Abstract
The human DNA polymerase ε catalytic subunit consists of a 140-kDa N‐terminal domain that contains the catalytic activity and a 120-kDa C-terminal domain that binds to the other subunits and to exogenous peptides, including PCNA and MDM2. We report here that recombinant human MDM2 purified from insect cells or Escherichia coli stimulated the activity of DNA polymerase ε up to 10- and 40-fold, respectively, but not those of DNA polymerase β or Klenow fragment of E.coli DNA polymerase I. Kinetic studies indicated that MDM2 increased the maximum velocity of the reaction, but did not change substrate affinities. The stimulation depended upon the interaction of the N‐terminal 166 amino acid residues of MDM2 with the C-terminal domain of the full-length catalytic subunit, since the deletion of 166 amino acids from N‐terminal of MDM2 or the removal of the C-terminal domain of DNA polymerase ε by trypsin digestion or competition for binding to it by the addition of excess C-terminal fragment eliminated the stimulation. Since DNA polymerase ε appears to be involved in DNA replication, recombination and repair synthesis, we suggest that MDM2 binding to DNA polymerase ε might be part of a reconfiguration process that allows DNA polymerase ε to associate with repair/recombination proteins in response to DNA damage.
INTRODUCTION
MDM2 was discovered in 1991 as the transforming gene of the tumorigenic 3T3 DM derivative of NIH 3T3 cells (1). Later studies demonstrated that MDM2 could transform cells in culture and in combination with an activated RAS gene could promote tumors in nude mice (2). Typical of several oncogenes, the transforming activity of MDM2 can be activated by overexpression of the gene in vitro and in vivo and in human tumors MDM2 protein levels are frequently abnormally high.
MDM2 can bind to and inhibit the transcriptional activation activity of p53, and thus it appeared likely that MDM2 tumorigenicity might result from the ability of MDM2 to compromise p53 function (3). Indeed, MDM2 is an E3 ubiquitin ligase with specificity for both p53 and itself and, as a result of this activity, MDM2 destabilizes p53 through the activity of proteosomes (4–6). MDM2 is itself a transcriptional target of p53, demonstrating an interplay between these genes and suggesting the existence of a negative feedback loop between MDM2 and p53 (7). Several lines of evidence suggest that MDM2 has functions in addition to its interplay with p53. For example, MDM2 binds a number of other proteins (8–11) and in transgenic mouse models, MDM2 has been shown to uncouple S-phase from mitosis in wild type, p53–/– and in E2F1–/– animals (12,13). Finally, forms of MDM2 that cannot bind to p53 retain both transforming potential and tumorigenicity (14).
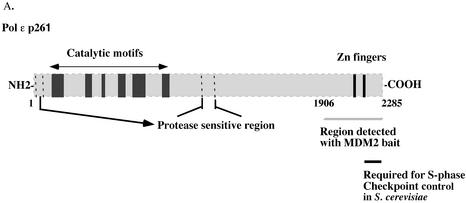
In a yeast two-hybrid screen for potential MDM2-binding partners, the C-terminus of DNA polymerase ε catalytic subunit (nt 5833–6984, numbering based on the human cDNA sequence) was suggested (15). DNA polymerase ε is one of 14 known human DNA template-directed DNA polymerases. It has been implicated in chromosomal DNA replication, DNA repair and recombination, and is composed of a 261-kDa catalytic subunit (p261) and three associated subunits of 59 (POLE2), 17 (POLE3) and 12 kDa (POLE4) (16–18). p261 can be proteolysed into a 140-kDa catalytically active N‐terminal domain containing six polymerase and five exonuclease motifs and a 120-kDa C-terminal domain (19) (see Fig. 6A below). Interestingly, the non-catalytic, C-terminal domain of p261, but not the catalytic N‐terminal domain is essential in Saccharomyces cerevisiae and Schizosaccharomyces pombe (20,21). p59 is required for the stability of the enzyme activity and it binds the C-terminal domain of p261 and PCNA, and is required for p17 and p12 binding (17,18). p17 and p12 contain complementary histone-fold motifs (18) that also are present in their homologs in S.cerevisiae (22,23) and S.pombe (18). Histone-fold motifs are involved in creating a protein–protein interaction surface and promoting protein–DNA interaction (24). p17 is also a component of human chromatin accessibility complex (huCHRAC), but its binding partner in that complex is huCHRAC p15, not DNA polymerase ε p12 (18,25). p12 and p17 heterodimer interact with both p261 and p59 or p261/p59 heterodimer (17,18).
Figure 6.
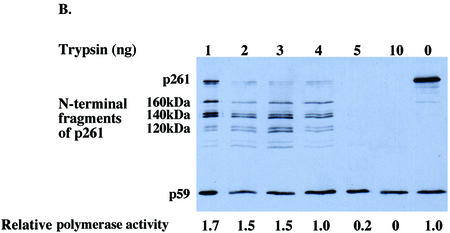
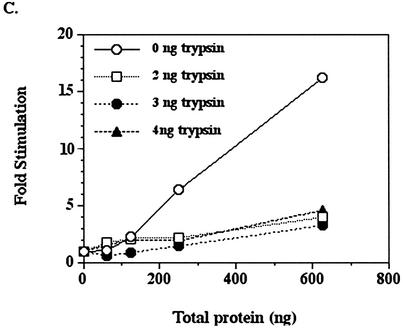
The C-terminal domain of intact DNA polymerase ε p261 is essential for stimulation by MDM2. (A) HeLa p261 can be separated into two domains which are linked by a protease-sensitive region. The N‐terminal domain contains the catalytic motifs, whereas the C-terminal domain binds other subunits and proteins. (B and C) Fifty nanogram aliquots of purified HeLa DNA polymerase ε were digested with various amounts of trypsin as indicated for 15 min at 37°C, after which 1 µg of soybean trypsin inhibitor was added to stop each reaction. Half of each product was separated by 7.5% SDS–PAGE and then transferred to a nitrocellulose filter which was probed with anti-p261 antibodies 3C5.1, which recognizes the N‐terminal domain of p261, and 3A5.6, which recognizes p59. The remainder of each sample was assayed for polymerase activity in the absence (B) or presence (C) of recombinant MDM2 expressed in E.coli.
Our previous study of MDM2 interaction with DNA polymerase ε (15) showed that DNA polymerase ε binds amino acid residues 50–166 of MDM2, a region of MDM2 that also interacts with p53, Numb (8), E2F1 (9), TFIIE (10) and Tag (11). Here we report that MDM2 stimulates the polymerase activity of DNA polymerase ε and that the stimulation requires the N‐terminal 166 amino acid residues of MDM2 and the C-terminal domain of DNA polymerase ε catalytic subunit.
