Abstract
By employing purified transcription factors and RNA polymerase III (pol III), we generated active pol III transcription complexes on the human 5S rRNA gene. These large complexes were separated by size exclusion chromatography from non- incorporated proteins. In addition, we succeeded in isolating specific intermediate stages of complex formation. Such isolated partial complexes require complementation with the missing activities for full transcription activity. One central finding is that a 5S DNA–TFIIIA–TFIIIC2–TFIIIBβ complex could be isolated which had been assembled in the absence of the general pol III transcription factor IIIC1. Thus TFIIIC1 is not an assembly factor for other transcription factors. Although pol III has the potential to bind unspecifically to DNA, such polymerase molecules cannot be rendered initiation competent by direct recruitment to a 5S DNA–TFIIIA–TFIIIC2– TFIIIBβ complex, but this process strictly requires additional TFIIIC1 activity. This clearly demonstrates that in contrast to yeast cells, hTFIIIB(β), although required, does not suffice for the functional recruitment of polymerase III. These data document that TFIIIC1 is the second transcription factor required for the recruitment of pol III in mammalian cells.
INTRODUCTION
The human gene encoding 5S rRNA is transcribed by RNA polymerase III (pol III). The three promoter elements forming the ‘internal control region’ (ICR) are located within the coding region of this gene [reviewed in Paule and White (1)].
The first step of the assembly pathway of a transcription complex on the 5S gene is the binding of the 42 kDa transcription factor IIIA (TFIIIA) to the ICR (2,3). Once assembled, this binary complex is enlarged by the 600 kDa multisubunit transcription factor IIIC2 that protects the DNA up to the start point of transcription (4).
Two other transcription factors are required for transcription of the 5S gene, the TBP–TAF complex TFIIIBβ, which is essential for all pol III genes regulated by internal promoters (5), and the general pol III transcription factor IIIC1 (4,6,7). TFIIIBβ interacts with subunits of pol III, and these interactions have been proposed to play an important role in the recruitment of the polymerase into the transcription complex (8).
While the molecular structures of TFIIIA, TFIIIC2 and TFIIIBβ are partly established (reviewed in Paule and White (1), Geiduschek and Kassavetis (9), Huang and Maraia (10)], the subunit composition of TFIIIC1, which is presumably 200–300 kDa in size (7), still remains undetermined. Although TFIIIC1 is required for the transcription of all pol III genes (4,6), its function within the transcription complex remains unclear. Nevertheless, we previously could show that TFIIIC1 plays an important role in the regulation of pol III transcription during differentiation of mouse F9 embryonal carcinoma cells (11).
In the experiments presented here, we have generated partial as well as fully assembled 5S transcription complexes from pre-purified components and have separated such complexes from non-incorporated factors using size exclusion chromatography. Such partial complexes can be complemented for full transcriptional activity with the missing activities and can be used to study the order in which individual components are integrated into the complex.
While TFIIIB is sufficient in yeast for the recruitment of pol III for multiple rounds of transcription (12), the data presented here demonstrate that in human cells, this process has gained a higher degree of complexity. The evolutionarily novel activity TFIIIC1 is not an ‘assembly factor’ for TFIIIBβ but is required together with TFIIIBβ for the functional recruitment of the polymerase to the 5S gene.
MATERIALS AND METHODS
Plasmids
The plasmid pBEhe5S wt carries the complete coding region of the human genomic 5S gene, as well as 75 bases of the downstream region beyond the terminator and 34 bases of the adjacent upstream region of the gene, including the D-box that is essential for an efficient 5S transcription (13). This plasmid was transcribed in comparison with the formerly used pBh5S and was found to be equally active in transcription (14).
Buffers
Buffer A contained: 20 mM HEPES pH 7.9, 10% (v/v) glycerol, 3 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF). Buffer B contained: 20 mM Tris–HCl pH 7.9, 10% (v/v) glycerol, 5 mM MgCl2, 3 mM DTT, 0.2 mM PMSF. All fractions employed were dialysed against buffer B including 60 mM KCl before use, except for the polymerase fraction (see below). After dialysis, the total protein concentration of each fraction was determined by a colorimetric protein assay (Bio-Rad).
Preparation of cytoplasmic extract (HEK S100) and purification of transcription factors
Cytoplasmic extract from human embryonic kidney cells (HEK S100) was prepared as described (15). The S100 extract was dialysed against buffer A including 100 mM KCl and subsequently was chromatographed through phosphocellulose (Whatman P11) into fractions PCA, PCB, PCC and PCD according to Seifart et al. (3) and Jahn et al. (16).
TFIIIA was purified from the PCA fraction by re-chromatography as described (3), and had a protein concentration of 0.3 mg/ml.
All further purification steps were conducted using buffer B with various KCl concentrations: TFIIIC1 and TFIIIC2 were purified from the PCC fraction by MonoQ chromatography as described (4). TFIIIC1 was purified further and concentrated by an EMD SO3– (Merck) chromatography step. TFIIIC1 eluted in the 400–600 mM KCl fraction, which contained 0.3 mg/ml protein.
To separate TFIIIC2 from possible impurities of polymerase, this fraction was loaded onto a 1 ml phosphocellulose column. TFIIIC2 eluted in the 450–600 mM step; the fraction contained 0.1 mg/ml protein and was free of polymerase activity or cross-contamination with other transcription factors.
The PCB fraction was separated into fractions containing pol III, TFIIIBβ and TFIIIBα as described previously by EMD DEAE Fractogel (Merck) chromatography (5). TFIIIBβ was purified further by MonoQ chromatography, eluting in the 280–300 mM fraction, which contained 0.4 mg/ml protein.
The polymerase fraction was purified by single-stranded DNA–cellulose as described (5) and diluted with glycerol to a final concentration of 50% (v/v) glycerol. The protein concentration was 0.1 mg/ml.
