Hanas,J.S. (1997) Molecular biology of vertebrate transcription factor IIIA: cloning and characterization of TFIIIA from channel catfish oocytes. Gene, 203, 103–112. [DOI] [PubMed] [Google Scholar]
- 8.Hanas J.S., Hocker,J.R., Cheng,Y.G., Lerner,M.R., Brackett,D.J., Lightfoot,S.A., Hanas,R.J., Madhusudhan,K.T. and Moreland,R.J. (2002) cDNA cloning, DNA binding and evolution of mammalian transcription factor IIIA. Gene, 282, 43–52. [DOI] [PubMed] [Google Scholar]
- 9.Archambault J., Milne,C.A., Schappert,K.T., Baum,B., Friesen,J.D. and Segall,J. (1992) The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J. Biol. Chem., 267, 3282–3288. [PubMed] [Google Scholar]
- 10.Schulman D.B. and Setzer,D.R. (2002) Identification and characterization of transcription factor IIIA from Schizosaccharomyces pombe. Nucleic Acids Res., 30, 2772–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao X. and Darby,M.K. (1993) A position-dependent transcription-activating domain in TFIIIA. Mol. Cell. Biol., 13, 7496–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polakowski N. and Paule,M.R. (2002) Purification and characterization of transcription factor IIIA from Acanthamoeba castellanii. Nucleic Acids Res., 30, 1977–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pittman R.H., Andrews,M.T. and Setzer,D.R. (1999) A feedback loop coupling 5S rRNA synthesis to accumulation of a ribosomal protein. J. Biol. Chem., 274, 33198–33201. [DOI] [PubMed] [Google Scholar]
- 14.Wyszko E. and Barciszewska,M. (1997) Purification and characterization of transcription factor IIIA from higher plants. Eur. J. Biochem., 249, 107–112. [DOI] [PubMed] [Google Scholar]
- 15.Wyszko E., Radlowski,M., Bartkowiak,S. and Barciszewska,M.Z. (1997) Maize TF IIIA—the first transcription factor IIIA from monocotyledons. Purification and properties. Acta Biochim. Pol., 44, 579–589. [PubMed] [Google Scholar]
- 16.Guddat U., Bakken,A.H. and Pieler,T. (1990) Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell, 60, 619–628. [DOI] [PubMed] [Google Scholar]
- 17.Deshmukh M., Tsay,Y.F., Paulovich,A.G. and Woolford,J.L.,Jr (1993) Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol., 13, 2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael W.M. and Dreyfuss,G. (1996) Distinct domains in ribosomal protein L5 mediate 5S rRNA binding and nucleolar localization. J. Biol. Chem., 271, 11571–11574. [DOI] [PubMed] [Google Scholar]
- 19.Rosorius O., Fries,B., Stauber,R.H., Hirschmann,N., Bevec,D. and Hauber,J. (2000) Human ribosomal protein L5 contains defined nuclear localization and export signals. J. Biol. Chem., 275, 12061–12068. [DOI] [PubMed] [Google Scholar]
- 20.Pieler T. and Rudt,F. (1997) Nucleocytoplasmic transport of 5S ribosomal RNA. Semin. Cell Dev. Biol., 8, 79–82. [DOI] [PubMed] [Google Scholar]
- 21.Steitz J.A., Berg,C., Hendrick,J.P., La Branche-Chabot,H., Metspalu,A., Rinke,J. and Yario,T. (1988) A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol., 106, 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allison L.A., North,M.T. and Neville,L.A. (1995) Differential binding of oocyte-type and somatic-type 5S rRNA to TFIIIA and ribosomal protein L5 in Xenopus oocytes: specialization for storage versus mobilization. Dev. Biol., 168, 284–295. [DOI] [PubMed] [Google Scholar]
- 23.Picard B., le Maire,M., Wegnez,M. and Denis,H. (1980) Biochemical research on oogenesis. Composition of the 42-S storage particles of Xenopus laevis oocytes. Eur. J. Biochem., 109, 359–368. [DOI] [PubMed] [Google Scholar]
- 24.Joho K.E., Darby,M.K., Crawford,E.T. and Brown,D.D. (1990) A finger protein structurally similar to TFIIIA that binds exclusively to 5S RNA in Xenopus. Cell, 61, 293–300. [DOI] [PubMed] [Google Scholar]
- 25.Dechampesme A.M., Koroleva,O., Leger-Silvestre,I., Gas,N. and Camier,S. (1999) Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J. Cell Biol., 145, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allison L.A., North,M.T., Murdoch,K.J., Romaniuk,P.J., Deschamps,S. and le Maire,M. (1993) Structural requirements of 5S rRNA for nuclear transport, 7S ribonucleoprotein particle assembly and 60S ribosomal subunit assembly in Xenopus oocytes. Mol. Cell. Biol., 13, 6819–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudt F. and Pieler,T. (1996) Cytoplasmic retention and nuclear import of 5S ribosomal RNA containing RNPs. EMBO J., 15, 1383–1391. [PMC free article] [PubMed] [Google Scholar]
- 28.Murdoch K.J. and Allison,L.A. (1996) A role for ribosomal protein L5 in the nuclear import of 5S rRNA in Xenopus oocytes. Exp. Cell Res., 227, 332–343. [DOI] [PubMed] [Google Scholar]
- 29.De Veylder L., de Almeida Engler,J., Burssens,S., Manevski,A., Lescure,B., Van Montagu,M., Engler,G. and Inze,D. (1999) A new D-type cyclin of Arabidopsis thaliana expressed during lateral root primordia formation. Planta, 208, 453–462. [DOI] [PubMed] [Google Scholar]
