Abstract
Ribosomal RNAs (rRNAs) are encoded by multicopy families of identical genes. In Dictyostelium and other protists, the rDNA is carried on extrachromosomal palindromic elements that comprise up to 20% of the nuclear DNA. We present the sequence of the 88 kb Dictyostelium rDNA element, noting that the rRNA genes are likely to be the only transcribed regions. By interrogating a library of ordered YAC clones, we provide evidence for a chromosomal copy of the rDNA on chromosome 4. This locus may provide master copies for the stable transmission of the extrachromosomal elements. The extrachromosomal elements were also found to form chromosome-sized clusters of DNA within nuclei of nocodazole-treated cells arrested in mitosis. These clusters resemble true chromosomes and may allow the efficient segregation of the rDNA during mitosis. These rDNA clusters may also explain the cytological observations of a seventh chromosome in this organism.
INTRODUCTION
The production of ribosomal RNAs (rRNAs) presents cells with a special challenge since their rate of synthesis must be much higher than that of even the most abundant messenger RNAs. The transcription of a single locus encoding the 35S rRNA precursor appears to be insufficient to supply all the new ribosomes needed for optimal growth rates, thus, most organisms have multiple copies of the rRNA genes. In most metazoa, the genes are present in multicopy arrays integrated into the chromosomes, but in the protists Tetrahymena, Dictyostelium and Physarum, they are present in specialized extrachromosomal elements (1–9).
Tetrahymena ciliates carry a transcriptionally inactive germ-line micronucleus, but, following conjugation, the micronucleus gives rise to the polyploid macronucleus, where genes are amplified, transcribed and replicated. The locus encoding rRNA in the micronucleus is copied into a mirror-symmetric 20 kb palindrome that is replicated in the macronucleus (9,10). Likewise, the genes encoding rRNA in the acellular slime mould Physarum polycephalum are carried on an extracellular palindrome (5). Several hundred copies of this 60 kb element are found in each nucleus. Replication of the palindrome is unsynchronized relative to the cell cycle, and is uneven in that some molecules replicate more than once in a cell cycle while others do not replicate (8,11). When two strains that can be distinguished by differences in the restriction endonuclease cleavage patterns of their rDNA are crossed and the resulting diploids induced to sporulate to generate haploids, progeny clones were found to have one of the parental rDNA types but not both, suggesting that only a single rDNA copy is replicated after meiosis (12). Moreover, continued growth of the diploids resulted in one or the other of the parental rDNA types predominating (12).
Dictyostelium amoebae also carry their genes encoding rRNA on an extrachromosomal palindrome. There are two transcription units on each arm of the mirror-symmetric 88 kb linear DNA; one encodes the 35S precursor of 17S, 5.8S and 26S rRNA and the other encodes the 5S rRNA (3,4,6,13). This organization is distinct from that of Tetrahymena, which does not carry a 5S gene on its amplified palindrome (14). Although the rRNA genes make up <20% of the 88 kb palindrome, the remainder is thought to be untranscribed (3,4,6). Each nucleus has approximately 100 copies of the palindrome providing 200 copies of the rRNA genes. The DNA in the palindromes is packaged into nucleosome-like higher order complexes observable under electron microscopy as ‘beads on a string’ (15). Susceptibility of specific sites to nuclease digestion changes during different stages of the asexual life cycle, correlated with changes in the rate of rRNA synthesis (16,17). Sequences at the ends of the palindrome have short sequence repeats similar to the telomeres characterized at the ends of chromosomes (18).
We have determined the sequence of the 88 kb extrachromosomal Dictyostelium rDNA palindrome and found that it is perfectly symmetrical except for a short central region. The sequences of the two arms appear to be identical to each other on the hundreds of copies in each nucleus implying that some rectification mechanism is at work. We present evidence for a locus on chromosome 4 that may be used to produce the extrachromosomal copies of the palindrome. We also suggest a solution to the long-standing question of whether there are six or seven chromosomes present in Dictyostelium by showing that clusters of the palindrome can form chromosome-like bodies that give the appearance of a nucleus that contains seven chromosomes.