In vitro transcription
Standard transcription without pre-incubation. The in vitro transcription mixtures contained the appropriate protein fractions, 0.5 µg of plasmid DNA, 600 µM ATP, CTP, UTP and 30 µM GTP, 3 µCi of [α-32P]GTP (Hartmann) and 15 U of RNase Block Ribonuclease Inhibitor (Eppendorf) in buffer B containing 60 mM KCl in a final volume of 70 µl. After 90 min incubation at 30°C, the RNA was purified and loaded onto a denaturing 7 M urea–6% polyacrylamide gel. The gel was analysed by autoradiography and by a Fuji FLA-3000 BioImaging Analyzer.
Isolation of transcription complexes or partly assembled transcription complexes
Complete or partly assembled transcription complexes were generated in several identical samples by incubating the fractions containing the appropriate transcription activities in optimal stoichiometry and 0.5 µg of plasmid DNA without nucleotides in a volume of 70 µl for 45 min. The samples were then combined and 900 µl of this input was loaded onto a HR 10/30 column containing Sephacryl S500 using an Äkta purifier (Pharmacia). The running buffer was buffer B containing 60 mM KCl. Fractions of 1 ml volume were collected and immediately employed for the following activity test.
To detect functionally active transcription complexes, 40–50 µl of the fractions from the Sephacryl column were supplemented, if necessary, with further transcription factors and/or pol III, mixed with nucleotides, 3 µCi of [α-32P]GTP and RNase Block, and transcribed as described above in a total volume of 70 µl for 1 h. No further template was added.
Western blot
The fractions of a S500 run were each concentrated by Strataclean Resin beads (Stratagene), electrophoresed on a 10% SDS–gel and semi-dry blotted on a PVDF membrane (Millipore) as described in (17). The membrane was cut horizontally into three parts, the parts were blocked, and subsequently treated with the appropriate antibodies for 2 h. Then a 125I-labelled second antibody was incubated for another 2 h as described (17). The analysis was performed by autoradiography and using a Fuji FLA-3000 BioImaging Analyzer.
RESULTS
We could show previously that functional pol III transcription complexes could be assembled on the VAI gene which subsequently could be isolated by glycerol gradient centrifugation (17). We attempted to develop an alternative method for the separation of such transcription complexes from non-incorporated proteins which provided better handling and less spreading of the isolated fractions. Therefore, we assembled transcription complexes on the 5S gene using purified transcription factors and pol III, and subsequently isolated the transcriptionally active complexes using S500 Sephacryl size exclusion chromatography. This material separates large macromolecules which elute in the separation range of the column (see below). Because of its size, however, plasmid DNA elutes in the void volume of the column (data not shown)
Isolation of 5S transcription complexes by size exclusion chromatography
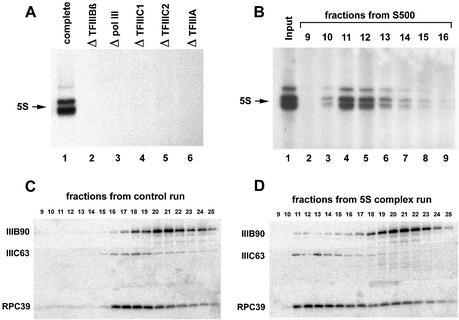
A prerequisite to conduct these experiments was to purify the required transcription factors and the pol III sufficiently. As shown in Figure 1A, these fractions were active and free from mutual cross-contamination. Only when pol III was used together with all four transcription factors, IIIA, IIIC1, IIIC2 and IIIBβ, could 5S rRNA be synthesized (lane 1). Omitting any one of these fractions abrogates the 5S signal (lanes 2–6).
Figure 1.
Isolation of active 5S transcription complexes from purified transcription factors and RNA polymerase III. (A) A standard in vitro transcription was performed using the following fractions: 2.5 µl of TFIIIA (protein concentration: 0.3 mg/ml), 2.5 µl of TFIIIBβ (0.4 mg/ml), 1 µl of pol III (0.1 mg/ml), 7.5 µl of TFIIIC1 (0.3 mg/ml) and 2.5 µl of TFIIIC2 (0.1 mg/ml). Lane 1, complete reaction; lanes 2–6, reactions without TFIIIBβ, pol III, TFIIIC1, TFIIIC2 or TFIIIA, respectively. (B) Complete transcription complexes were assembled using the same fractions and the same stoichiometry as in (A), isolated and tested for transcription activity as described in Materials and Methods. Lane 1, input; lanes 2–9, fractions 9–16 from the S500 column. (C) Western blot. S500 elution profile of TFIIIC2, TFIIIBβ and pol III not bound to DNA. (D) Western blot of the fractions stemming from the S500 column with 5S transcription complexes from (B).
Figure 1B shows the analysis of isolated 5S transcription complexes that were assembled from the purified transcription factors TFIIIA, TFIIIBβ, TFIIIC1 and TFIIIC2 together with the polymerase on a DNA plasmid containing the 5S gene in the absence of nucleotides and then applied to a column containing S500 Sephacryl. The resulting fractions were tested for transcriptional activity in an in vitro transcription.
The input (lane 1) and fractions 9–16, eluted from the S500 column (lanes 2–9), were supplemented with nucleotides but no additional DNA before the second incubation. Input and fractions 10–14 (lanes 3–7) show specific transcriptional activity culminating in fraction 11 (lane 4), which corresponds to the void volume of the column.
A western blot experiment documented that even the large multiprotein complexes TFIIIC2, TFIIIBβ or pol III eluted in the separation range of the S500 with sufficient distance from the fractions containing transcription complexes. The fractions resulting from a control run without DNA show that these non-incorporated protein complexes are detectable beyond fraction 15 in the separation range of the column, while the void volume (fractions 11 and 12) is free of these proteins (Fig. 1C). In contrast, a corresponding experiment containing the analysis of a S500 run with the 5S rDNA transcription complexes (Fig. 1D) reveals subunits of all three protein complexes in the fractions containing the 5S template, representing the factors and the polymerase associated with DNA.