MATERIALS AND METHODS
Purification of recombinant human MDM2 and Δ1–166 MDM2 from insect cells and Escherichia coli
For expression in insect cells, full-length human MDM2 cDNA or MDM2 cDNA lacking the coding region for the N‐terminal 166 amino acids were cloned into a pBacPAK8 transfer vector using conventional restriction digest/ligation reactions. The construction and amplification of the recombinant baculoviruses were carried out according to the BacPAK Expression System protocol of BD Biosciences (Clontech). For expression of full-length MDM2, 1500 ml of Sf9 cells were grown to 1 × 106 cells/ml in Ex-Cell 400 medium supplemented with 2% fetal bovine serum, infected with the recombinant virus and harvested 72 h post infection by centrifugation. Cells were washed once with phosphate-buffered saline (PBS) and then resuspended in 100 mM Tris–HCl (pH 8.0), 20% glycerol, 4 mM DTT, 0.3 M potassium glutamate and protease inhibitor cocktail tablet complete™ EDTA-free (Roche Molecular Biochemicals). All purification procedures were carried out at 4°C. The presence of MDM2 was monitored by immunoblots with monoclonal antibody SMP-14 (Santa Cruz Biotechnology, Inc.) throughout the purification.
Cells were lysed by sonication and then Brij35 was added to a final concentration of 0.01%. After stirring on ice for 1 h, the lysate was centrifuged at 47 800 g for 30 min. The supernate was brought to 60% saturated ammonium sulfate, and after at least 3 h on ice it was centrifuged for 30 min at 27 000 g. The pellet was dissolved in 50 ml of Buffer A [50 mM Tris–HCl (pH 8.0), 10% glycerol, 4 mM DTT, 0.15 M NaCl, 0.01% Brij35] and dialyzed against Buffer A. The dialyzed material, containing 440 mg of protein, was applied to a 200-ml DEAE–Sephacel column (Amersham Biosciences) pre-equilibrated with Buffer A. The column was washed with 400 ml of Buffer A and then the bound proteins were eluted with a 1200-ml linear gradient from 0.15 to 1 M NaCl in Buffer A. MDM2 eluted near 350 mM NaCl in a pool containing 111 mg of protein. A 20-mg aliquot in 30 ml was mixed with 300 µg of monoclonal antibody SMP-14 covalently crosslinked to protein A–Sepharose beads (Amersham Biosciences) and incubated with gentle mixing overnight at 4°C. The beads were washed with 20 ml of Buffer A and then bound proteins were eluted with 100 mM glycine (pH 2.5), 0.15 M NaCl. Fractions (0.5 ml) were collected into tubes containing 200 µl of 1 M Tris–HCl (pH 8.0), 70 µl 80% glycerol, 2 µl 1 M DTT and 1 µl 10% Brij35. Fractions containing MDM2 were pooled and dialyzed against Buffer A. The immunopurification procedure was repeated twice more to yield 8 mg of a protein pool, which was loaded onto a 1-ml HiTrapQ column (Amersham Biosciences) equilibrated with 50 mM HEPES–KOH (pH 7.5), 10% glycerol, 4 mM DTT, 0.01% Brij35 (Buffer B). A 15-ml linear gradient from 0 to 1 M NaCl in Buffer B and then 5 ml of 1 M NaCl in Buffer B were successively applied to the column. Fractions containing MDM2 eluting around 0.6 M NaCl were pooled, dialyzed against Buffer B, and concentrated with a Millipore Centriprep YM-50 to yield roughly 0.1 mg of MDM2 protein.
Expression and purification of Δ1–166 MDM2 in insect cells were carried out with 1 l of infected cells. Extracts were prepared as described for full-length MDM2 and then the dialyzed cell free extract was directly purified over monoclonal antibody 2A10 (Santa Cruz Biotechnology, Inc.) crosslinked to protein A–Sepharose beads. The presence of Δ1–166 MDM2 was monitored by immunoblots with monoclonal antibody 2A10 and rabbit polyclonal antibody H211 (Santa Cruz Biotechnology, Inc.).
Recombinant N‐terminal His6-tagged MDM2 was expressed in E.coli and purified using QIAexpress Protein Purification System (Qiagen) including a Ni-NTA column according to the manufacturers’ protocols. The purified protein was dialyzed against 50 mM HEPES–KOH (pH 7.5), 10% glycerol, 150 mM NaCl2 and 0.1% Triton X-100.
DNA polymerases
DNA polymerase ε was purified from 40 l of HeLa cells essentially as described by Nishida et al. (26), except that a HiLoad 20/60 Superdex 200 column (Amersham Biosciences) was used in place of glycerol gradient sedimentation. Human DNA polymerase β was expressed in E.coli and purified using DEAE–cellulose and phosphocellulose column chromatography. Klenow fragment of E.coli DNA polymerase I was purchased from Invitrogen.
DNA polymerase assays
DNA polymerase ε activity was assayed as described (26) in 50-µl reactions containing 50 mM HEPES–KOH (pH 7.5), 15 mM MgCl2, 10 mM DTT, 20% glycerol, 0.2 mg/ml acetylated BSA, 50 µM [3H]dTTP and 40 µM poly(dA) hybridized to 4 µM oligo(dT)16 (nucleotides residues; Midland Certified Reagent Co.) according to Crute et al. (27). DNA polymerase β and E.coli DNA polymerase I were assayed as described previously (28) in 50-µl reaction mixtures containing 12.5 µg of activated calf thymus DNA (29), 50 mM Tris–HCl (pH 7.5), 7.5 mM MgCl2, 0.5 mM DTT, 0.2 mg/ml acetylated BSA and 50 µM each of dATP, dCTP, dGTP and [3H]dTTP. Ultrapure dNTPs were from Amersham Biosciences and [methyl-3H]dTTP (70–90 Ci/mmol) was from Perkin-Elmer Life Sciences. All polymerase assays were incubated at 37°C for 30 min, except in the kinetic studies for which the incubation time was 10 min. Kinetic constants were estimated from Lineweaver–Burke and Eadie–Hofstee plots. DNA polymerase assays containing MDM2 were preceded by incubating the polymerase with MDM2 for 30 min or 1 h on ice prior to the addition of the remaining reactants and incubation at 37°C.
Preparation of the isolated C-terminal domain of DNA polymerase ε p261
The cDNA corresponding to p261 C-terminal amino acids 1306–2285 was subcloned into the pFastBac HTa donor plasmid (Bac-to-Bac, Invitrogen) by PCR. The resulting construct (pFBHTapolεC) encoded a His6-tag on the 5′-end of the cDNA and was transformed by heat shock into BH10Bac cells in order to transpose the cDNA into the viral bacmid. After selection, bacmid DNA was prepared from four white colonies according to the Bac-to-Bac manual protocol. Two of the colonies contained bacmid DNA of the appropriate size that also encoded C-terminal sequence of DNA polymerase ε as judged by PCR and sequencing. This DNA was transfected into Sf9 insect cells using lipofectin (Invitrogen), and the resulting virus was harvested and amplified.
