Abstract
Plautia stali intestine virus (PSIV) has an internal ribosome entry site (IRES) at the intergenic region of the genome. The PSIV IRES initiates translation with glutamine rather than the universal methionine. To analyze the mechanism of IRES-mediated initiation, binding of IRES RNA to salt-washed ribosomes in the absence of translation factors was studied. Among the three pseudoknots (PKs I, II and III) within the IRES, PK III was the most important for ribosome binding. Chemical footprint analyses showed that the loop parts of the two stem–loop structures in Domain 2, which are highly conserved in related viruses, are protected by 40S but not by 60S ribosomes. Because PK III is close to the two loops, these structural elements were considered to be important for binding of the 40S subunit. Competitive binding analyses showed that the IRES RNA does not bind poly(U)-programmed ribosomes preincubated with tRNAPhe or its anticodon stem– loop (ASL) fragment. However, Domain 3-deleted IRES bound to programmed ribosomes preincubated with the ASL, suggesting that Domains 1 and 2 have roles in IRES binding to 40S subunits and that Domain 3 is located at the ribosome decoding site.
INTRODUCTION
Internal ribosome entry is a mode of translation initiation in which protein synthesis occurs independently of the 5′ mRNA cap (1,2). Tertiary mRNA structures upstream of some coding sequences promote ribosome entry. Such an RNA region responsible for ribosome entry is called an internal ribosome entry site (IRES) (3,4).
The intergenic region (IGR) of the positive-strand RNA virus, Plautia stali intestine virus (PSIV), contains an IRES (5). PSIV is a member of the cricket paralysis-like viruses (CrP-like viruses), which have a dicistronic, positive-stranded RNA genome (6,7). An outstanding feature of IRES-mediated translation initiation is that methionine is not the amino acid used for initiation (6,8,9). Usually, a ternary complex containing eIF2, GTP and initiator methionine transfer RNA (tRNAiMet) is absolutely required for translation initiation of normal mRNAs (10). However, in PSIV IRES-mediated translation initiation, a pseudoknot (PK) is formed at the initiation site and polypeptide synthesis starts with glutamine (8).
To date, genome sequences of 10 CrP-like viruses have been reported (7,11,12). Sequence alignments and predicted secondary structures of IGR elements from these viruses suggest that they are highly conserved and that they initiate capsid protein translation in a similar manner (13). The IGR elements of CrP-like viruses consist of about 200 nt containing four stem–loops and three PKs. Because IRES-mediated translation initiation does not use methionine, it has been generally believed that the eIF2/GTP/tRNAiMet ternary complex is not necessary for initiation. Analysis of purified ribosome binding to the cricket paralysis virus (CrPV) IRES shows assembly of an 80S monosome in the absence of eukaryotic translation initiation factors (eIFs) (9). Toeprint analysis shows that a PK is located at the P site of the ribosome (9). This strongly suggests that the ternary complex is not required for IRES-mediated translation initiation. Indeed, an increase in IRES-mediated translation is observed in yeast and mammalian cell lines when eIF2α phosphorylation is promoted by GCN2, PKR and PKRK kinases and normal translation is suppressed (14,15). In addition, when viral structural protein synthesis is maximal in CrPV-infected cells, eIF2α is heavily phosphorylated (16). However, molecular details of interactions between the IRES and ribosomes have not been resolved. To approach this question, we have performed in vitro binding experiments using PSIV IRES transcripts and high salt-washed ribosomes, and revealed that structurally conserved elements in CrP-like viruses are responsible for its binding to the 40S ribosomal subunit.
MATERIALS AND METHODS
Plasmid
RNA containing the PSIV IRES, nucleotides 6005–6192 of the PSIV sequence, was synthesized from the dicistronic plasmid pT7CAT-5375 (5) by PCR using primers 5′-GGG TATGTGATCTTATTAAAATTAGG-3′ and 5′-AACTGAG ATTCTTTTCGCACAACA-3′. The underlined nucleotides denote mutations introduced into the PSIV IRES, AC6006–6007→ GG and C6190→G. The amplified fragment was ligated with a vector fragment obtained by PCR using pT7Blue (Novagen) as a template and primers 5′-GACTCTAGAGGATCTACTAG-3′ and 5′-TATAGTGAGTCGTATTAGAGCTT-3′. To conserve base pair interactions in the IRES element, two further mutations were introduced into the resultant plasmid, GU6070–6071→CC and G6165→C, by PCR-based mutagenesis, to generate pT7D1-2-3. This plasmid has the sequence GGG immediately downstream of the T7 promoter to facilitate transcription and a HincII site at nucleotide 6190. Thus T7 transcription of HincII-linearized pT7D1-2-3 produces a cognate IRES element containing all four mutations (Fig. 1A). Variant plasmids containing disrupted or restored base pairs in each PK and truncated IRES sequences were constructed by PCR-based mutagenesis using pT7D1-2-3 as a template. For studies of chemical probes, PSIV nucleotides 5961–6230 were amplified from pT7CAT-5375 with primers having HindIII and EcoRI sites at their 5′ ends. The amplified fragment was digested with HindIII and EcoRI, and ligated into those sites of pT7Blue. For translation activity studies, plasmids that produce capsid–luciferase fusion proteins were constructed. Nucleotides 5951–6241 of the PSIV sequence were amplified from pT7CAT-5375 using primers having HindIII and NcoI sites at their 5′ ends. The amplified fragments were digested with HindIII, phosphorylated and cloned into pT7Blue treated with HindIII and HincII. Mutations in the IRES were introduced into the resultant plasmid by PCR mutagenesis and replacement of nucleotides was confirmed by sequencing. These plasmids were digested with HindIII and NcoI to yield 300 bp fragments containing mutated IRESs. The fragments were cloned into HindIII and NcoI sites of pT7-luc into which had been inserted a luciferase coding sequence (obtained from pSP-luc+; Promega) in pT7Blue using HindIII and EcoRI.