Assembly and isolation of partial 5S transcription complexes
Until now, the order of assembly of human pol III transcription complexes and the functional roles of the involved transcription factors have been incompletely understood.
In particular, the assembly of TFIIIBβ into the complex and a possible involvement of TFIIIC1 in this process remain to be investigated. The strategy we used here was to isolate incomplete subcomplexes. It was then attempted to convert these subcomplexes into complete, functionally active transcription complexes by subsequent complementation with the initially omitted activities.
At first, we succeeded in isolating a 5S rDNA–TFIIIA binary complex eluting in the fractions representing the void volume of the column that could be complemented with TFIIIC2, IIIC1, IIIBβ and the polymerase after S500 chromatography, resulting in efficient 5S rRNA transcription (data not shown).
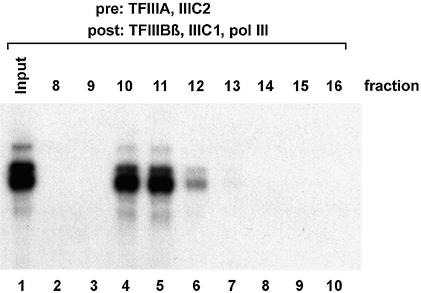
It was also possible to isolate a stable 5S rDNA–TFIIIA– TFIIIC2 ternary subcomplex that was convertible to a complete transcription complex (Fig. 2). After appropriate incubation and chromatography, the input (lane 1) and the fractions from the S500 column (lanes 2–10) were transcribed in the presence of TFIIIBβ, TFIIIC1 and pol III. This resulted in efficient transcription activity in fractions 10–12 (lanes 4–6).
Figure 2.
Isolation of a ternary 5S–TFIIIA–TFIIIC2 complex. A 5 µl aliquot of TFIIIA and 2.5 µl of TFIIIC2 were pre-incubated with the 5S plasmid. The resulting complexes were isolated by S500 chromatography as described before. The input and the fractions from the column were complemented with TFIIIC1, TFIIIBβ and pol III in the same amounts as in Figure 1 and tested for transcription activity. Lane 1, input; lanes 2–10, S500 fractions 8–16.
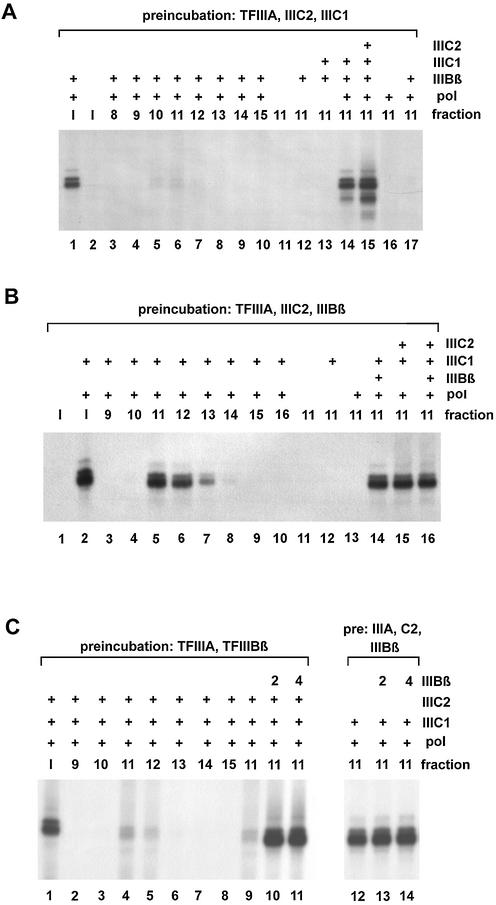
While it is clearly established that TFIIIC2 associates with the 5S rRNA gene after TFIIIA is bound to the ICR, the further steps, i.e. the order in which TFIIIBβ and TFIIIC1 are assembled into the complex, are not yet characterized. To address this issue, we performed two further experiments. TFIIIA, TFIIIC2 and 5S rDNA were pre-incubated together with either TFIIIC1 or TFIIIBβ to find out whether one of the two (or both) quaternary complexes could be isolated.
In Figure 3A, the 5S gene was pre-incubated with TFIIIA, TFIIIC2 and TFIIIC1, but without TFIIIBβ and polymerase. Control lane 1 shows that the addition of TFIIIBβ and polymerase to the input led to active 5S transcription, while the input without further supplementation (lane 2) did not. Remarkably, the supplementation of the S500 fractions 8–15 (lanes 3–10) with TFIIIBβ and pol III did not lead to efficient transcription activity. Only an extremely weak signal could be detected in fraction 11 (lane 6). However, this scarcely active fraction could be stimulated strongly upon addition of further TFIIIC1 together with TFIIIBβ and polymerase (compare lanes 6 and 14). Additional TFIIIC2 only slightly increased the transcription activity of fraction 11 (lane 15). These results indicate that it was not possible to isolate a stable quaternary 5S rDNA–TFIIIA–TFIIIC2–TFIIIC1 complex. Most of the isolated complexes represent ternary DNA–TFIIIA–TFIIIC2 complexes and possibly also a small portion of even lower order complexes.
Figure 3.