A total of 12 l of Sf9 insect cells were infected with the virus for 48–72 h and harvested on five different days over a 2-week period. Cultures were centrifuged at 1000 g for 15 min and the cell pellets were washed once with cold PBS, centrifuged as above and then stored at –80°C. The frozen pellets (128 g) were thawed, resuspended in 500 ml of Resuspension Buffer [50 mM Tris–HCl (pH 8.5), 100 mM KCl, 5 mM 2-mercaptoethanol, 1% NP-40, Complete Protease Inhibitor] and centrifuged at 10 000 g for 20 min. The cell pellet was resuspended in 100 ml of Resuspension Buffer by stirring for several hours at 4°C, and the suspension was sonicated 3 × 10 s with a Branson Sonifier at maximum power in a glass beaker on ice and then centrifuged at 10 000 g for 20 min. The pellet was discarded and the supernatant was centrifuged at 100 000 g for 1 h. One milliliter of Ni-NTA resin that had been equilibrated in Resuspension Buffer was added to the supernatant and the suspension was rotated for 2 days at 4°C. The resin was packed into a FPLC column (Pharmacia) and washed with 10 ml of Start Buffer [20 mM Tris–HCl (pH 8.5), 500 mM KCl, 20 mM imidazole, 5 mM 2-mercaptoethanol, 10% glycerol]. The DNA polymerase ε fragment was eluted with a 10-ml gradient from 20 to 500 mM imidazole and 95 0.4-ml fractions were collected. Fractions 34–41 were pooled and dialyzed into Immunoprecipitation Buffer [20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, 10% glycerol].
Glycerol gradient sedimentation
Two hundred microliter samples were layered on top of four 5-ml glycerol gradients [25–45% (v/v)] containing 130 mM potassium phosphate (pH 7.5), 4 mM DTT, 0.05% Triton X-100 and centrifuged for 30 h at 49 000 r.p.m. in a Beckman SW 50.1 rotor at 4°C. Gradient 1 was layered with 0.5 U of HeLa DNA polymerase ε and 0.5 mg of hemoglobin, gradient 2 with MDM2 and 2.5 mg of catalase, gradient 3 with 0.5 U of HeLa DNA polymerase ε premixed with MDM2, and gradient 4 with 2.5 mg of catalase and 0.5 mg of hemoglobin. Twenty-drop fractions were collected from the bottom of the tubes. The positions of protein markers were detected by protein assay and A410; that of DNA polymerase ε by assay and by immunoblots with HeLa DNA polymerase ε antibodies 3C5.1 and 3A5.6. The positions of MDM2 were determined by immunoblots with the SMP-14 antibody.
Protein estimations
Total protein was determined by the method of Bradford (30). The amounts of individual MDM2, C-terminal p261 fragment, and HeLa DNA polymerase ε p261 protein were estimated from SDS–acrylamide gels stained with Brilliant Blue G-Colloidal (Sigma). BSA was used as a standard for comparison. Gels were scanned and images were quantitated using ImageQuant v. 1.2 for Macintosh.
RESULTS
Properties of purified recombinant human MDM2 from insect cells
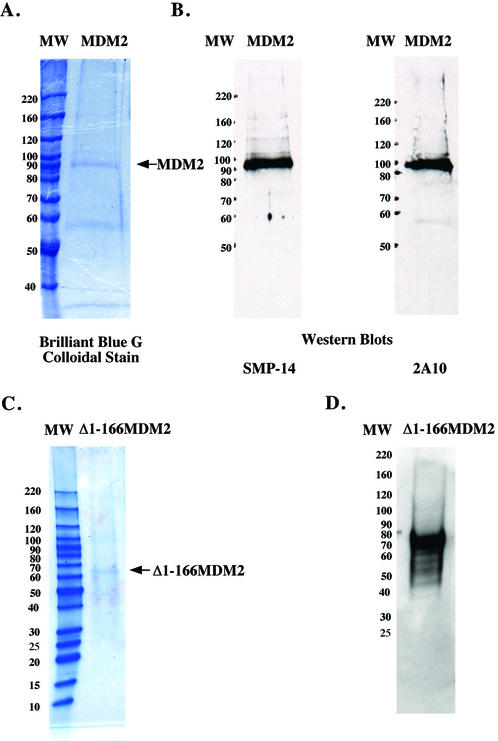
MDM2 was overexpressed and purified from Sf9 insect cells as described in Materials and Methods. The MDM2 was monitored for purification by SDS–PAGE and immunoblots with antibody SMP-14 that recognizes an epitope between amino acids 154 and 167 (Fig. 1). The recombinant MDM2 represented roughly 30% of the protein in the preparation. The two smaller proteins corresponding to 59 and 34 kDa that are visible in the gel stained with Brilliant Blue G Colloidal (Fig. 1A) are unlikely to be degradation products of MDM2, since they are not recognized by the SMP-14 or 2A10 antibodies, whose epitopes lie between amino acids 154–167 and 295–330, respectively (Fig. 1B). The MDM2 migrated to a position corresponding to 90 kDa during SDS–PAGE (Fig. 1). Although the human MDM2 gene codes for a protein of 491 amino acids with a theoretical molecular weight of ∼55 kDa, its acidic domain causes the protein to migrate to a position corresponding to 90 kDa during SDS–PAGE (31).
Figure 1.
SDS–PAGE of human MDM2 and Δ1–166 MDM2 expressed in insect cells. Purified MDM2 (roughly 0.6 µg of total protein) was subjected to SDS–PAGE on 7.5% polyacrylamide gels and then stained with Brilliant Blue G Colloidal stain (A) or electroblotted onto a nitrocellulose filter and probed with anti-MDM2 antibodies SMP-14 or 2A10 whose epitopes are amino acid 154–167 and 295–330, respectively (B). Purified Δ1–166 MDM2 was subjected SDS–PAGE on 4–20% linear gradient polyacrylamide gel and stained with Brilliant Blue G Colloidal stain (roughly 0.96 µg of total protein) (C) or electroblotted onto a nitrocellulose filter and probed with antibody 2A10 (roughly 0.48 µg of total protein) (D). Molecular weight markers were the Invitrogen BenchMark™ Protein Ladder.