MATERIALS AND METHODS
Shotgun sequencing
Nuclear DNA was purified from strain AX4 (19) by detergent lysis and CsCl gradient separation (20), and high molecular weight DNA was prepared in agarose as described previously (21). The 88 kb palindromic elements were separated from high molecular weight chromosomal DNA on pulsed field gels, sheared into fragments of 1.5–2.0 kb and cloned into modified M13 vectors using the previously described double-adaptor strategy (21,22). Automated sequencing was carried out on ABI-377 sequencing machines (Applied Biosystems) and the sequence assembled using the programs phred and phrap (http://www.phrap.org). Contig inspection and editing was done with the Staden package (http://www.mrc-lmb.cam.ac.uk/pubseq/). The two terminal 2.8 kb sequences were deposited in DDBJ/EMBL/GenBank under the accession nos AY171065 and AY171067, and the 79.3 kb central segment under accession no. AY171066.
Sequencing the central region of asymmetry
Nuclear DNA was digested with BamHI to generate the 2.8 kb fragment known to contain the central region of the palindrome (4). Since there are 100 copies of the palindrome per nucleus, the 2.8 kb BamHI fragment could be visualized with ethidium bromide following electrophoresis on a 0.8% agarose gel, isolated and electroeluted. This DNA was sheared to 100 bp fragments and cloned into M13 vectors for sequencing. The sequence of the assembled central region was validated by standard PCR techniques using primers based on the sequence of the asymmetric region.
Sequencing the termini
Nuclear DNA was digested with SalI to generate the 10 kb fragments known to occur at the termini of the palindrome. The fragments were isolated following the same protocol used for the central region and sequenced directly using 33P-labeled dideoxy-terminators and the Thermosequenase™ kit (USB). This same fragment was also used as a template to sequence across the one remaining gap (Fig. 1). These sequencing reactions failed precisely at the gap border.
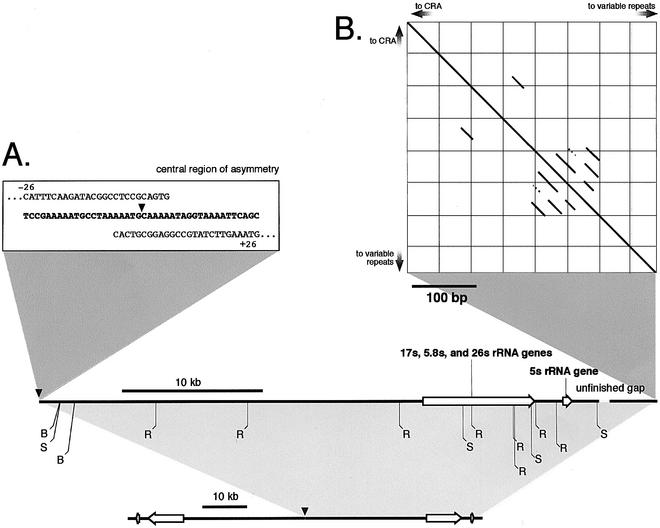
Figure 1.
Overview of the sequence of the rDNA palindrome. Only one arm is presented here. The other arm is an exact complement that extends to the left of the arm shown. The predicted restriction map agrees with previously published maps (B: BamHI, R: EcoRI, S: SalI). (A) The 42 bp central region of asymmetry (CRA) is shown in detail. (B) A Pustell DNA matrix (44) of the terminal sequences indicating the presence of a series of short repeats.
Chromosomal mapping
Sequences from the palindrome were mapped to specific yeast strains harboring the YAC library that has been ordered along the six chromosomes (21,23). Strains were arrayed in 96-well trays, and pools of strains were prepared from rows and columns prior to being grown. DNA samples prepared from the different yeast pools were used as the substrate for high-throughput PCR detection of sequence-tagged sites. Specific YACs identified through this process were confirmed with DNA isolated from the individual host yeast strains.