Stable quaternary 5S–TFIIIA–TFIIIC2–TFIIIBβ can be isolated, but not a 5S–TFIIIA–TFIIIC2–TFIIIC1 complex. (A) The pre-incubation was performed with TFIIIA, TFIIIC2 and TFIIIC1. The input and the fractions from the S500 column were supplemented with activities as indicated. Lanes 1–2, input (I); lanes 3–10, S500 fractions 8–15, supplemented with TFIIIBβ and pol III; lanes 11–17, fraction 11 supplemented with different activities as indicated. (B) Pre-incubation with TFIIIA, TFIIIC2 and TFIIIBβ; prior to post-incubation, activities were supplemented as indicated. Lanes 1 and 2, input; lanes 3–10, S500 fractions 9–16 supplemented with TFIIIC1 and pol III; lanes 11–16, fraction 11 supplemented with different activities as indicated. (C) The 5S gene was incubated together with either TFIIIA and TFIIIBβ (first run, left panel, lanes 1–11), or TFIIIA, TFIIIC2 and TFIIIBβ (second run, right panel, lanes 12–14). After S500 chromatography, the resulting fractions were supplemented either with TFIIIC2, TFIIIC1 and pol III (left panel) or with TFIIIC1 and pol III (right panel). Where indicated, 2 or 4 µl of TFIIIBβ were added. Lane 1; input (run 1); lanes 2–8, S500 fractions 9–15 (run 1); lanes 9–11, fraction 11 (run 1) with 0, 2 and 4 µl of TFIIIBβ; lanes 12–14, fraction 11 (run 2) with 0, 2 and 4 µl of TFIIIBβ.
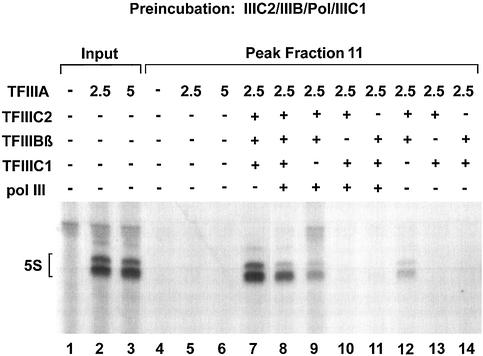
In Figure 3B, fractions 9–16 of the S500 chromatography of the proposed TFIIIA–TFIIIC2–TFIIIBβ complexes were supplemented with TFIIIC1 and polymerase (lanes 3–10). This led to efficient transcription in fractions 11–13. As a control, the input was transcribed either alone, showing no transcription activity (lane 1), or with added TFIIIC1 and polymerase, resulting in active 5S rRNA transcription (lane 2). Further controls are shown in lanes 11–16, in which peak fraction 11 was incubated in the presence of different factors and/or the polymerase. Transcription did not occur after addition of TFIIIC1 or polymerase alone (lanes 12 and 13). Interestingly, the addition of TFIIIBβ together with TFIIIC1 and polymerase did not increase the transcription activity of the purified complexes further (compare lanes 5 and 14). Moreover, neither further addition of TFIIIC2 nor the addition of all activities except for TFIIIA showed stimulating effects (lanes 15 and 16). This suggests that most of the purified complexes are indeed quaternary complexes containing DNA, TFIIIA, TFIIIC2 and TFIIIBβ.
To verify the specificity of the TFIIIBβ assembly into the 5S–TFIIIA–TFIIIC2 complex in the absence of TFIIIC1, we performed an additional control experiment consisting of two parallel assembly procedures (Fig. 3C). On the one hand, TFIIIA and TFIIIBβ were incubated with the plasmid in the absence of TFIIIC2 which should not lead to a functional complex of higher order than the 5S DNA–TFIIIA complex. On the other hand, as a control, TFIIIA, TFIIIC2 and TFIIIBβ were present during the assembly phase, which corresponded exactly to the experiment presented in Figure 3B.
Protein–DNA complexes were isolated and tested for transcriptional activity. After addition of polymerase, TFIIIC1 and TFIIIC2 to the TFIIIC2-lacking run, the input was transcriptionally active (Fig. 3C, lane 1). However, the addition of the same activities to fractions 9–15 (lanes 2–8) resulted only in a very weak transcription signal in fractions 11 and 12 (lanes 4 and 5). This activity, which may be due to a weak interaction of TFIIIBβ with TFIIIA, is negligibly small, as control lanes 9–11 show. The supplementation with additional TFIIIBβ, TFIIIC2, TFIIIC1 and polymerase resulted in a 10-fold increase in the activity (lanes 10 and 11) compared with the control reaction without additional TFIIIBβ (lane 9).
In the reference run with TFIIIC2 (right panel), we again observed full transcription activity after addition of TFIIIC1 and polymerase (lane 12), while additional supplementation with TFIIIBβ did not lead to significant further stimulation (lanes 13 and 14).
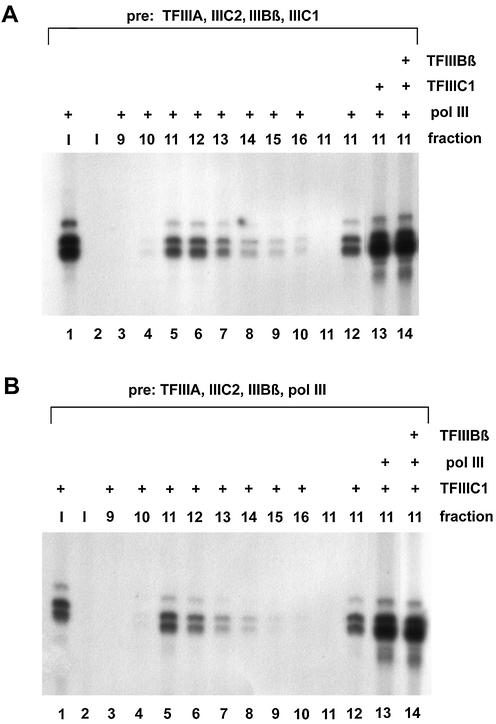
Collectively, these results show that we succeeded in generating a 5S rDNA–TFIIIA–TFIIIC2–TFIIIBβ complex in the absence of TFIIIC1, showing a high degree of formation rate and stability after S500 chromatography. This complex could be complemented with TFIIIC1 and pol III for full transcriptional activity. In contrast, TFIIIC1 does not assemble stably into a 5S complex consisting of TFIIIA and TFIIIC2 in the absence of TFIIIBβ. Therefore, the question was addressed of whether TFIIIC1 can be stably assembled into the quaternary 5S–TFIIIA–TFIIIC2–TFIIIBβ complex, and whether this is a prerequisite for the functional recruitment of the polymerase. To answer this question, we tried to assemble quintary complexes of all the required transcription factors lacking only the polymerase and to convert these into functional transcription complexes. Alternatively, we conducted a control run in which the polymerase was incubated together with TFIIIA, TFIIIC2 and TFIIIBβ, but lacking TFIIIC1.