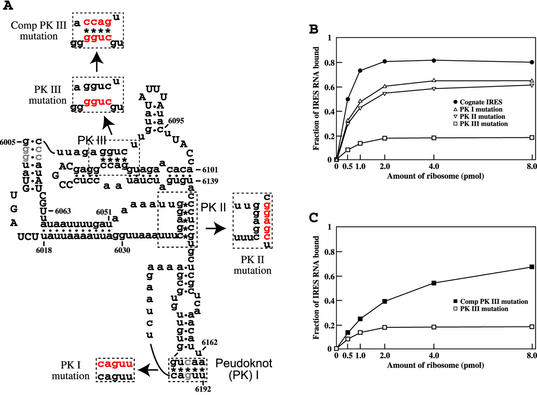
Figure 1.
Effect of base pairing in IRES PKs on 80S ribosome binding. (A) Secondary structure model of PSIV IRES RNA. Asterisks and dots indicate base pairing in PKs and helical stems, respectively. Highly conserved nucleotides in CrP-like viruses are capitalized. Half-tones denote nucleotides mutated in these studies to produce the cognate IRES (see Materials and Methods). Nucleotides mutated in PKs, to disrupt base pairing and to restore base pairing by compensatory mutations, are in red. (B) Binding of 32P-labeled IRES RNAs to 80S ribosomes. The fraction of IRES RNA bound is the ratio of [32P]RNA retained on the filter to that of the input [32P]RNA. (C) Effect of the compensatory mutations restoring base pairing in PK III on ribosome binding.
Ribosome isolation
Puromycin-treated and high salt-washed 80S ribosomes were prepared from the posterior part of silkworm silk glands (17). Ribosomal 40S and 60S subunits were prepared from rabbit reticulocyte lysate (Promega) as previously described (18,19).
RNA synthesis and renaturation
Plasmids linearized with restriction enzymes or PCR- amplified fragments containing a T7 promoter were used for transcription. RNAs labeled with 32P or 33P were synthesized and purified as described (20). Non-labeled RNAs were synthesized using a T7 Ribomax large scale RNA production system (Promega). The isolated RNA fragments (in a solution containing 2 mM MgCl2, 100 mM KCl, 0.2 mM dithiothreitol and 50 mM Tris–HCl, pH 7.5) were heated at 70–75°C for 3 min and then cooled to room temperature.
Filter binding assay
A ribosome sample was incubated with 1 pmol labeled RNA in 50 µl binding buffer (0.5–5 mM MgCl2, 100 mM KCl, 0.2 mM dithiothreitol and 50 mM Tris–HCl, pH 7.5) at 37°C for 5 min. The mixture was filtered through a nitrocellulose membrane (type HA, 0.45 µm pore size; Millipore). The membrane was washed with 1 ml binding buffer. Radio activity retained by the filter was counted using a liquid scintillation counter (Aloka). For competitive binding assays with programmed ribosomes, 50 pmol 80S ribosomes were preincubated with 60 µg poly(U), 200 pmol tRNAPhe (Sigma) or 500 pmol of the synthesized anticodon stem–loop (ASL), 5′-GGGGAUUGAAAAUCCCC-3′, at 37°C for 15 min as described by Iwasaki and Kaziro (21).
Sedimentation analysis
Renatured 33P-labeled RNAs were incubated with 40S or 60S ribosomes in binding buffer (2 mM MgCl2, 100 mM KCl and 20 mM Tris–HCl, pH 7.4) at 30°C for 5 min. This mixture then was layered on a 15–30% linear sucrose gradient in binding buffer and centrifuged at 35 000 r.p.m. at 4°C for 3.5 h using an SW 60 rotor (Beckman). Fractions (0.2 ml) were recovered from the bottom of each tube and one-twentieth of each fraction volume was transferred to a solid phase scintillator Ready Cup (Beckman).
Chemical modification and primer extension
Chemical probing of RNAs was done using dimethyl sulfate (DMS), 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT) and hydroxyl radical modification. Preliminary experiments for DMS and CMCT modification were carried out according to the methods of Brunel et al. (22), and conditions were optimized for PSIV IRES RNA. Since Mg2+ affects RNA folding and tertiary structure (23,24), Mg2+ concentrations from 0 to 10 mM were tested and 2 mM Mg2+ selected for these experiments, because results were not significantly affected when Mg2+ exceeded 2 mM. Prior to chemical modification, 0.4 pmol renatured RNA was incubated at room temperature for 5 min with or without 1.8 pmol rabbit ribosomes. DMS modification was carried out at 20°C for 30 min in a buffer containing 50 mM potassium cacodylate (pH 7.2), 2 mM MgCl2, 100 mM KCl and 40 mM DMS. CMCT modification was carried out at 20°C for 40 min in a buffer containing 50 mM sodium borate (pH 7.4), 2 mM MgCl2, 100 mM KCl and 20 mM CMCT. Hydroxyl radical modification was performed as described by Merryman and Noller (25). Two picomoles of renatured IRES RNA was incubated at room temperature for 5 min with or without 6 pmol ribosomal subunits in 21 µl of 96 mM K–HEPES (pH 7.8), 2 mM MgCl2, 120 mM NH4Cl and 6 mM DTT. In a separate tube, 4 µl of 50 mM Fe(NH4)2(SO4)2, 4 µl of 100 mM EDTA, 4 µl of 250 mM ascorbic acid and 4 µl of 2.5% H2O2 were mixed sequentially, and immediately 4 µl of the mixture was added to the RNA sample solution. The reaction was done at 0°C for 10 min, and quenched by adding an equal volume of 1 M thiourea followed by ethanol precipitation.