In situ hybridization
In situ hybridization was carried out essentially according to Bandyopadhyay et al. (24). Chromosome spreads were prepared from AX4 cells that were grown in axenic media (HL-5) to 2 × 107 cells/ml (25). To increase the proportion of cells with condensed chromosomes, nocodozole was added directly to cultures at 10 µg/ml for 2 h with shaking at 180 r.p.m. (26). Nocodazole-treated or untreated cells (200 µl) were then pelleted onto microscope slides treated with 3-aminopropyltriethoxysilane, as described by Henderson (27), at 2000 r.p.m. for 10 min using a cytospin centrifuge. Cells were then fixed in freshly prepared, ice-cold ethanol/glacial acetic acid (3:1) for 1 h, fixed a second time for 10 min and then air-dried at 37°C. Slides were stored at –20°C for future use.
The rDNA was visualized by FISH with a mixture of labeled templates derived from the sequencing libraries. Equimolar mixture of 12 templates (IIAIP2421, IICBP34344, IICBP46896, IICBP29965, IIAIP0564, IICBP45880, IICBP34683, IIAFP2537, IICBP45847, IIAIP3922, IICCP1088 and IICBP43759) that correspond to the rRNA-coding genes and totaling 1 µg were labeled with biotin- or digoxygenin-modified nucleotides. In situ hybridization was carried out according to Bandyopadhyay et al. (24). The standard denaturation conditions of 70°C for 2 min in 50% formamide and 2× SSC were used to examine interphase nuclei and produced the best FISH signals. To preserve the structure of the large DNAs observed in the nuclei of nocodazole-treated cells, the denaturation temperature was lowered to 55°C. After hybridization, coverslips were mounted on glass slides in 3 µl of Vectashield containing DAPI (10 µg/ml). Preparations were visualized on a Delta Vision deconvolution microscope and images were processed with the SoftWoRx (version 2.5) software package (Applied Precision, Issaquah, WA).
RESULTS
Using conventional M13 shotgun techniques and automated assembly of the reads from the linear extrachromosomal ele ment, a 38 kb contig was assembled with 16-fold average clone depth and with no ambiguous base calls. The sequence is avail able at http://dictygenome.bcm.tmc.edu/extrachromosomal/rDNA/sequencedRegion.html. Since there was no evidence of variations on one arm of the palindrome or the other, the 88 kb linear element appears to be a perfect palindrome in this region and all copies of the element within cells must be identical. A 5 kb contig representing the termini of the element failed to join the 38 kb contig leaving a gap that is probably <1 kb. We made numerous attempts to close this gap without success. These efforts included surveying the available worldwide genome sequence data, various PCR strategies, small insert libraries produced from the terminal 10 kb SalI fragment and direct sequencing of genomic DNA.
Sequencing the central region of the rDNA palindrome proved to be a challenge since we could not recover the central region as an intact clone in bacterial vectors, possibly due to the formation of secondary structures in the DNA. To circumvent this problem, we took advantage of the high relative abundance of the rDNA palindromic element and purified the 2.8 kb BamHI restriction fragment containing the central region directly from genomic DNA by gel electrophoresis. We sheared it into small pieces (∼100 bp) to reduce the likelihood that any fragment would form a secondary structure that might be targeted by bacterial restriction systems, and cloned the fragments in an M13 vector. This small insert library was sequenced to generate 10-fold depth of coverage and a robust 1.4 kb contig was assembled. One end of this contig overlapped the previously described 38 kb contig, while the other end contained a 42 bp non-palindromic stretch of sequence, followed by the beginning of the other arm of the palindrome. This 42 bp segment forms the central region of asymmetry that divides the two halves of the palindrome (CRA; Fig. 1).