The input of the pol III-lacking run (Fig. 4A) showed no transcriptional activity (lane 2), while the mere addition of pol III led to efficient transcription (lane 1). Likewise, after addition of pol III alone to the isolated complexes, active transcription could be observed (lanes 5–7). The supplementation of fraction 11 with TFIIIC1 in addition to pol III enhanced this transcription activity (compare lanes 13 and 12), while TFIIIBβ is not limiting in the isolated complex fraction (compare lanes 14 and 13). This indicates that a major part of the isolated complexes were in fact quaternary 5S rDNA– TFIIIA–TFIIIC2–TFIIIBβ complexes.
Figure 4.
Recruitment of TFIIIC1 or polymerase III into the TFIIIA– TFIIIC2–TFIIIBβ complex. The procedure was similar to the previous experiments, with the following modifications. (A) Pre-incubation of the 5S gene with TFIIIA, TFIIIC2, TFIIIBβ and TFIIIC1. The input (I) (lanes 1 and 2) and the resulting fractions from the S500 chromatography (lanes 3–14) were supplemented with additional activities as indicated. (B) Pre-incubation of the 5S gene with TFIIIA, TFIIIC2, TFIIIBβ and pol III. The input (I) (lanes 1 and 2) and the resulting fractions from the S500 chromatography (lanes 3–14) were supplemented with additional activities as indicated.
Thus we conclude that in the presence of TFIIIBβ, it was possible to isolate a quintary complex including TFIIIC1, although the efficiency of formation of this complex and/or its stability were only moderate. Given the fact that a TFIIIA–TFIIIC2–TFIIIC1 complex could not be formed in the absence of TFIIIBβ (Fig. 3A), this clearly shows that the assembly of TFIIIBβ is a strict prerequisite for the binding of TFIIIC1 to the 5S transcription complex.
Figure 4B shows the parallel experiment in which TFIIIA, TFIIIC2, TFIIIBβ and pol III were pre-incubated without TFIIIC1. Importantly, the void volume of the column, representing the complexes formed under these conditions, yielded no transcription signal (lane 11), indicating that polymerase was not functionally incorporated into the complex in the absence of TFIIIIC1. The addition of TFIIIC1 to the fractions of the S500 chromatography led, interestingly, to an efficient transcription, indicating that DNA-associated pol III was contained in the void volume. Further addition of pol III to the TFIIIC1-deficient run led to a significant stimulation of transcription (compare lanes 13 and 12), while TFIIIBβ was obviously not limiting (compare lanes 14 and 13).
A possible explanation for the observations made in Figure 4 could be that pol III and TFIIIC1 are assembled into the TFIIIA–TFIIIC2–TFIIIBβ complex simultaneously but independently from each other. It is known, however, that RNA polymerases are able to bind to DNA efficiently (see Discussion). Thus it was conceivable that the activity in the void fractions of Figure 4B after TFIIIC1 complementation was due to polymerase unspecifically associated with DNA and subsequently recruited into the nascent transcription complex on the same plasmid. This aspect was investigated in more detail in the experiments shown in Figure 5. We incubated TFIIIC2, TFIIIBβ, TFIIIC1 and pol III with the 5SrDNA plasmid in the absence of TFIIIA. Without TFIIIA, the input, as expected, yielded no specific signal (lane 1) but, with added TFIIIA, 5S rRNA was synthesized efficiently (lanes 2 and 3). After S500 chromatography, the central fraction of the void volume likewise yielded no transcription without adding TFIIIA (lane 4). Importantly, the addition of TFIIIA in this case did not restore transcription activity (lanes 5 and 6). These controls clearly show that in the absence of TFIIIA, no functional complexes had been formed on the 5S gene. Moreover, these controls reinforce the findings in the previous experiments that the observed transcription activities were in fact due to specific bona fide complexes and were not just the result of a fortuitous co-elution of transcription activities with the DNA.
Figure 5.
Pre-incubation of 5S plasmid DNA with transcription factors TFIIIC2, TFIIIBβ, TFIIIC1 and pol III, and subsequent analysis of potential complexes. After S500 chromatography, the input (lanes 1–3) or the peak fraction 11 of the void volume (lanes 4–14) were incubated with 2.5 or 5 µl of TFIIIA, 2.5 µl of TFIIIC2, 2.5 µl of TFIIIBβ, 7.5 µl of TFIIIC1 or 1 µl of pol III as indicated.
Addition of TFIIIA, TFIIIC2, TFIIIBβ and TFIIIC1, but no pol III, resulted in an active transcription (lane 7), indicating that during the pre-incubation, polymerase molecules had bound to the DNA. These polymerases obviously were available for subsequent integration into specific 5S transcription complexes and were sufficient in number to serve the nascent complexes.
Interestingly, in this experimental procedure, subsequently added pol III did not enhance transcription, and even led to a slight decrease of transcription activity (lane 8). This addition obviously increased the number of polymerase molecules bound to the DNA in such a way that the de novo assembly of transcription complexes was interfered with significantly.
In lanes 9–14, each of the transcription factors TFIIIC1, TFIIIBβ and TFIIIC2 was omitted singly from the post-incubation, either with additional polymerase (lanes 9–11) or without polymerase (lanes 12–14). Both reactions without TFIIIC1 led to a very weak, but detectable pol III transcription, indicating that a small portion of TFIIIC1 may initially have bound to the DNA and subsequently was recruited into the nascent transcription complex (lanes 9 and 12). Note, however, that the additional supplementation of TFIIIC1 led to a drastic stimulation of transcription (compare lane 12 with 7, and lane 9 with 8). The reactions without TFIIIBβ (lanes 10 and 13) or TFIIIC2 (lanes 11 and 14) did not display any transcription activity.