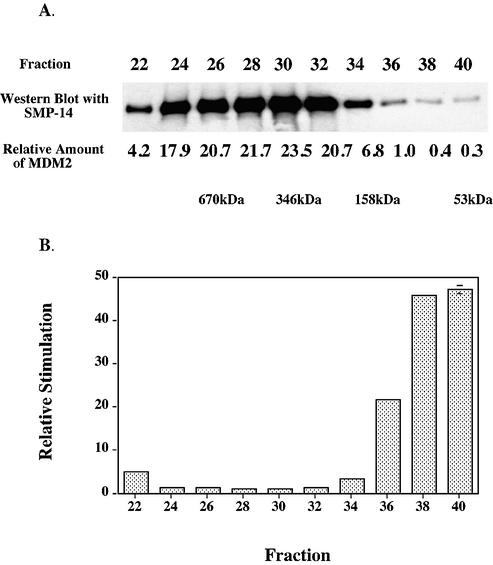
When a sample of recombinant MDM2 purified through DEAE–Sephacel was chromatographed upon an analytical Superdex 200 HR 10/30 column, it eluted as a broad band with calculated molecular weights (assuming globular shapes for the marker and sample proteins) of the major fractions ranging from 192 000 to 106 with the peak fraction corresponding to 346 kDa, as judged by immunoblotting with the SMP-14 antibody (Fig. 2A). Clearly most of the purified MDM2 was aggregated (see also sedimentation velocity studies below). When immunopurified MDM2 was similarly chromatographed, a similar elution pattern was obtained (data not shown).
Figure 2.
Chromatography of recombinant MDM2 upon Superdex. MDM2 expressed in insect cells was purified through chromatography upon DEAE–Sephacel as described in Materials and Methods, and fractions containing MDM2 were concentrated using a Millipore Centriprep YM-50 and then chromatographed upon Superdex 200 HR 10/30 (Amersham Biosciences). (A) Equal volumes of samples were analyzed by immunoblots using antibody SMP-14. (B) The Superdex fractions were assayed for DNA polymerase ε simulating activity such that each fraction stimulated between 2- and 4-fold. The amounts of MDM2 in each fraction were quantitated from the immunoblot (with a lesser exposure time than the one shown) using the program ImageQuant for Macintosh v. 1.2. The fold-stimulations were then normalized for the amount of MDM2 antigen in each fraction and then those ratios were normalized to that of fraction 30 which had the highest amount of MDM2 antigen. The values are averages of duplicate assays and error bars were too small to show except for fraction 40. The Bio-Rad gel filtration standard (Catalog number 151-1901) was used to calibrate the column.
Human MDM2 expressed in insect cells stimulates HeLa DNA polymerase ε in vitro
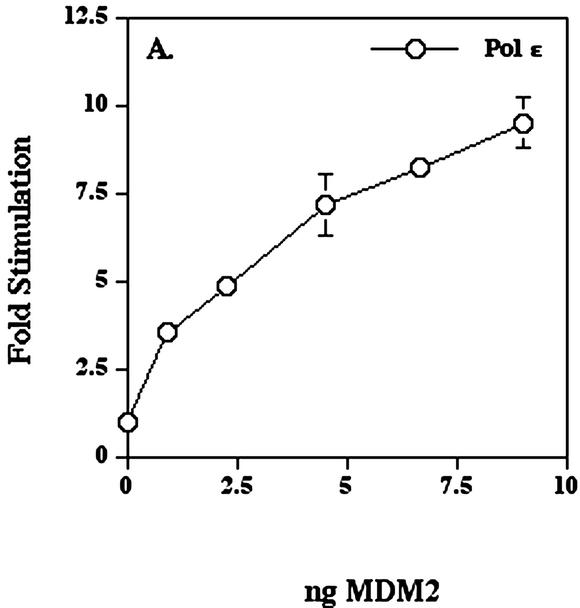
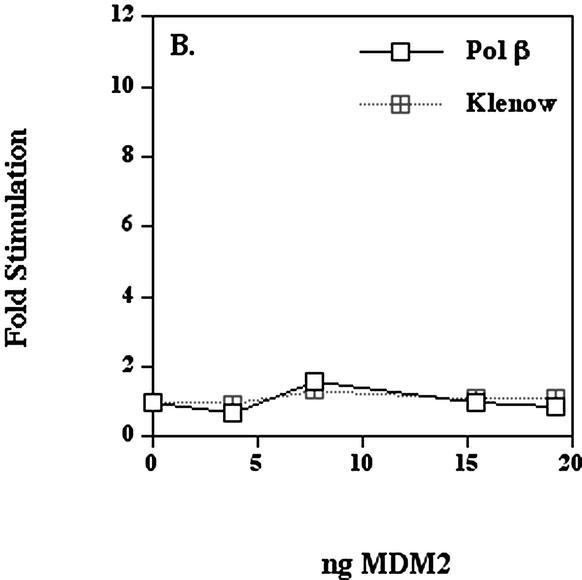
When various amounts of immunopurified human MDM2 were mixed with HeLa DNA polymerase ε on ice for 1 h and then assayed for polymerase activity, the DNA polymerase ε activity increased up to 10-fold in a dose-dependent manner (Fig. 3A). MDM2 fractions, when assayed alone, did not have significant polymerase activity (data not shown). The molar ratio of MDM2 to HeLa pol ε p261 was approximately 60:1 at a 10-fold stimulation. This apparently large molar excess of MDM2 over p261 could have a number of explanations. For example, the MDM2 eluted from the Superdex sizing column in aggregated forms (Fig. 2), and very little monomer MDM2 is present which may be the most effective stimulatory form (see also sedimentation velocity studies below). Alternatively, since MDM2 is eluted from protein A antibodies at pH 2.5, it may be partially unfolded. Finally, at these protein concentrations of DNA polymerase ε, MDM2 binding might require higher concentrations due to mass action constraints. The recombinant MDM2 purified from insect cells had very small effects upon the activities of either human DNA polymerase β expressed in E.coli or Klenow fragment of E.coli DNA polymerase I (Fig. 3B).
Figure 3.
Human MDM2 expressed in insect cells stimulates human DNA polymerase ε. (A) DNA polymerase ε (5.8 × 10–3 units) was pre-incubated as indicated with the preparation of MDM2 shown in Figure 1A for 1 h on ice and then the activity was assayed as described in Materials and Methods. (B) The MDM2 was pre-incubated for 1 h on ice with either recombinant human DNA polymerase β expressed in E.coli (7.7 × 10–3 units) or Klenow fragment of E.coli DNA polymerase I (8.8 × 10–3 units) and then assayed. This preparation of MDM2 stimulated the activity of DNA polymerase ε up to 11-fold in the range of 4–20 ng. Values represent the mean of duplicate assays and in most cases error bars would be smaller than the data symbols.