Modified RNAs were treated twice with phenol–chloroform, precipitated with ethanol and then used for primer extension. Primers complementary to PSIV nucleotides 6230–6211 and 6170–6151 were used for DMS and CMCT probing, and to nucleotides 6170–6151 and 6118–6096 for hydroxyl radical probing. Modified samples were mixed with 0.1 pmol 33P-labeled primers, and primer extension was carried out at 50°C for 30 min using reverse transcriptase (ReverTra Ace; Toyobo). The resulting cDNA was analyzed on a 6% (w/v) acrylamide sequencing gel.
In vitro translation
RNAs transcribed from linearized plasmids were quantified and translated using a wheatgerm extract (Promega). Luciferase activity in the translation mixtures was measured using the Stedy-Glo Luciferase Assay System (Promega).
RESULTS AND DISCUSSION
Mutational analysis of IRES pseudoknots and ribosome binding
Previous mutational analyses showed that RNA base pairing in PKs I, II and III is essential for IRES translation initiation activity (8,13). To examine the contribution of base pairing in each PK to ribosome binding, base pairs in each PK were disrupted by mutations (Fig. 1A) and ribosome binding was measured. 32P-labeled cognate and mutated IRES RNAs were incubated with increasing concentrations of 80S ribosomes and ribosome-bound IRES RNA was quantitated by filter binding assays. The amount of ribosome-bound IRES RNA increased with increasing ribosome concentration and reached a maximum at high ribosome concentrations (Fig. 1B). For cognate IRES RNA, 80% of the RNA bound at high ribosome concentrations (Fig. 1B). Mutations disrupting IRES PK base pairing decreased ribosome binding, with mutations in PK III having the greatest effect. At high ribosome concentrations, less than 20% of IRES RNA with mutations in PK III was ribosome bound and about 60% with mutations in PKs I or II was ribosome bound. To confirm the role of base pairing in ribosome binding, compensatory mutations were introduced in disrupted PK III RNA to restore base pairing (Fig. 1A). The PK III construct with compensatory mutations showed an increase in ribosome binding compared to PK III with disrupted base pairing (Fig. 1C), but the binding affinity was not as high as that of the cognate IRES. Similar compensatory mutations to restore base pairing in disrupted PKs I and II had no marked effects on ribosomal binding (data not shown). The modest effects of mutations in both PKs I and II on ribosome binding are consistent with the results of recent binding experiments by Jan and Sarnow (26), in which binding affinity between the CrPV IRES and 40S ribosomes was assayed in a gel shift system. However, mutations in PK III were not tested in the CrPV system. Therefore, these results indicate that the structure of each IRES PK plays a role in ribosome binding, with PK III structure having the most significant role.
Chemical probing of IRES structure
A secondary structure model of the PSIV IRES (Figs 1A and 4A) was generated using the MFOLD program (27) and subsequently updated based on results of PSIV mutation experiments and comparison of CrP-like virus IGR sequences (13). To facilitate discussion, the PSIV IRES structure has been divided into three domains (13): Domain 1 contains PK II and the largest stem–loop, Domain 2 (further divided into Domains 2a and 2b) contains PK III and two stem–loops and Domain 3 contains PK I and one stem–loop. However, the detailed structure of this model has not been confirmed. Therefore, IRES structure was probed using DMS and CMCT to modify IRES RNAs, with and without added ribosomal subunits, and analyzing these RNAs by primer extension for termination at modified bases. DMS reacts with unpaired adenine, cytosine and guanine, although the guanine modification cannot be detected by primer extension, while CMCT reacts with unpaired uracil and guanine (28,29).
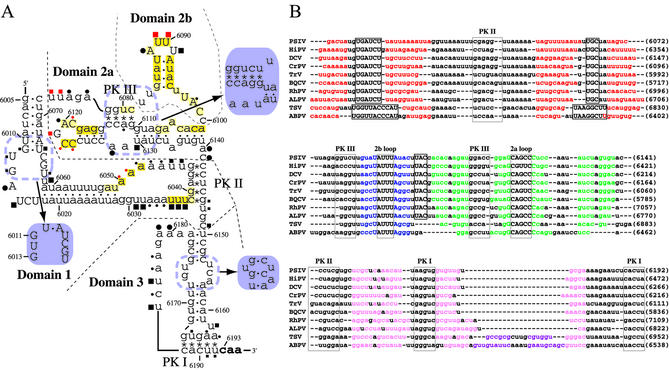
Figure 4.
Summary of results of chemical probing and structural similarity analyses of the CrP-like virus IRESs. (A) Secondary structure model of the PSIV IRES showing the results of chemical probe studies. Filled circles and squares mark nucleotides modified by DMS and CMCT, respectively, and red symbols mark nucleotides protected from these modifications in the presence of 40S ribosomes. The sizes of these symbols denote band intensity in primer extension assays. The deep yellow shaded nucleotides mark sites protected from hydroxyl radicals in the presence of 40S ribosomes, and pale yellow shaded nucleotides mark sites with weak protection. Refined structures of some regions based on the results of chemical probing described in the text are shown in purple. (B) Manually aligned IGR sequences of CrP-like viruses. Accession numbers: PSIV, AB006531; HiPV (Himetobi P virus), AB017037; DCV (Drosophila C virus), AF014388; CrPV, AF218039; TrV (Triatoma virus), AF178440; BQCV (Black queen cell virus), AF183905; RhPV (Rhopalosiphum padi virus), AF022937; ALPV (Aphid lethal paralysis virus), AF536531; TSV (Taura syndrome virus), AF277675; ABPV (Acute bee paralysis virus), AF150629. Red, blue, green and pink letters denote nucleotides in stems in Domains 1, 2b, 2a and 3, respectively. Purple letters denote an additional putative stem in Domain 3 of TSV and ABPV. PK I, PK II, PK III, Domain 2a loop and Domain 2b loop nucleotides are in half-tone lines. Conserved short nucleotide sequences among these viruses are in boxes. Nucleotide positions in viral genomes are shown at right in parentheses.