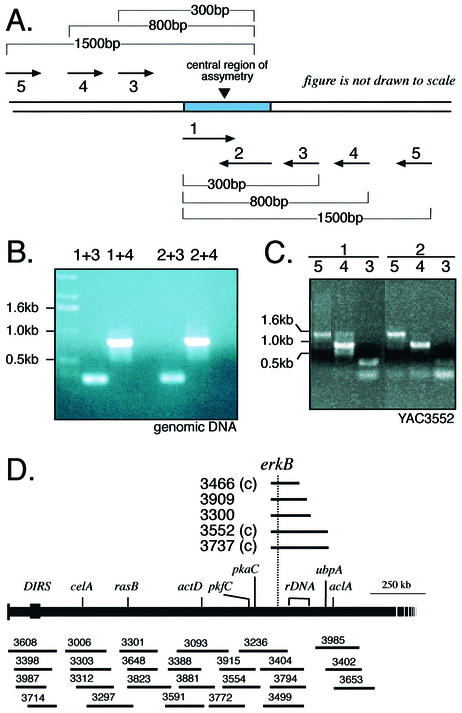
To validate the structure of the CRA, we designed a set of primers complementary to different strands within the region of asymmetry and paired them with flanking primers at known distances (Fig. 2A). Although the two CRA primers were distinct in sequence and targeted opposite strands, they generated identical PCR products with the flanking primers, consistent with the consensus sequence from the assembly and the symmetrical form of the palindrome (Fig. 2B).
Figure 2.
Validating the sequence of the central non-palindromic region of the rDNA element. (A) Oligonucleotides corresponding to each half of the central region were paired with oligonucleotides corresponding to sequences at predicted distances into the arms of the elements. (B) PCR amplification produced the predicted, identical DNA fragments. (C) The same predicted PCR fragments were amplified from YAC DNA derived from the chromosome 4 YACs. (D) Region of chromosome 4 containing the rDNA locus. The rDNA-containing YACs are shown above the chromosome map. The rDNA is located between the erkB and ubpA.
When a YAC library representing the Dictyostelium genome was probed with cloned fragments of the gene encoding the 35S rRNA precursor, eight YACs were identified, six of which also carried the erkB locus that is on chromosome 4 (21,23). The other two YACs carried genes from chromosome 2 and presumably contain some rDNA from that chromosome (28). We used PCR primers that specifically identify the CRA to interrogate the YAC library and found that all of the erkB containing YACs also carry the central region of the palindrome. To determine if the chromosomal copy of the palindrome is present as a half-element, as has been shown to be the case in Tetrahymena micronuclear rDNA (10,29), we used the DNA of YACs 3552 and 3737 as templates for a series of PCRs that map the extent of the symmetry around the CRA. We were able to reliably recognize sequences of the palindrome 1.6 kb from the CRA (Fig. 2C). Although we used primers to opposite strands, no evidence for heterogeneity in the arms was seen. Thus, the chromosome 4 rDNA locus has at least 3.2 kb of palindromic sequence surrounding the CRA as well as more distal sequences encoding rRNA. A complete copy of the palindromic sequence may be present on chromosome 4 between erkB and ubpA (Fig. 2D). Since the sequence of this chromosome has not yet been completed, the structures at the ends of this chromosome-embedded copy of the palindrome remain to be determined.
Emery and Weiner (18) previously analyzed the terminal EcoRI fragment of the palindrome. They found four nearly perfect, tandem repeats of a 29 bp sequence followed by variable tracts with the repeat unit AG1–8. We sequenced the terminal 10 kb SalI fragment after isolating it directly from genomic DNA and confirmed the previously reported sequence including the tandem repeats 48 bp upstream of three copies of the sequence AG7 (Fig. 1B). We were also able to extend the sequence of the palindrome such that only a small gap, estimated to be between 300 and 1000 bp in length, separates the 39 kb palindromic sequence from the 3.5 kb telomeric sequence (Fig. 1).