To distinguish whether the pH 2.5 elution from the antibody column affected the stimulation and/or caused aggregation, and to study the effectiveness of aggregated forms upon stimulation, the cell free extract was purified by chromatography only over DEAE–Sephacel, then concentrated, and chromatographed upon Superdex 200 HR 10/30 column (Fig. 2). Those fractions which were most dilute and contained smaller proteins clearly showed the greatest stimulation when normalized for the amount of MDM2 protein, with the monomer form more than 45-fold more effective than the peak of the aggregated fractions. Hence we believe that aggregation is somewhat reversible in dilute fractions, and that the monomer and/or dimer forms are the most effective in stimulating DNA polymerase ε.
The stimulatory effect prior to purification of the MDM2 was unstable and some variability was observed among different preparations of MDM2. This variation of the stimulating activity was also observed with MDM2 expressed in E.coli (see below). However, for both sources, once MDM2 was purified, its stimulating activity was relatively stable.
Human MDM2 expressed in E.coli also stimulates HeLa DNA polymerase ε
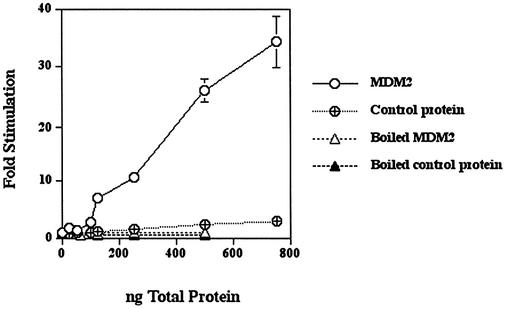
In order to control for stimulation being due to contaminants from the insect cells used for the MDM2 overexpression and purification, recombinant MDM2 was also prepared as an N‐terminal His-tagged protein in E.coli and then purified under denaturing conditions (guanidine–HCl) over a Ni-NTA column. MDM2 was 5–10% of the protein in this preparation. Control E.coli protein was prepared from E.coli that did not contain the MDM2 construct by passing a cell lysate through a Ni-NTA column and eluting the bound protein under the same conditions. The recombinant MDM2 stimulated HeLa DNA polymerase ε up to 40-fold, whereas the control protein had relatively little effect (Fig. 4). Moreover, when the recombinant MDM2 was heated at 100°C, no stimulation occurred. MDM2 expressed in E.coli also did not have polymerase activity when assayed alone (data not shown). This stimulatory effect has been obtained with two different preparations of recombinant human MDM2 from E.coli.
Figure 4.
Human MDM2 expressed in E.coli stimulates HeLa pol ε. Pol ε (4.4–6 × 10–3 U was pre-incubated with the indicated amounts of MDM2, control protein or boiled MDM2 for 30 min on ice and then assayed as described in Materials and Methods. The MDM2 protein was about 5–10% of the total protein in the preparation. Values represent the mean of duplicate assays. In most cases, the error bars were smaller than the data symbols.
Recombinant MDM2 protein from both E.coli (Fig. 4) and insect cells (Fig. 3A) exhibited approximately 10-fold stimulation when an estimated 10 ng of either MDM2 protein (not total protein of the preparation) was added to 6 × 10–3 units of DNA polymerase ε. This similarity makes it unlikely that the stimulation of DNA polymerase ε was due to contaminating protein(s) or other factors from the source cells.
The N‐terminal 166 amino acids of MDM2 are essential for stimulation of human DNA polymerase ε
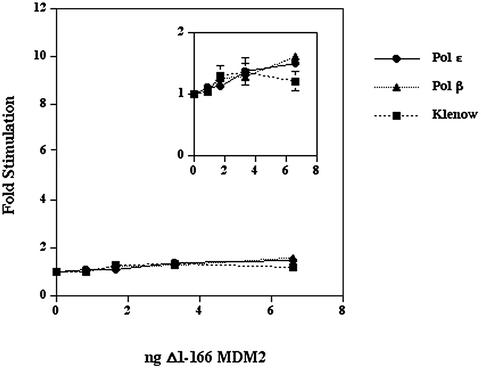
Our previous study showed that the first 166 amino acid residues of MDM2 were sufficient to interact with the C-terminal region of DNA polymerase ε in a yeast two hybrid assay. To confirm that the stimulation of human DNA polymerase ε depended upon the same region of MDM2, we constructed and expressed in insect cells MDM2 which lacked the N‐terminal 166 amino acids. The purification of Δ1–166 MDM2 was monitored by SDS–PAGE and immunoblots with antibodies 2A10 and H221 whose epitopes correspond to amino acids 295–330 and 100–320 of human MDM2, respectively. The purified Δ1–166 MDM2 was ∼30–40% of the protein in the preparation as judged by a gel stained with Brilliant Blue G Colloidal (Fig. 1C) and an immunoblot probed with antibody 2A10 (Fig. 1D). The Δ1–166 MDM2 migrated on SDS–PAGE as a protein of 68 kDa. When this Δ1–166 MDM2 was assayed for stimulation of polymerase activity, it had little effect with DNA polymerase ε, DNA polymerase β or Klenow fragment (Fig. 5). The DNA polymerase ε preparation used in this experiment was stimulated 14-fold by 20 ng of full-length MDM2.
Figure 5.
Effect of Δ1–166 MDM2 upon polymerase activities. DNA polymerase ε (12 × 10–3 U), DNA polymerase β (32 × 10–3 U) or Klenow fragment of E.coli DNA polymerase I (19 × 10–3 units) were pre-incubated with the indicated amounts of purified Δ1–166 MDM2 on ice for 1 h and then assayed. The inset is a magnification of the data. Values represent the mean of duplicate assays for DNA polymerase β and Klenow fragment and the mean of triplicate assays for DNA polymerase ε. Error bars were generally smaller than the data symbols.
MDM2 has little effect upon the apparent KM values of human DNA polymerase ε
In order to study how MDM2 stimulated the activity of HeLa DNA polymerase ε, the effects of MDM2 upon the apparent KM values for dTTP and poly(dA)·oligo(dT) primer/template were determined. The KM for dTTP remained unchanged at 3–4 µM and that for the primer/template was also little changed between 7 and 14 µM (total residues of dAMP plus dTMP). The latter value varied depending upon the particular polymer pair, presumably because of their somewhat varied chain lengths. Therefore, it appears that the effect of MDM2 is directly upon the velocity of the reaction rather than upon the substrate affinities.
Interaction with the C-terminal domain of full-length DNA polymerase ε p261 is essential for the stimulation by MDM2
The region between the N- and C-terminal domains of p261 is susceptible to proteolysis by trypsin (19), caspase III during apoptosis (32) and endogenous proteases (Fig. 6A). In fact, limited trypsin digestion forms a catalytic 122-kDa N‐terminal domain with roughly twice the activity of the full-length protein (Fig. 6B) (19,32). Since the sequences of human DNA polymerase ε p261 that are necessary and sufficient for interaction with MDM2 were mapped to the 380 amino acids closest to the C-terminus (residues 1906–2285) (Fig. 6A) (15), the effect of MDM2 upon the N‐terminal active domain cleaved from the C-terminal domain was tested.