Nucleotides that reacted with DMS and CMCT in the absence of added 40S ribosomes (Fig. 2A, lanes 3 and 6) are shown in Figure 4A. Most of these are in single-stranded regions of the predicted model. However, several reactive nucleotides (U6066, A6112, U6130, G6146, AA6163–6164, U6166 and UU6191–6192) are in helical regions. This suggests a re-evaluation of these nucleotides. (i) U6066 reacted strongly with CMCT but G6011, the nucleotide to which it is base paired in the model, was only slightly modified (Fig. 2A), suggesting that these residues are not base paired (Fig. 4A). (ii) A6112 was reactive with DMS (Fig. 2A and B) so the A6112–U6080 pair may be unstable. (iii) U6130 showed strong reactivity with CMCT although in the model it is base paired with G6110. Since neither U6082 nor U6083 was reactive with CMCT and sequence alignment of CrP-like viruses suggests a single U or G nucleotide between PK III and the stem–loop in Domain 2b (Fig. 4B), G6110 may be base paired with U6082 rather than U6130 (Fig. 4A). (iv) A band corresponding to C6149 was detected in the CMCT-modified sample even though CMCT should not react with cytosine. This suggests that the band at C6149 is due to non-specific termination. Also, a band at G6146 is probably due to non-specific termination since preliminary experiments found that reverse transcription sometimes terminates at nucleotides 6146–6149 when the samples contain 40S ribosomes (data not shown). (v) AA6163–6164, U6166 and UU6191–6192 are in PK I in the model. Since PK I is essential for translation initiation in CrP-like viruses (8,9,30), it is unlikely that these residues remain single-stranded throughout the initiation process. The weak reactivity at these residues in PK I may be due to some IRES RNA molecules having an incompletely folded Domain 3 or to Domain 3 being unstable in these experimental conditions.
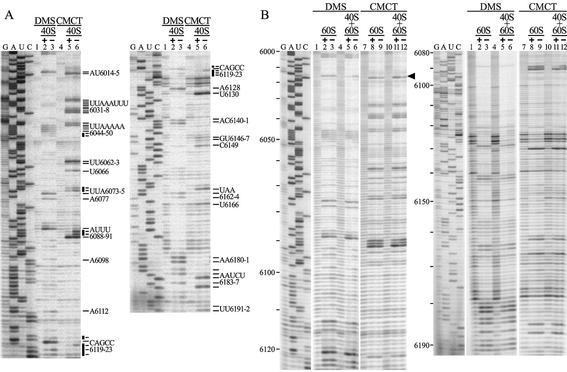
Figure 2.
DMS and CMCT probing of the PSIV IRES. To detect modified bases, DMS- and CMCT-modified RNAs were analyzed by primer extension. (A) Modification with DMS (lanes 2 and 3) or CMCT (lanes 5 and 6) in the presence (lanes 2 and 5) or absence (lanes 3 and 6) of 40S ribosomes. Lanes 1 and 4 are the unmodified controls to detect non-specific termination. Bold bars mark IRES nucleotides protected from modification by 40S ribosomes. Nucleotide positions are shown at right. (B) Modification with DMS (lanes 2, 3, 5 and 6) or CMCT (lanes 8, 9, 11 and 12) in the presence of 60S (lanes 2 and 8) or 40S and 60S (lanes 5 and 11) ribosomes. Lanes 1, 4, 7 and 10 are the unmodified controls. The arrowhead marks A6014, which was only protected in the presence of both 40S and 60S ribosomes.
Sequence alignments indicate that the CrP-like viruses CrPV, Rhopalosiphum padi virus and Black queen cell virus form a helical region without an internal bulge in Domain 3 (Fig. 4B). In agreement with this model, UCA6154–6156 and GU6173–6174 in PSIV Domain 3 did not show obvious reactivity to DMS and CMCT (Fig. 2A). Also, using measurement of translation activity of a capsid–luciferase fusion protein, replacement of G6173 with C decreased luciferase activity to 40% of that of wild-type. Translation activity was completely restored by the compensatory replacement of C6155 with G to restore the base pair (Table 1). Also, in DMS probe studies there was a band corresponding to nucleotide 6173 in the C6173 mutant and this band was missing in RNA with the additional compensatory mutation (data not shown). Thus, the model of internal bulges in Domain 3 should be refined (Fig. 4A).
Table 1. Luciferase activity of PSIV IRES variants with mutations in the internal bulge of Domain 3.
| Sequencea | Relative light unitsb | Relative luciferase activity | |
|---|---|---|---|
| Wild-type | CUCAA6153–6157 | 48 445 ± 9843 | 100% |
| GUGU6175–6172 | |||
| C6173 | CUCAA6153–6157 | 19 275 ± 1465 | 39.8% |
| GUCU6175–6172 | |||
| C6173 + G6155 | CUGAA6153–6157 | 51 923 ± 3964 | 107.2% |
| GUCU6175–6172 |
aUnderlining indicates mutated nucleotides.
bStandard deviations are also indicated.
The DMS and CMCT probes did not detect several nucleotides that were predicted to be in single-stranded regions of the model (Fig. 4A), such as CU6016–6017, GG6029–6030, GC6064–6065, G6076, A6105, A6129, A6135, A6179 and G6182. Therefore, some of these nucleotides may be involved in base pair interactions to form tertiary structures not in the model.