- 30.Moffatt B.A., McWhinnie,E.A., Agarwal,S.K. and Schaff,D.A. (1994) The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene, 143, 211–216. [DOI] [PubMed] [Google Scholar]
- 31.Mathieu O., Yukawa,Y., Sugiura,M., Picard,G. and Tourmente,S. (2002) 5S rRNA genes expression is not inhibited by DNA methylation in Arabidopsis. Plant J., 29, 313–323. [DOI] [PubMed] [Google Scholar]
- 32.von Arnim A.G., Deng,X.W. and Stacey,M.G. (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene, 221, 35–43. [DOI] [PubMed] [Google Scholar]
- 33.Siemering K.R., Golbik,R., Sever,R. and Haseloff,J. (1996) Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol., 6, 1653–1663. [DOI] [PubMed] [Google Scholar]
- 34.Mathur J., Koncz,C. and Szabados,L. (1995) A simple method for isolation, liquid culture, transformation and regeneration of Arabidopsis thaliana protoplasts. Plant Cell Rep., 14, 221–226. [DOI] [PubMed] [Google Scholar]
- 35.Kim S.H., Darby,M.K., Joho,K.E. and Brown,D.D. (1990) The characterization of the TFIIIA synthesized in somatic cells of Xenopus laevis. Genes Dev., 4, 1602–1610. [DOI] [PubMed] [Google Scholar]
- 36.Vrana K.E., Churchill,M.E., Tullius,T.D. and Brown,D.D. (1988) Mapping functional regions of transcription factor TFIIIA. Mol. Cell. Biol., 8, 1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fridell R.A., Fischer,U., Luhrmann,R., Meyer,B.E., Meinkoth,J.L., Malim,M.H. and Cullen,B.R. (1996) Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1 Rev. Proc. Natl Acad. Sci. USA, 93, 2936–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Rio S., Menezes,S.R. and Setzer,D.R. (1993) The function of individual zinc fingers in sequence-specific DNA recognition by transcription factor IIIA. J. Mol. Biol., 233, 567–579. [DOI] [PubMed] [Google Scholar]
- 39.Rowland O. and Segall,J. (1996) Interaction of wild-type and truncated forms of transcription factor IIIA from Saccharomyces cerevisiae with the 5S RNA gene. J. Biol. Chem., 271, 12103–12110. [DOI] [PubMed] [Google Scholar]
- 40.Engelke D.R., Ng,S.Y., Shastry,B.S. and Roeder,R.G. (1980) Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell, 19, 717–728. [DOI] [PubMed] [Google Scholar]
- 41.Fan H. and Sugiura,M. (1995) A plant basal in vitro system supporting accurate transcription of both RNA polymerase II- and III-dependent genes: supplement of green leaf component(s) drives accurate transcription of a light-responsive rbcS gene. EMBO J., 14, 1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yukawa Y., Sugita,M. and Sugiura,M. (1997) Efficient in vitro transcription of plant nuclear tRNA(Ser) genes in a nuclear extract from tobacco cultured cells. Plant J., 12, 965–970. [DOI] [PubMed] [Google Scholar]
- 43.Grebenok R.J., Pierson,E., Lambert,G.M., Gong,F.C., Afonso,C.L., Haldeman-Cahill,R., Carrington,J.C. and Galbraith,D.W. (1997) Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J., 11, 573–586. [DOI] [PubMed] [Google Scholar]
- 44.Stauber R., Gaitanaris,G.A. and Pavlakis,G.N. (1995) Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology, 213, 439–449. [DOI] [PubMed] [Google Scholar]
- 45.Theunissen O., Rudt,F., Guddat,U., Mentzel,H. and Pieler,T. (1992) RNA and DNA binding zinc fingers in Xenopus TFIIIA. Cell, 71, 679–690. [DOI] [PubMed] [Google Scholar]
- 46.Moorefield B. and Roeder,R.G. (1994) Purification and characterization of human transcription factor IIIA. J. Biol. Chem., 269, 20857–20865. [PubMed] [Google Scholar]
- 47.Brown T.R., Scott,P.H., Stein,T., Winter,A.G. and White,R.J. (2000) RNA polymerase III transcription: its control by tumor suppressors and its deregulation by transforming agents. Gene Expr., 9, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowland O. and Segall,J. (1998) A hydrophobic segment within the 81-amino-acid domain of TFIIIA from Saccharomyces cerevisiae is essential for its transcription factor activity. Mol. Cell. Biol., 18, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottlieb E. and Steitz,J.A. (1989) The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J., 8, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gottlieb E. and Steitz,J.A. (1989) Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J., 8, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt-Zachmann M.S. and Nigg,E.A. (1993) Protein localization to the nucleolus: a search for targeting domains in nucleolin. J. Cell Sci., 105, 799–806. [DOI] [PubMed] [Google Scholar]
- 52.Gall J.G., Bellini,M., Wu,Z. and Murphy,C. (1999) Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell, 10, 4385–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy C., Wang,Z., Roeder,R.G. and Gall,J.G. (2002) RNA polymerase III in Cajal bodies and lampbrush chromosomes of the Xenopus oocyte nucleus. Mol. Biol. Cell, 13, 3466–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]