Purified DNA polymerase ε was treated with varying amounts of trypsin, then part of each reaction mixture was separated by SDS–PAGE and the proteins were transferred to a nitrocellulose filter which was probed with an antibody against the N‐terminal domain of p261 (Fig. 6B). The N‐terminal domain remained intact and the catalytic activity was greater or equal to that of the uncleaved p261 after digestion by 1–4 ng of trypsin. However, when the polymerase activity of these truncated proteins was assayed in the presence of recombinant human MDM2 expressed in E.coli, the trypsin-digested DNA polymerase ε was only slightly stimulated by MDM2, in contrast to the more than 15-fold stimulation of the full-length protein (Fig. 6C). This observation clearly demonstrates that the stimulation of HeLa DNA polymerase ε by MDM2 is dependent upon the C-terminal domain of DNA polymerase ε catalytic subunit being joined to the catalytic, N‐terminal domain.
Free C-terminal domain of DNA polymerase ε p261 is able to overcome stimulation of intact p261 by MDM2
To verify that MDM2 binding to the C-terminal domain of intact p261 was truly necessary for stimulation, the C-terminal domain of DNA polymerase ε p261 (amino acids 1306–2285) was expressed as an N‐terminal His-tagged protein in insect cells and purified over Ni-NTA resin. This purified C-terminal fragment was pre-incubated with MDM2 on ice for 30 min prior to addition of DNA polymerase ε. The mixture was incubated on ice for an additional 30 min and then assayed for polymerase activity (Table 1). Indeed, the C-terminal DNA polymerase ε p261 fragment was able to suppress the stimulation by MDM2 in a dose-dependent manner. This result, combined with the result of trypsin digestion of HeLa DNA polymerase ε (Fig. 6), further confirms the finding of Vlatkovic et al. (15) that the MDM2 interacts with the C-terminal domain of DNA polymerase ε p261.
Table 1. Free C-terminal domain can overcome the stimulation of intact p261 by MDM2.
| DNA polymerase ε | 10 ng MDM2 | ng C-terminal domain | pmol dNTP incorporated |
|---|---|---|---|
| – | – | 85 | <0.5 |
| – | + | – | <0.5 |
| + | – | – | 1.1 ± 0.5 |
| + | + | – | 14.7 ± 0.4 |
| + | + | 51 | 11.3 ± 0.4 |
| + | + | 68 | 7.5 ± 0.6 |
| + | + | 85 | 4.6 ± 0.6 |
| + | – | 85 | 6.0 ± 0.5 |
Human MDM2 protein expressed in insect cells (10 ng) was mixed with various amounts of C-terminal p261 fragment on ice for 30 min, then native DNA polymerase ε was added and the mixture was incubated further on ice for 30 min, and the samples were assayed as described in Materials and Methods. (Protein amounts are estimated MDM2 protein, not total protein.) The values represent the mean of duplicates in each of two independent experiments.
It is interesting to note that the C-terminal domain stimulated DNA polymerase ε in the absence of MDM2 (Table 1). While we do not have a precise explanation for this effect, DNA polymerase ε generally exists as a dimer (16,33) and should this dimerization be mediated through the C-terminal domain of p261 (directly or indirectly), excess C-terminal domain could monomerize DNA polymerase ε and/or possibly sequester away its other subunits so as to stimulate the enzyme.
Sedimentation of HeLa DNA polymerase ε with MDM2
In order to estimate the approximate size of the stimulated MDM2–DNA polymerase ε complex, recombinant human MDM2 and DNA polymerase ε were incubated on ice for 1 h and then sedimented through a glycerol gradient. DNA polymerase ε alone, MDM2 alone and protein markers were sedimented through parallel gradients. DNA polymerase ε activity sedimented at a rate of roughly 10.4 S when sedimented alone as reported for its dimeric form (16), while MDM2 alone sedimented at 10.8 S, confirming the considerable aggregation noted during Superdex chromatography. When DNA polymerase ε and MDM2 were mixed together then sedimented, the polymerase activity peaked at 10.5 S and immunoblot results confirmed that the MDM2 center of mass sedimented to a slightly heavier position (roughly 11.3 S). Approximately 0.5 U of HeLa DNA polymerase ε were loaded onto each gradient (as assayed without MDM2) and approximately 0.28 and 0.85 U of activity were recovered from the gradients without and with MDM2, respectively. Together, these results imply that MDM2 does not bind to DNA polymerase ε as a large aggregate, but in a form which adds little to the apparent mass of the DNA polymerase ε dimer.
DISCUSSION
The physical interaction between human DNA polymerase ε and human MDM2 had previously been established by a yeast two hybrid screen, an in vitro binding assay and co-immunoprecipitation (15). We have now shown that human MDM2 expressed in and purified from either E.coli or insect cells stimulates the activity of DNA polymerase ε in vitro. Moreover, the C-terminal domain of DNA polymerase ε p261, to which MDM2 binds, and the N‐terminal 166 amino acids of MDM2 to which DNA polymerase ε binds are both essential for the stimulation.
What could be the biological significance of an interaction between MDM2 and mammalian DNA polymerase ε? Human DNA polymerase ε was first isolated as a factor required for long-patch repair synthesis in UV-irradiated permeabilized human diploid fibroblasts (26). DNA polymerase ε has been reported to participate in NER in mammalian cells (34,35) and BER in S.cerevisiae (36) and it is a component of a recombination protein complex (RC-1) from calf thymus nuclei that catalyzes the recombinational repair of in vitro gaps and deletions in DNA (37,38). Additionally, the involvement of DNA polymerase ε in chromosomal DNA replication in yeast and higher eukaryotes has been established (39–44). Saccharomyces cerevisiae DNA polymerase ε also acts as a sensor of DNA damage and other replication blocks, resulting in activation of the S phase checkpoint in a process that requires the C-terminus of the catalytic subunit (45,46). Strangely, however, in S.cerevisiae, mutants lacking the entire N‐terminal domain are viable (though point mutations in the domain are lethal), whereas deletion of the C-terminal domain is lethal (20,21,47). Mutations within the C-terminal zinc finger region block DNA replication, are sensitive to the DNA replication inhibitor hydroxyurea and to the DNA cross-linking agent MMS, and are defective in the dimerization and the assembly of the DNA polymerase ε holoenzyme (21,48). This C-terminal region is also the site to which MDM2 binds the human DNA polymerase ε homolog, so that the MDM2 binding might be part of a process for the regulation of DNA replication and checkpoint responses in mammalian cells that is functionally similar to that found in yeast.