Footprint analysis
To investigate IRES–ribosome binding, DMS and CMCT chemical probing of IRES RNA was done in the presence of ribosomal subunits. Modified 40S ribosome-bound IRES RNA had reduced band intensities at nucleotides 6049–6050, 6073–6074, 6089–6090, 6119 and 6121–6123 (Fig. 2A, lanes 2 and 5 for DMS and CMCT, respectively) compared to band intensities for IRES RNA in the absence of added ribosome (Fig. 2A, lanes 3 and 6 for DMS and CMCT, respectively). This indicates that 40S ribosomes protect these nucleotides from chemical modification. Two of these nucleotides, UU6089–6090, are in the loop part of the stem–loop structure in Domain 2b, which is conserved in all CrP-like viruses (Fig. 4B). C6119 and GCC6121–6123 are in the loop part of the stem–loop structure in Domain 2a, which is also highly conserved in CrP-like viruses. In particular, C6119 and C6123 are conserved in all these viruses and, therefore, probably have an important role in 40S ribosome binding. However, A6120 was not significantly protected by 40S ribosomes from chemical modification in PSIV and is replaced by a G in the HiPV sequence, so it probably is not important for 40S ribosome binding.
Similar chemical modification experiments were carried out in the presence of the 60S subunit (Fig. 2B). However, no effect on primer extension profiles was detected. This indicates that 60S ribosomes do not interact with single-stranded regions of IRES RNA. When both 40S and 60S ribosomes were included in the incubation mixture, the protection pattern was almost identical to that for 40S ribosomes except that A6014 was also protected (Fig. 2B). A6014 is in the conserved bulge sequence in Domain 1. It was not protected by either 40S or 60S ribosomes alone, but was protected in the presence of both 40S and 60S ribosomes. This suggests that A6014 is in a part of the IRES close to an inter-subunit bridge or that the conformation of the IRES changes in the presence of the 60S ribosomes.
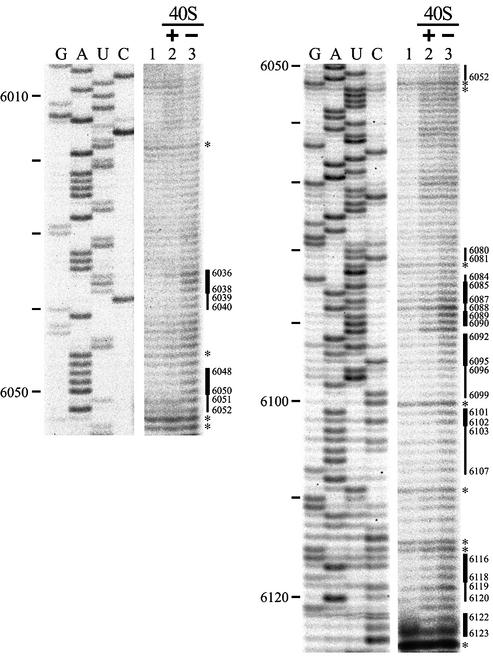
To investigate PSIV IRES ribosomal binding sites further, IRES RNA in the presence and absence of 40S ribosomes was treated with hydroxyl radicals. Since hydroxyl radicals attack ribose in the nucleic acid backbone with little sequence specificity, this is a useful tool for probing interactions between nucleic acids and ligands (31,32). Protection from hydroxyl radical attack was monitored by primer extension. An unmodified control (with 40S ribosomes added to unmodified RNA before RNA isolation) was used to identify non-specific termination caused either by IRES tertiary structure or by the presence of 18S ribosomal RNA (Fig. 3, lane 1). Such non-specific terminations were observed at nucleotides 6018, 6046, 6053–6054, 6082, 6100, 6109, 6114–6115 and 6124 (Fig. 3, lanes 1–3). Band intensities at these non-specific termination sites were almost identical for unmodified control IRES RNA, IRES RNA modified in the presence of 40S ribosomes and IRES RNA modified in the absence of 40S ribosomes (Fig. 3, lanes 1–3, respectively), indicating that experimental conditions for primer extension were equivalent in these samples. Therefore, differences in band intensities between IRES RNA treated in the presence and absence of 40S ribosomes (Fig. 3, lanes 2 and 3, respectively) represent protection of IRES RNA from hydroxyl radical cleavage due to 40S ribosome binding. These data show band intensity decreases (i.e. 40S ribosome protection) at nucleotides 6036–6038, 6048–6050, 6085– 6087, 6089–6090, 6092–6095, 6101–6102, 6116–6118 and 6122–6123 and small intensity decreases (i.e. weak protection) at nucleotides 6039–6040, 6051–6052, 6080–6081, 6084, 6088, 6096–6099, 6103–6107 and 6119–6120 (compare Fig. 3, lanes 2 and 3). These protected residues were observed in repeated experiments although the edges of the weakly protected regions varied by a few nucleotides. These results are shown in the PSIV IRES structural model (Fig. 4A). The data suggest that nucleotides 6065–6067, 6083, 6091, 6108, 6110–6113 and 6121 also might be weakly protected (Fig. 3), but these nucleotides are not marked in Figure 4A because of uncertainties about the differences in their band intensities. To investigate 40S ribosome protection downstream of nucleotide 6124, hydroxyl radical footprint analysis was done using a primer corresponding to nucleotides 6230–6211, but no protection was observed (data not shown).
Figure 3.
Hydroxyl radical probing of PSIV IRES RNA. The RNA was exposed to hydroxyl radicals in the presence (lane 2) or absence (lane 3) of 40S ribosomes. Lane 1 is the unmodified control, but 40S ribosomes were added to the sample before phenol–chloroform extraction to detect non- specific terminations caused by 18S rRNA as well as the sample in lane 3. Bold bars denote locations of protected residues and narrow bars denote sites of weak protection. Asterisks mark sites of non-specific terminations, as listed in the text. Nucleotide positions of the PSIV sequence are shown at left.
The distribution of sites protected from hydroxyl radicals and DMS and CMCT treatments by 40S ribosomes indicates that the two stem–loop structures in Domain 2 and the loop of the stem–loop structure in Domain 1 interact with 40S ribosomes (Fig. 4A). In contrast, nucleotides in Domain 3 were not significantly protected from chemical probes by 40S ribosomes, suggesting that they have little or no role in ribosome binding.