DISCUSSION
Isolation of complete and partly assembled 5S transcription complexes
Four different transcription factors, TFIIIA, TFIIIC2, TFIIIBβ and TFIIIC1, together with pol III are involved in the transcription of the human 5S gene [reviewed in Paule and White (1), Geiduschek and Kassavetis (9), Huang and Maraia (10)]. Together, these activities form a complex of ∼1800 kDa of protein on the DNA. While cloning of the subunits of individual transcription factors has helped to unravel some of the mechanisms involved in complex formation, total reconstitution of functional complexes from recombinant proteins hitherto has not been successful. Therefore, it remains important to generate and isolate functional complexes from pre-purified components. Such complexes could form the basis for future biochemical, structural and electron microscopic analyses.
We succeeded in isolating complete 5S transcription complexes, via Sephacryl S500 size exclusion chromatography, which showed efficient and specific 5S RNA transcription (Fig. 1). Moreover, this new separation method enabled us to isolate intermediate stages of the assembly pathway of 5S transcription complexes, which served to determine the sequence in which factors enter the nascent complex.
The successful isolation of a ternary 5S rDNA–TFIIIA– TFIIIC2 complex (Fig. 2) served as the starting point for further investigations concerning the recruitment of the transcription factors TFIIIC1 and TFIIIBβ as well as pol III into the human 5S transcription complex. Previous investigations by Lassar et al. (18) and Bieker et al. (19) concerning the order of complex assembly on the 5S gene had been performed before TFIIIC1 was discovered by Yoshinaga et al. in 1987 (7), and therefore in those reports the role of this factor could not be taken into account.
Integration of TFIIIBβ into the nascent 5S transcription complex
The pol III transcription system in Saccharomyces cerevisiae consists of only three transcription factors, TFIIIA, TFIIIC and TFIIIB, which are sufficient for the transcription of all pol III genes (1,9,10). The human pol III transcription system displays a much greater variety of factors, and most of them, including TFIIIC2, are specific for different types of pol III genes (see below). Furthermore, with TFIIIC1, a new general pol III transcription factor has evolved (4,6,7) which may represent an important target for the regulation of pol III transcription (11). However, up to now, the role of TFIIIC1 within the assembly pathway of the 5S transcription complex is not well understood.
It has been reported that TFIIIC1 enhances the binding of the primarily binding transcription factors TFIIIA, TFIIIC2 and PBP to the 5S, VAI and U6 genes, respectively (4,20). It was also proposed that TFIIIC1 enlarges the TFIIIC2 footprint over the A-box of the promoters of tRNA-like genes (7), but in the meantime it could be shown that TFIIIC2 is able to protect the whole promoter region of a tRNAMet gene alone (4). Moreover, the isolation of a ‘holoTFIIIC’ complex by co-immunoprecipitation has been reported, which displayed TFIIIC2 and TFIIIC1 activity (21). Collectively, these data suggested a model in which TFIIIC1 interacts with the transcription complex at an early stage of assembly in conjunction with TFIIIC2, possibly providing the prerequisite for the recruitment of TFIIIBβ into the transcription complex.
In contrast, other data suggested the direct recruitment of TFIIIBβ mediated by TFIIIC2. TFIIIC2 consists of five subunits (1,9,10) and largely diverges from yeast TFIIIC because only hTFIIIC63 and hTFIIIC102 display significant homologies to subunits of yTFIIIC. Using GST pull-down experiments, it could be shown that both hTFIIIC63 and hTFIIIC102 together with the non-conserved hTFIIIC90 displayed direct interactions with hTFIIIB90 and hTBP (22,23). Moreover, recent evidence could show that the interaction of hTFIIIC102 and hTFIIIB90 depends on the activity of CK2 (24).
However, all these data were accrued in experiments in the absence of DNA, and therefore it had to be clarified in a functional assay whether the TFIIIA–TFIIIC2 complex on the 5S gene by itself is capable of recruiting TFIIIBβ into the 5S transcription complex or whether TFIIIC1 also plays an important role in this process.
In an important study based on template commitment experiments with the VAI and tRNAArg genes, Dean and Berk (25) presented a model in which TFIIIC2 binds primarily to this type of promoter. These authors left open the question of whether TFIIIC1 or TFIIIB(β) would be assembled next to the complex, and they proposed that direct characterization of the intermediates in the assembly of the pre-initiation complex would be required to confirm and extend their model.
Figure 3A demonstrates that TFIIIC1 cannot interact stably with a ternary 5S–TFIIIA–TFIIIC2 complex. In contrast, it could be shown in Figure 3B that TFIIIBβ is integrated into such a 5S–TFIIIA–TFIIIC2 complex with high efficiency. This integration is not enhanced in the presence of TFIIIC1 (compare Figs 3B and 4B with 4A). Thus we conclude that TFIIIC1 does not play a role as an ‘assembly factor’ for TFIIIBβ on the human 5S gene.
Conversion of a 5S–TFIIIA–TFIIIC2–TFIIIBβ complex into a complete functional pol III transcription complex
The previously discussed results demonstrate that a TFIIIA– TFIIIC2 complex on the 5S promoter is sufficient for the assembly of TFIIIBβ. Nevertheless, adding pol III to an isolated quaternary 5S–TFIIIA–TFIIIC2–TFIIIBβ complex does not suffice for transcription, but the further addition of TFIIIC1 is absolutely required (see Fig. 3B, lane 12).
This finding is in good agreement with the data presented by Kober et al. (26), who used an artificial TATA box-containing promoter, with which TFIIIBβ primarily associates under certain conditions. This bound TFIIIBβ alone was not sufficient for transcription and required additional TFIIIC1 for a functional recruitment of pol III.