It is currently proposed that in budding yeast the signal for the intra-S checkpoint is generated only when replication forks encounter DNA damage, whereas in fission yeast and mammals the same response requires two signaling pathways, one that detects stalled replication forks and another that detects damaged sites (49). It is currently believed that DNA polymerase ε is both a component of the replication machinery and a sensor of stalled replication forks. Presumably, when DNA polymerase ε encounters damage or when a replication fork is otherwise blocked, the replication complex would be reorganized with the recruitment of recombination/repair proteins. DNA polymerase ε foci in human cells are adjacent to PCNA and BrdU pulse foci early in S phase, but become colocalized with PCNA and sites of DNA synthesis late in S phase (44). In view of this observation, it was proposed that DNA polymerase ε is involved in replication-associated repair that either precedes or follows replication forks early in S phase and then takes part in the replication of heterochromatin late in S phase (44). Since DNA polymerase ε appears to be involved in both DNA replication and in DNA repair synthesis, it would be reasonable to propose that it is a stable factor in this transition, being part of both processes. MDM2 binding might then be part of this transition process, perhaps displacing other proteins from DNA polymerase ε so as to allow reconfiguration from a replication to a repair complex. Ultimately, alterations in post-translational modifications of MDM2 might then alter its binding to DNA polymerase ε or lead to its diminution so as to allow the re-establishment of DNA polymerase ε-containing replication complexes.
Roles proposed for DNA polymerase ε include DNA repair, recombination, replication, damage sensing and chromatin remodeling. Thus there may be as many as five distinct functional DNA polymerase ε complexes, making dissection of the compositions, interconversions and roles of the complexes containing DNA polymerase ε particularly difficult. Nevertheless, the results presented here suggest that in response to stress, MDM2 may have an important role to play in regulating or modifying DNA polymerase ε function.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ms Ann Fischer for expert tissue culture aid. This research was supported in part by Grants 1RO1GM30415, 1RO1GM59424, 1R29CA70165 and P30ES08196 from the National Institutes of Health and Grant 26/C15088 from the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Fakharzadeh S.S., Trusko,S.P. and George,D.L. (1991) Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J., 10, 1565–1569. [DOI] [PMC free article] [PubMed]
- 2.Finlay C.A. (1993) The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol. Cell. Biol., 13, 301–306. [DOI] [PMC free article] [PubMed]
- 3.Oliner J.D., Kinzler,K.W., Meltzer,P.S., George,D.L. and Vogelstein,B. (1992) Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature, 358, 80–83. [DOI] [PubMed]
- 4.Maya R., Balass,M., Kim,S.T., Shkedy,D., Leal,J.F., Shifman,O., Moas,M., Buschmann,T., Ronai,Z., Shiloh,Y., Kastan,M.B., Katzir,E. and Oren,M. (2001) ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev., 15, 1067–1077. [DOI] [PMC free article] [PubMed]
- 5.Haupt Y., Maya,R., Kazaz,A. and Oren,M. (1997) Mdm2 promotes the rapid degradation of p53. Nature, 387, 296–299. [DOI] [PubMed]
- 6.Kubbutat M.H., Ludwig,R.L., Levine,A.J. and Vousden,K.H. (1999) Analysis of the degradation function of Mdm2. Cell Growth Differ., 10, 87–92. [PubMed]
- 7.Wu X., Bayle,J.H., Olson,D. and Levine,A.J. (1993) The p53-mdm-2 autoregulatory feedback loop. Genes Dev., 7, 1126–1132. [DOI] [PubMed]
- 8.Juven-Gershon T., Shifman,O., Unger,T., Elkeles,A., Haupt,Y. and Oren,M. (1998) The Mdm2 oncoprotein interacts with the cell fate regular Numb. Mol. Cell. Biol., 18, 3972–3982. [DOI] [PMC free article] [PubMed]
- 9.Martin K., Trouche,D., Hagemeier,C., Sorensen,T.S., LaThangue,N.B. and Kousarides,T. (1995) Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature, 375, 691–694. [DOI] [PubMed]
- 10.Thut C.J., Goodrich,J.A. and Tjian,R. (1997) Repression of P53-mediated transcription by MDM2: a dual mechanism. Genes Dev., 11, 1974–1986. [DOI] [PMC free article] [PubMed]
- 11.Brown D.R., Deb,S., Munoz,R.M., Subler,M.A. and Deb,S.P. (1993) The tumor suppressor p53 and the oncoprotein simian virus 40 T antigen bind to overlapping domains on the MDM2 protein. Mol. Cell. Biol., 13, 6849–6857. [DOI] [PMC free article] [PubMed]
- 12.Lundgren F., Montes de Oca Luna,R., McNeill,Y.D., Emerick,D.P., Spencer,B., Barfield,C.R., Lozano,G., Rosenberg,M.P. and Finlay,C.A. (1997) Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development indepsndent of p53. Genes Dev., 11, 714–725. [DOI] [PubMed]
- 13.Reinke V., Borner,D.M., Amelse,L.L., Lundgren,K., Rosenberg,M.P., Finlay,C.A. and Lozano,G. (1999) Overproduction of MDM2 in vivo disrupts S phase independent of E2F1. Cell Growth Differ., 10, 147–154. [PubMed]
- 14.Sigalas I., Calvert,A.H., Anderson,J.J., Neal,D.E. and Lunec,J. (1996) Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat. Med., 2, 912–917. [DOI] [PubMed]
- 15.Vlatkovic N., Guerrera,S., Li,Y., Linn,S., Haines,D.S. and Boyd,M.T. (2000) MDM2 interacts with the C-terminus of the catalytic subunit of DNA polymerase epsilon. Nucleic Acids Res., 28, 3581–3586. [DOI] [PMC free article] [PubMed]
- 16.Syvaoja J. and Linn,S. (1989) Characterization of a large form of DNA polymerase delta from HeLa cells that is insensitive to proliferating cell nuclear antigen. J. Biol. Chem., 264, 2489–2497. [PubMed]
- 17.Li Y., Asahara,H., Patel,V.S., Zhou,S. and Linn,S. (1997) Purification, cDNA cloning, and gene mapping of the small subunit of human DNA polymerase epsilon. J. Biol. Chem., 272, 32337–32344. [DOI] [PubMed]
- 18.Li Y., Pursell,Z.F. and Linn,S. (2000) Identification and cloning of two histone fold motif-containing subunits of HeLa DNA polymerase epsilon. J. Biol. Chem., 275, 23247–23252. [DOI] [PubMed]