Sedimentation analysis of Domain 2–ribosome binding
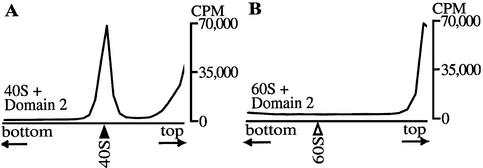
Footprint analyses indicated that the two stem–loop structures in IRES Domain 2 are the major regions protected by 40S ribosomes. To examine whether Domain 2 RNA (IRES nucleotides 6078–6139) alone can bind ribosomes, sucrose gradient sedimentation analyses were carried out on 33P-labeled Domain 2 RNA that had been incubated with purified rabbit 40S or 60S ribosomes. Domain 2 RNA co-sedimented with 40S ribosomes (Fig. 5A), but not with 60S ribosomes (Fig. 5B). Therefore, IRES Domain 2 RNA binds 40S but not 60S ribosomes.
Figure 5.
Sedimentation profiles of Domain 2 RNA and ribosomal subunits. 33P-labeled Domain 2 RNA was incubated with purified (A) 40S or (B) 60S ribosomes and centrifuged in sucrose gradients. Fractions were collected and the radioactivity in each fraction was measured. Black and white arrowheads indicate positions of 40S and 60S subunits, respectively, which were monitored by absorbance at 254 nm.
Effect of mutations in IRES conserved sequences on translation activity
Protection of the loop sequences in the stem–loop structures in PSIV IRES Domains 2a and 2b by 40S ribosomes from chemical probes suggested that these sequences are important for IRES activity. Therefore, the effects of mutations in these loop sequences on translation activity were studied using a capsid–luciferase fusion protein translation system. The translation activity of a PSIV variant with two mutations in the Domain 2a loop, C6119→G and C6123→G, decreased to 12% of that of the wild-type, and the activity of a variant with mutations in the Domain 2b loop, AUUU6088–6091→UAAA, decreased to 62% (Table 2). This result for PSIV was different from that reported for CrPV: the same mutation in the CrPV IRES Domain 2b loop sequence as described here for PSIV decreases CrPV IRES translation activity to less than 10% of wild-type (26). Translation activity of the CrPV IRES is measured using dicistronic constructs. On the other hand, that of PSIV was monocistronic. Therefore, we constructed two additional dicistronic uncapped RNAs similar to CrPV using Renilla and firefly luciferases. One contained the wild-type PSIV IRES sequence and the other contained a UAAA mutation in the loop sequence in PSIV IRES Domain 2b. The UAAA mutant showed 23% activity of the wild-type. Thus, we considered that the discrepancy of the results in PSIV and CrPV probably came from the difference of constructs, monocistronic and dicistronic. These results indicate that conserved sequences in the loops of the stem–loop structures in Domains 2a and 2b are important for IRES activity. Although the effect of these mutations in PSIV loops on 40S ribosome binding has not been studied, similar constructs in CrPV slightly decrease binding (26).
Table 2. Luciferase activity of PSIV IRES variants with mutations in the loops of the stem–loop structures in Domains 2a and 2b.
| Sequencea | Relative light unitsb | Relative luciferase activity | |
|---|---|---|---|
| Wild-type | CAGCC6119–6123 | 48 445 ± 9843 | 100% |
| AUUU6088–6091 | |||
| Domain 2a | GAGCG6119–6123 | 5689 ± 1687 | 11.7% |
| Domain 2b | UAAA6088–6091 | 29 834 ± 3187 | 61.6% |
aUnderlining indicates mutated nucleotides.
bStandard deviations are also indicated.
The single-strand sequence that connects helical segments in Domains 2a and 2b (UUACC6096–6100) is highly conserved among CrP-like viruses (Fig. 4B). Since replacement of ACC6098–6100 with GGG has been shown to abolish translation (13), this region must have an essential role in IRES-mediated translation. However, obvious modifications by the chemical probes DMS and CMCT were not observed at nucleotides 6096–6100 (Fig. 2), suggesting that this region might base pair in the IRES structure. Since the refined structure model (Fig. 4A) suggested that G6011 should be single strand, base pair interactions between GUG6011–6013 and CAU6099–6097 were considered. To examine this, mutational analyses were carried out using three variants with mutations to disrupt and restore possible base pairs between these sequences: ACC6098–6100→UUU, UGUG6010–6013→AAAA and combined mutation of both sequences to restore possible base pair formation. However, recovery of translation activity was not observed using the capsid–luciferase fusion protein translation system (data not shown), suggesting that the GUG6011–6013 and CAU6099–6097 sequences do not interact.
IRES binding to programmed ribosomes
The experiments above identified IRES structural elements involved in 40S ribosome binding and showed that Domain 2 has a crucial role in this binding. However, since no obvious protection in Domain 3 was detected in chemical probe studies, the function of Domain 3 in ribosome binding is unknown. To investigate a possible role for Domain 3, the binding of programmed 80S ribosomes to complete IRES RNA and to truncated IRES RNA, from which Domain 3 was deleted, was studied. Poly(U)-programmed 70S ribosomes are frequently used for translation analysis in prokaryotic systems (33). It is known that tRNAPhe binds to the A, P and E sites of poly(U)-charged prokaryotic 70S ribosomes at high Mg2+ concentrations (34) and that the ASL domain of tRNA binds to the decoding site of poly(U)-programmed ribosomes (35,36). Therefore, it was assumed that binding of IRES RNA to 80S ribosomes would be inhibited by poly(U), tRNAPhe or ASL, if the IRES binds to a region at or near the tRNA binding sites of eukaryotic 80S ribosomes in vitro.