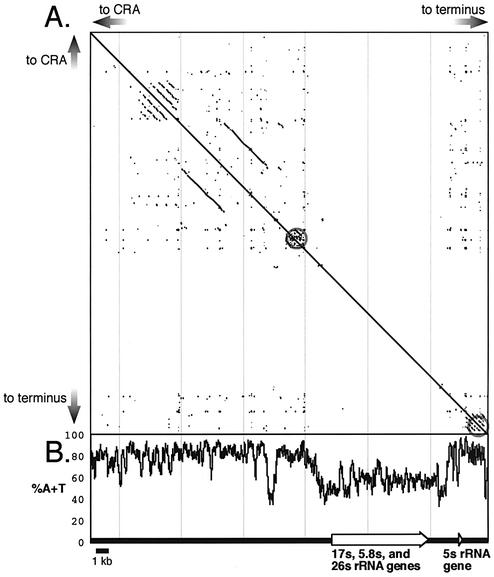
Although the sequence of the palindrome is generally very A/T-rich, sequences encoding rRNA are significantly more G/C-rich, consistent with the bimodal base composition of Dictyostelium DNA, where coding regions have higher G/C content than non-coding regions (Fig. 3). The palindrome is rife with short simple repeats that would seem to preclude it from encoding any genes outside of the rRNA genes. Upstream of the gene encoding the 35S rRNA precursor, there is a region of relatively high G/C content that may regulate rRNA transcription (30).
Figure 3.
Structural features of the extrachromosomal rDNA element. (A) Pustell dot-matrix autocomparison of one arm of the palindrome shows the prevalence of short repeated regions throughout except for the region transcribed into rRNA (44). Longer repeats are found towards the center of the palindrome. Two packets of highly repetitive DNA bracket the transcribed region (gray circles). The upstream and downstream sequences are unrelated. (B) The A+T content is high except in the transcribed regions and a short region upstream.
We used fluorescence in situ hybridization (FISH) with rDNA probes to examine the nuclear distribution of the rDNA palindromes. Within interphase nuclei the rDNA appeared as organized structures that surrounded regions devoid of DNA, presumably the multi-lobed nucleoli (Fig. 4A) (31). These structures are very similar to ones previously observed by FISH as well as by silver staining, and suggest that multiple copies of the rDNA palindrome associate non-covalently in exponentially growing cells (16). We also treated cells with the microtubule-destabilizing drug nocodazole to examine arrested mitotic nuclei (26). The nuclei of nocodazole-treated AX4 cells displayed seven chromosome-sized DNA bodies, as widely reported in the literature (Fig. 4B). However, we found that heating these samples in formamide at 70°C to denature the DNA in preparation for FISH resulted in only six of the chromosome-sized DNA bodies remaining intact; the seventh one dispersed into many smaller fragments. These smaller fragments always hybridized with an rDNA-specific FISH probe, indicating that these are the nuclear rDNA molecules (Fig. 4C).
Figure 4.
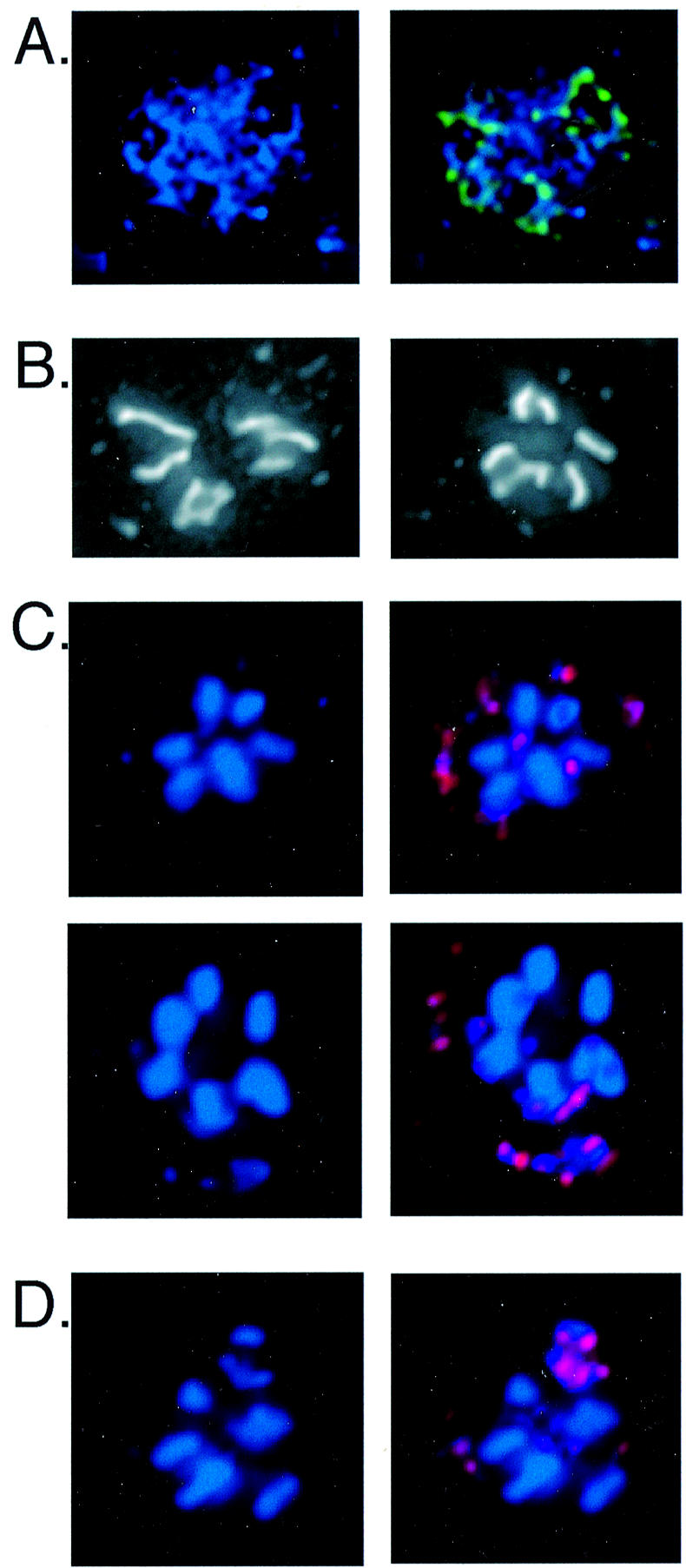