- 19.Kesti T. and Syvaoja,J.E. (1991) Identification and tryptic cleavage of the catalytic core of HeLa and calf thymus DNA polymerase epsilon. J. Biol. Chem., 266, 6336–6341. [PubMed]
- 20.Kesti T., Flick,K., Keranen,S., Syvaoja,J.E. and Wittenberg,C. (1999) DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell, 3, 679–685. [DOI] [PubMed]
- 21.Dua R., Levy,D.L. and Campbell,J.L. (1999) Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem., 274, 22283–22288. [DOI] [PubMed]
- 22.Araki H., Hamatake,R.K., Morrison,A., Johnson,A.L., Johnston,L.H. and Sugino,A. (1991) Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res., 19, 4867–4872. [DOI] [PMC free article] [PubMed]
- 23.Ohya T., Maki,S., Kawasaki,Y. and Sugino,A. (2000) Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res., 28, 3846–3852. [DOI] [PMC free article] [PubMed]
- 24.Baxevanis A.D. and Landsman,D. (1997) Histone and histone fold sequences and structures: a database. Nucleic Acids Res., 25, 272–273. [DOI] [PMC free article] [PubMed]
- 25.Poot R.A., Dellaire,G., Hulsmann,B.B., Grimaldi,M.A., Corona,D.F., Becker,P.B., Bickmore,W.A. and Varga-Weisz,P.D. (2000) HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J., 19, 3377–3387. [DOI] [PMC free article] [PubMed]
- 26.Nishida C., Reinhard,P. and Linn,S. (1988) DNA repair synthesis in human fibroblasts requires DNA polymerase delta. J. Biol. Chem., 263, 501–510. [PubMed]
- 27.Crute J.J., Wahl,A.F. and Bambara,R.A. (1986) Purification and characterization of two new high molecular weight forms of DNA polymerase delta. Biochemistry, 25, 26–36. [DOI] [PubMed]
- 28.Linn S., Kairis,M. and Holliday,R. (1976) Decreased fidelity of DNA polymerase activity isolated from aging human fibroblasts. Proc. Natl Acad. Sci. USA, 73, 2818–2822. [DOI] [PMC free article] [PubMed]
- 29.Schlabach A., Fridlender,B., Bolden,A. and Weissbach,A. (1971) DNA-dependent DNA polymerases from HeLa cell nuclei. II. Template and substrate utilization. Biochem. Biophys. Res. Commun., 44, 879–885. [DOI] [PubMed]
- 30.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed]
- 31.Chen J., Marechal,V. and Levine,A.J. (1993) Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol., 13, 4107–4114. [DOI] [PMC free article] [PubMed]
- 32.Liu W. and Linn,S. (2000) Proteolysis of the human DNA polymerase varepsilon catalytic subunit by caspase-3 and calpain specifically during apoptosis. Nucleic Acids Res., 28, 4180–4188. [DOI] [PMC free article] [PubMed]
- 33.Dua R., Edwards,S., Levy,D.L. and Campbell,J.L. (2000) Subunit interactions within the Saccharomyces cerevisiae DNA polymerase epsilon (pol epsilon) complex. Demonstration of a dimeric pol epsilon. J. Biol. Chem., 275, 28816–28825. [DOI] [PubMed]
- 34.Aboussekhra A., Biggerstaff,M., Shivji,M.K., Vilpo,J.A., Moncollin,V., Podust,V.N., Protic,M., Hubscher,U., Egly,J.M. and Wood,R.D. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. [DOI] [PubMed]
- 35.Shivji M.K., Podust,V.N., Hubscher,U. and Wood,R.D. (1995) Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry, 34, 5011–5017. [DOI] [PubMed]
- 36.Wang Z., Wu,X. and Friedberg,E.C. (1993) Nucleotide-excision repair of DNA in cell-free extracts of the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 4907–4911. [DOI] [PMC free article] [PubMed]
- 37.Jessberger R., Podust,V., Hubscher,U. and Berg,P. (1993) A mammalian protein complex that repairs double-strand breaks and deletions by recombination. J. Biol. Chem., 268, 15070–15079. [PubMed]
- 38.Jessberger R., Chui,G., Linn,S. and Kemper,B. (1996) Analysis of the mammalian recombination protein complex RC-1. Mutat. Res., 350, 217–227. [DOI] [PubMed]
- 39.Budd M.E. and Campbell,J.L. (1993) DNA polymerases delta and epsilon are required for chromosomal replication in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 496–505. [DOI] [PMC free article] [PubMed]
- 40.Zlotkin T., Kaufmann,G., Jiang,Y., Lee,M.Y., Uitto,L., Syvaoja,J., Dornreiter,I., Fanning,E. and Nethanel,T. (1996) DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. EMBO J., 15, 2298–2305. [PMC free article] [PubMed]
- 41.D’Urso G. and Nurse,P. (1997) Schizosaccharomyces pombe cdc20+ encodes DNA polymerase epsilon and is required for chromosomal replication but not for the S phase checkpoint. Proc. Natl Acad. Sci. USA, 94, 12491–12496. [DOI] [PMC free article] [PubMed]
- 42.Pospiech H., Kursula,I., Abdel-Aziz,W., Malkas,L., Uitto,L., Kastelli,M., Vihinen-Ranta,M., Eskelinen,S. and Syvaoja,J.E. (1999) A neutralizing antibody against human DNA polymerase epsilon inhibits cellular but not SV40 DNA replication. Nucleic Acids Res., 27, 3799–3804. [DOI] [PMC free article] [PubMed]
- 43.Waga S., Masuda,T., Takisawa,H. and Sugino,A. (2001) DNA polymerase varepsilon is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc. Natl Acad. Sci. USA, 98, 4978–4983. [DOI] [PMC free article] [PubMed]
- 44.Fuss J. and Linn,S. (2002) Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem., 277, 8658–8666. [DOI] [PubMed]
- 45.Navas T.A., Zhou,Z. and Elledge,S.J. (1995) DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell, 80, 29–39. [DOI] [PubMed]
- 46.Navas T.A., Sanchez,Y. and Elledge,S.J. (1996) RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev., 10, 2632–2643. [DOI] [PubMed]
- 47.Ohya T., Kawasaki,Y., Hiraga,S.I., Kanbara,S., Nakajo,K., Nakashima,N., Suzuki,A. and Sugino,A. (2002) The DNA polymerase domain of Pol epsilon is required for rapid, efficient and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem., 277, 28099–28108. [DOI] [PubMed]
- 48.Dua R., Levy,D.L. and Campbell,J.L. (1998) Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase epsilon in DNA replication and the S/M checkpoint pathway. J. Biol. Chem., 273, 30046–30055. [DOI] [PubMed]
- 49.Marchetti M.A., Kumar,S., Hartsuiker,E., Maftahi,M., Carr,A.M., Freyer,G.A., Burhans,W.C. and Huberman,J.A. (2002) A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc. Natl Acad. Sci. USA, 99, 7472–7477. [DOI] [PMC free article] [PubMed]