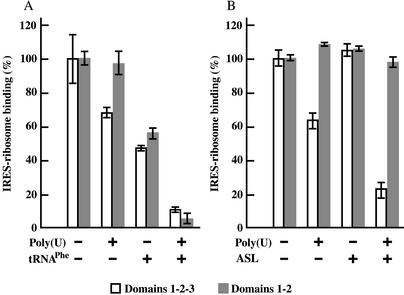
IRES–ribosome binding was measured by filter binding assays. Preincubation of 80S ribosomes with poly(U) decreased the binding of 32P-labeled complete IRES RNA (containing Domains 1–2–3) to 60–70% of that of controls without poly(U), but did not affect binding of truncated IRES RNA (containing only Domains 1–2) (Fig. 6A and B). When ribosomes were preincubated with tRNAPhe, binding of both IRES RNAs decreased to 45–55% of that of controls without tRNAPhe (Fig. 6A). When ribosomes were preincubated with poly(U) and tRNAPhe together, binding of both IRES RNAs decreased to less than 10% (Fig. 6A). In contrast to the results using tRNAPhe, when only ASL was used for preincubation, binding of both IRES RNAs to ribosomes was not inhibited (Fig. 6B), but this can be explained by the instability of ASL on ribosomes without mRNA. However, preincubation of ribosomes with poly(U) and ASL together decreased binding of complete IRES RNA (Domains 1–2–3) to about 20%, but had no effect on the binding of truncated IRES RNA (Domains 1–2) (Fig. 6B). These results suggest that part of Domain 3 competes with the ASL when IRES RNA binds to poly(U)-charged ribosomes. Binding of truncated IRES RNA (Domains 1–2) to poly(U)-charged ribosomes was also inhibited by tRNAPhe (Fig. 6A). Since the ASL lacks the tRNA molecule acceptor stem and D and T stem–loops, part of Domains 1–2 would occupy the ribosome sites for the tRNA acceptor stem and D or T stem–loop interactions, but not for the ribosome tRNA ASL site. In addition, the filter binding experiments showed that complete and truncated IRES RNAs bind to salt-washed ribosomes with similar affinities (Kd 21–23 nM). From these observations, it appears that Domain 3 does not have a significant role in IRES RNA–ribosome binding.
Figure 6.
Filter binding analyses of complete IRES RNA (Domains 1–2–3) and truncated IRES RNA (Domains 1–2) binding to programmed 80S ribosomes. (A) Effects of poly(U) and/or tRNAPhe programming on RNA binding. (B) Effects of poly(U) and/or 17mer tRNAPhe ASL programming on RNA binding. 32P-labeled IRES RNAs were incubated with 80S ribosomes that had been preincubated with poly(U), tRNAPhe and/or ASL. Binding for samples containing 1 pmol RNA and 4 pmol ribosomes [without poly(U), tRNAPhe or ASL] was taken as 100% binding. Results are the average of three independent experiments. Bars represent standard errors.
The studies presented here allow several conclusions regarding the molecular events in IRES-mediated translation initiation. Purified 40S ribosomes interact with the loop sequences in the stem–loop structures in PSIV IRES Domains 2a and 2b. The two loop sequences are highly conserved among CrP-like viruses (Fig. 4B) and mutations in these sequences decreased PSIV IRES translation activity, indicating that these loops have essential roles in IRES function. Mutational disruption of base pair interactions in PK III had deleterious effects on 40S ribosome binding (Fig. 1). Since PK III is near the two Domain 2 loops, PK III may affect the relative positions of these two loops for efficient 40S ribosome binding. Binding of complete IRES RNA (Domains 1–2–3), but not truncated IRES RNA (Domains 1–2), to 80S ribosomes was inhibited by ASL. This suggests that Domain 3 probably interacts with part of the ribosome decoding site. These results imply that Domains 1–2 and Domain 3 have distinct roles in IRES-mediated translation initiation. Since Domain 2 alone can bind 40S ribosomes (Fig. 5A), interactions between 40S ribosomes and the Domain 2 loops are probably one of the initial steps in IRES-mediated translation initiation. Although when and how 60S ribosomes engage the IRES–40S complex is unknown, PK II may have a role in promoting the interaction of Domain 3 and the ribosome decoding site, presumably in the presence of the 60S ribosomes. CrPV IRES-mediated translation elongation, in the absence of any eIFs, has recently been shown by toeprinting and peptidyl-puromycin formation analysis using purified elongation factors and acylated tRNAs (37). This also means that binding between ribosomes and CrP-like virus IRESs plays a key role for translation initiation.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Pioneer Program, MAFF and PROBRAIN, Japan to N.N., and by grants-in-aid for scientific research (14035222) from the Ministry of Education, Science, Sports and Culture of Japan to T.U.
REFERENCES
- 1.Pelletier J. and Sonenberg,N. (1988) Internal initiation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature, 334, 320–325. [DOI] [PubMed]
- 2.Jang S.K., Krausslich,H.G., Nicklin,M.J.H., Duke,G.M., Palmenberg,A.C. and Wimmer,E. (1988) A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol., 62, 2636–2643. [DOI] [PMC free article] [PubMed]
- 3.Hellen C.U.T. and Sarnow,P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev., 15, 1593–1612. [DOI] [PubMed]
- 4.Vagner S., Galy,B. and Pyronnet,S. (2001) Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep., 2, 893–898. [DOI] [PMC free article] [PubMed]
- 5.Sasaki J. and Nakashima,N. (1999) Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol., 73, 1219–1226. [DOI] [PMC free article] [PubMed]
- 6.Wilson J.E., Powell,M.J., Hoover,S.E. and Sarnow,P. (2000) Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol., 20, 4990–4999. [DOI] [PMC free article] [PubMed]
- 7.Liljas L., Tate,J., Lin,T., Christian,P. and Johnson,J.E. (2002) Evolutionary and taxonomic implications of conserved structural motifs between picornaviruses and insect picorna-like viruses. Arch. Virol., 147, 59–84. [DOI] [PubMed]
- 8.Sasaki J. and Nakashima,N. (2000) Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl Acad. Sci. USA, 97, 1512–1515. [DOI] [PMC free article] [PubMed]
- 9.Wilson J.E., Pestova,T.V., Hellen,C.U. and Sarnow,P. (2000) Initiation of protein synthesis from the A site of the ribosome. Cell, 102, 511–520. [DOI] [PubMed]
- 10.Jackson R.J. (2000) A comparative view of initiation site selection mechanisms. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 127–183.