Consequently, human TFIIIBβ cannot be regarded as the sole ‘initiation factor’ for transcription, as has been firmly established for TFIIIB in yeast by Kassavetis et al. (12). Nevertheless, human TFIIIBβ and yeast TFIIIB show a significant degree of structural conservation. Similarly to yTFIIIB, hTFIIIBβ is a TBP–TAF complex consisting of TBP, TFIIIB90 (hBRF) and TFIIIB150 (hB′′) [reviewed in Geiduschek and Kassavetis (9) and Huang and Maraia (10)] which show sequence homologies to the yeast TAFs yTFIIIB70 (BRF) and yTFIIIB90 (B′′), respectively (27–29). Moreover, hTFIIIB90 interacts with particular unique pol III subunits, implicating that TFIIIBβ plays an important role in polymerase recruitment (8).
The integration of TFIIIC1 into the nascent 5S transcription complex depends on the prior assembly of TFIIIBβ (compare Figs 3A and 4A). Thus it is obvious that in the human 5S transcription system, a new step of transcription complex assembly has evolved, which takes place after the evolutionarily conserved integration of TFIIIB(β). Based on the observation that the further addition of TFIIIC1 to isolated 5S–TFIIIA–TFIIIC2–TFIIIBβ–TFIIIC1 complexes led to a significant increase of activity, we conclude that TFIIIC1 does not bind to the complex very tightly, even when TFIIIBβ is assembled properly. Alternatively, the chromatographic procedure could have partly diminished the activity of this rather sensitive transcription factor.
In the second part of the experiment in Figure 4, we also observed a lack of transcription when TFIIIA, TFIIIC2, TFIIIBβ and polymerase were pre-incubated. Only after the isolated complexes subsequently were supplemented with TFIIIC1 could active transcription be detected. It is conceivable that TFIIIC1 and pol III are integrated into the 5S–TFIIIA–TFIIIC2–TFIIIBβ complex at the same time but independently from each other, mediated by interactions with TFIIIBβ. The control experiment in Figure 5, however, provides strong arguments for another explanation. It is well known that pol III has a strong affinity for DNA and is even able to initiate transcription unspecifically (30). We can show accordingly that a sizeable portion of pol III binds to the 5S plasmid DNA prior to S500 isolation, and these DNA-associated polymerase molecules can be recruited specifically into a nascent 5S transcription complex. Recently it could be shown with the scanning force microscope (SFM) that RNA polymerase from Escherichia coli reaches its promoter by prior sliding along the DNA. This one-dimensional seeking probably facilitates promoter recognition compared with a three-dimensional recognition by diffusion without prior DNA contact (31). It is conceivable that such a mechanism is also used by pol III. Sliding along the DNA, the polymerase could be ‘captured’ by the assembled transcription factors and is recruited to complete the transcription complex on the same plasmid.
Thus our new model of the assembly pathway of the human 5S transcription complex implies that after the primary association of TFIIIA with the ICR and the subsequent enlargement of the complex by TFIIIC2, TFIIIBβ is definitely the third transcription factor to enter the growing complex without any requirement for TFIIIC1 for this process. Most interestingly, the 5S–TFIIIA–TFIIIC2–TFIIIBβ complex is not able to functionally recruit the polymerase but depends on TFIIIC1, which is integrated into the complex as the last transcription factor. The combined action of TFIIIBβ and TFIIIC1 is then sufficient for the recruitment of the polymerase and/or the activation of the pre-initiation complex.
In the vertebrate pol III transcription system, a clear separation in the subset of transcription factors has evolved between those genes with internally organized promoters and the 5′-regulated genes such as the U6 and 7SK genes (1,9,10,32). Several investigations showed that TFIIIC1 is also an essential transcription factor for the U6 gene (4,6,11). Chong et al. (33) could reconstitute an active U6 transcription system from recombinant TBP, rSNAPC, rB′′ and rBRFU together with a partly purified RNA polymerase. Since the authors explicitly state, however, that their polymerase preparation could contain TFIIIC1 activity, these data do not exclude that TFIIIC1 is a general pol III transcription factor. The previous finding that TFIIIC1 plays an important role in the downregulation of all types of pol III genes in F9 embryonal carcinoma cells during differentiation into parietal endoderm cells (11) suggests that TFIIIC1 might be a junction between pol III transcription and the cell cycle. Future investigations will have to reveal the exact function of TFIIIC1 during the final phase of the assembly of human pol III transcription complexes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ulla Kopiniak and Sarah Fehl for expert technical assistance, Sabine Wagner for helpful discussions and proofreading of the manuscript, and Robert G. Roeder for antiserum against TFIIIC2 and RNA polymerase III. This work was supported by grants from the Deutsche Forschungsgemein schaft and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Paule M.R. and White,R.J. (2000) Survey and summary: transcription by polymerase I and III. Nucleic Acids Res., 28, 1283–1298. [DOI] [PMC free article] [PubMed]
- 2.Moorefield B. and Roeder,R.G. (1994) Purification and characterisation of human transcription factor IIIA. J. Biol. Chem., 269, 20857–20865. [PubMed]
- 3.Seifart K.H., Wang,L., Waldschmidt,R., Jahn,D. and Wingender,E. (1989) Purification of human transcription factor IIIA and its interaction with a chemically synthesised gene encoding human 5S rRNA. J. Biol. Chem., 264, 1702–1709. [PubMed]
- 4.Oettel S., Härtel,F., Kober,I., Iben,S. and Seifart,K.H. (1997) Human transcription factors IIIC2, IIIC1 and a novel component IIIC0 fulfil different aspects of DNA binding to various pol III genes. Nucleic Acids Res., 25, 2440–2447. [DOI] [PMC free article] [PubMed]
- 5.Teichmann M. and Seifart,K.H. (1995) Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J., 14, 5974–5983. [DOI] [PMC free article] [PubMed]
- 6.Yoon J.-B., Murphy,S., Bai,L., Wang,Z. and Roeder,R.G. (1995) Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol., 15, 2019–2027. [DOI] [PMC free article] [PubMed]
- 7.Yoshinaga S.K., Boulanger,P.A. and Berk,A.J. (1987) Resolution of human transcription factor TFIIIC into two functional components. Proc. Natl Acad. Sci. USA, 84, 3585–3589. [DOI] [PMC free article] [PubMed]
- 8.Wang Z. and Roeder R.G. (1997) Three human polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev., 11, 1315–1326. [DOI] [PubMed]
- 9.Geiduschek E.P. and Kassavetis,G.A. (2001) The RNA polymerase III transcription apparatus. J. Mol. Biol., 310, 1–26. [DOI] [PubMed]
- 10.Huang Y. and Maraia,R.J. (2001) Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res., 29, 2675–2690. [DOI] [PMC free article] [PubMed]
- 11.Meissner W., Thomae,R. and Seifart,K.H. (2002) The activity of transcription factor IIIC1 is impaired during differentiation of F9 cells. J. Biol. Chem., 277, 7148–7156. [DOI] [PubMed]
- 12.Kassavetis G.A., Braun,B.R., Nguyen,L.H. and Geiduschek,E.P. (1990) S.cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell, 60, 235–245. [DOI] [PubMed]
- 13.Nielsen J.N., Hallenberg,C., Frederiksen,S., Sorensen,P.D. and Lomholt,B. (1993) Transcription of human 5S rRNA genes is influenced by an upstream DNA sequence. Nucleic Acids Res., 21, 3631–3636. [DOI] [PMC free article] [PubMed]
- 14.Weser S. (2000) Analyse einzelner Phasen des RNA Polymerase III Transkriptionszyklus. PhD thesis, Philipps Universität, Marburg, Germany.