- 11.Mari J., Poulos,B.T., Lightner,D.V. and Bonami,J.R. (2002) Shrimp Taura syndrome virus: genomic characterization and similarity with members of the genus Cricket paralysis-like viruses. J. Gen. Virol., 83, 915–926. [DOI] [PubMed]
- 12.van Munster M., Dullemans,A.M., Verbeek,M., van den Heuvel,J.F., Clerivet,A. and van der Wilk,F. (2002) Sequence analysis and genomic organization of Aphid lethal paralysis virus: a new member of the family Dicistroviridae. J. Gen. Virol., 83, 3131–3138. [DOI] [PubMed]
- 13.Kanamori Y. and Nakashima,N. (2001) A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA, 7, 266–274. [DOI] [PMC free article] [PubMed]
- 14.Thompson S.R., Gulyas,K.D. and Sarnow,P. (2001) Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl Acad. Sci. USA, 98, 12972–12977. [DOI] [PMC free article] [PubMed]
- 15.Fernandez J., Yaman,I., Sarnow,P., Snider,M.D. and Hatzoglou,M. (2002) Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem., 277, 19198–19205. [DOI] [PubMed]
- 16.Bushell M. and Sarnow,P. (2002) Hijacking the translation apparatus by RNA viruses. J. Cell Biol., 158, 395–399. [DOI] [PMC free article] [PubMed]
- 17.Uchiumi T., Nomura,T., Shimizu,T., Katakai,Y., Mita,K., Koike,Y., Nakagaki,M., Taira,H. and Hachimori,A. (2000) A covariant change of the two highly conserved bases in the GTPase-associated center of 28 S rRNA in silkworms and other moths. J. Biol. Chem., 275, 35116–35121. [DOI] [PubMed]
- 18.Blobel G. and Sabatini,D. (1971) Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl Acad. Sci. USA, 68, 390–394. [DOI] [PMC free article] [PubMed]
- 19.Pestova T.V., Hellen,C.U.T. and Shatsky,I.N. (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol., 16, 6859–6869. [DOI] [PMC free article] [PubMed]
- 20.Uchiumi T., Sato,N., Wada,A. and Hachimori,A. (1999) Interaction of the sarcin/ricin domain of 23 S ribosomal RNA with proteins L3 and L6. J. Biol. Chem., 274, 681–686. [DOI] [PubMed]
- 21.Iwasaki K. and Kaziro,Y. (1979) Polypeptide chain elongation factors from pig liver. Methods Enzymol., 60, 657–676. [DOI] [PubMed]
- 22.Brunel C., Romby,P., Westhof,E., Ehresmann,C. and Ehresmann,B. (1991) Three-dimensional model of Escherichia coli ribosomal 5 S RNA as deduced from structure probing in solution and computer modeling. J. Mol. Biol., 221, 293–308. [DOI] [PubMed]
- 23.Kieft J.S., Zhou,K., Jubin,R., Murray,M.G., Lau,J.Y. and Doudna,J.A. (1999) The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol., 292, 513–529. [DOI] [PubMed]
- 24.Rook M.S., Treiber,D.K. and Williamson,J.R. (1999) An optimal Mg2+ concentration for kinetic folding of the Tetrahymena ribozyme. Proc. Natl Acad. Sci. USA, 96, 12471–12476. [DOI] [PMC free article] [PubMed]
- 25.Merryman C. and Noller,H.F. (1998) Footprinting and modification-interference analysis of binding sites on RNA. In Smith,C.W.J. (ed.), RNA:Protein Interactions. Oxford University Press, Oxford, UK, pp. 237–253.
- 26.Jan E. and Sarnow,P. (2002) Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol., 324, 889–902. [DOI] [PubMed]
- 27.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed]
- 28.Inoue T. and Cech,T.R. (1985) Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl Acad. Sci. USA, 82, 648–652. [DOI] [PMC free article] [PubMed]
- 29.Moazed D. and Noller,H.F. (1986) Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell, 47, 985–994. [DOI] [PubMed]
- 30.Domier L.L., McCoppin,N.K. and D’Arcy,C.J. (2000) Sequence requirements for translation initiation of Rhopalosiphum padi virus ORF2. Virology, 268, 264–271. [DOI] [PubMed]
- 31.Latham J.A. and Cech,T.R. (1989) Defining the inside and outside of a catalytic RNA molecule. Science, 245, 276–282. [DOI] [PubMed]
- 32.Darsillo P. and Huber,P.W. (1991) The use of chemical nucleases to analyze RNA-protein interactions. The TFIIIA-5 S rRNA complex. J. Biol. Chem., 266, 21075–21082. [PubMed]
- 33.Schilling-Bartetzko S., Franceschi,F., Sternbach,H. and Nierhaus,K.H. (1992) Apparent association constants of tRNAs for the ribosomal A, P and E sites. J. Biol. Chem., 267, 4693–4702. [PubMed]
- 34.Moazed D. and Noller,H.F. (1989) Interaction of tRNA with 23S rRNA in the ribosomal A, P and E sites. Cell, 57, 585–597. [DOI] [PubMed]
- 35.Joseph S., Weiser,B. and Noller,H.F. (1997) Mapping the inside of the ribosome with an RNA helical ruler. Science, 278, 1093–1098. [DOI] [PubMed]
- 36.Ogle J.M., Brodersen,D.E., Clemons,W.M.,Jr, Tarry,M.J., Carter,A.P. and Ramakrishnan,V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science, 292, 897–902. [DOI] [PubMed]
- 37.Pestova T.V. and Hellen,C.U.T. (2003) Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev., 17, 181–186. [DOI] [PMC free article] [PubMed]