- 15.Weil P.A., Segall,J., Ng,S.Y. and Roeder R.G. (1979) Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J. Biol. Chem., 254, 6163–6173. [PubMed]
- 16.Jahn D., Wingender,E. and Seifart,K.H. (1987) Transcription complexes for various class III genes differ in parameters of formation and stability towards salt. J. Mol. Biol., 193, 303–313. [DOI] [PubMed]
- 17.Weser S., Bachmann,M., Seifart,K.H. and Meissner,W. (2000) Transcription efficiency of human polymerase III genes in vitro does not depend on the RNP-forming autoantigen La. Nucleic Acids Res., 28, 3935–3942. [DOI] [PMC free article] [PubMed]
- 18.Lassar A.B., Martin,P.L. and Roeder,R.G. (1983) Transcription of class III genes: formation of preinitiation complexes. Science, 222, 740–748. [DOI] [PubMed]
- 19.Bieker J.J., Martin,P.L. and Roeder,R.G. (1985) Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell, 40, 119–127. [DOI] [PubMed]
- 20.Wang Z. and Roeder,R.G. (1996) TFIIIC1 acts through a downstream region to stabilise TFIIIC2 binding to RNA polymerase III promoters. Mol. Cell. Biol., 16, 6841–6850. [DOI] [PMC free article] [PubMed]
- 21.Wang Z. and Roeder,R.G. (1998) DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol. Cell, 1, 749–757. [DOI] [PubMed]
- 22.Hsieh Y.-J., Wang,Z., Kovelman,R. and Roeder,R.G. (1999) Cloning and characterisation of two evolutionary conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol. Cell. Biol., 19, 4944–4952. [DOI] [PMC free article] [PubMed]
- 23.Hsieh Y.-J., Kundu,T.K., Wang,Z., Kovelman,R. and Roeder,R.G. (1999) The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol. Cell. Biol., 19, 7697–7704. [DOI] [PMC free article] [PubMed]
- 24.Johnston I.M., Allison,S.J., Morton,J.P., Schramm,L., Scott,P.H. and White,R.J. (2002) CK2 forms a stable complex with TFIIIB and activates RNA polymerase III transcription in human cells. Mol. Cell. Biol., 22, 3757–3768. [DOI] [PMC free article] [PubMed]
- 25.Dean N. and Berk,A.J. (1988) Ordering promoter binding of class III transcription factors TFIIIC1 and TFIIIC2. Mol. Cell. Biol., 8, 3017–3025. [DOI] [PMC free article] [PubMed]
- 26.Kober I., Teichmann,M. and Seifart,K.H. (1998) hTFIIIBβ stably binds to pol II promoters and recruits RNA polymerase III in a hTFIIIC1 dependent way. J. Mol. Biol., 284, 7–20. [DOI] [PubMed]
- 27.Schramm L., Pendergrast,P.S., Sun,Y. and Hernandez,N. (2000) Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev., 14, 2650–2663. [DOI] [PMC free article] [PubMed]
- 28.Teichmann M., Wang,Z. and Roeder,R.G. (2000) A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50 and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc. Natl Acad. Sci. USA, 97, 14200–14205. [DOI] [PMC free article] [PubMed]
- 29.Wang Z. and Roeder,R.G. (1995) Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc. Natl Acad. Sci. USA, 92, 7026–7030. [DOI] [PMC free article] [PubMed]
- 30.Sklar V.E.F., Schwartz,L.B. and Roeder,R.G. (1975) Distinct molecular structures of nuclear class I, II and III DNA-dependent RNA polymerases. Proc. Natl Acad. Sci. USA, 72, 348–352. [DOI] [PMC free article] [PubMed]
- 31.Guthold M., Zhu,X., Rivetti,C., Yang,G., Thomson,N.H., Kasas,S., Hansma,H.G., Smith,B., Hansma,P.K. and Bustamante,C. (1999) Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys. J., 77, 2284–2294. [DOI] [PMC free article] [PubMed]
- 32.Hernandez N. (2001) Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem., 276, 26733–26736. [DOI] [PubMed]
- 33.Chong S.S., Hu,P. and Hernandez,N. (2001) Reconstitution of transcription from the human U6 small nuclear RNA promoter with eight recombinant polypeptides and a partially purified RNA polymerase III complex. J. Biol. Chem., 276, 20727–20734. [DOI] [PubMed]