Nuclear distribution of extrachromosomal rDNA in AX4 cells. (A) An interphase nucleus stained with DAPI and combined on the right with the signal obtained by FISH with a mixture of rDNA probes (green). (B) Two examples of condensed nuclear DNA in nocodazole-treated cells stained with DAPI displaying seven chromosome-sized DNA bodies. (C) Two examples of rDNA FISH of nuclei from nocodazole-treated cells with the denaturation step carried out at 70°C. DAPI staining displaying six chromosome-sized DNAs along with many smaller DNA fragments (left panels) and the FISH signal (red) that identifies the smaller fragments as rDNA (right panels). (D) An example of rDNA FISH of a nocodazole- treated cell, with denaturation carried out at 55°C, in which the rDNA appears as chromosome-sized DNAs.
We could not find denaturing conditions that preserved the DNA structures seen in Figure 4B and also produced reliable FISH signals. However, lowering the denaturation temperature to 55°C caused less dispersion of the DNA and still produced robust FISH signals with the rDNA probes. Under these conditions we often observed chromosome-sized DNA bodies that hybridized with the rDNA probes (Fig. 4D). These results suggest that the extrachromosomal rDNA elements can form non-covalently associated chromosome-sized aggregates in cells that have been arrested in mitosis by nocodazole.
Discussion
Sequencing the linear extrachromosomal element that carries the rRNA genes confirmed its general palindromic structure and showed that the two arms are indistinguishable. The only region of asymmetry is a 42 bp segment at the center of the palindrome. Small A/T-rich asymmetric segments have also been reported to occur at the center of the palindromes of two Tetrahymena species, but they have no significant sequence similarity with the 42 bp Dictyostelium segment (32,33). A Pustell dotplot matrix illustrates that the Dictyostelium rDNA palindrome is peppered with a large number of direct and inverted, small, low-complexity repeats, as well as pockets of large direct repeats (Fig. 3). The 10 kb regions coding for the rRNA genes and a 500 bp stretch 2.5 kb upstream of these 35 S gene are relatively free of repeats and have a lower A+T content, and are bracketed by a number of complex direct repeats (Fig. 3). The upstream region includes the mapped nuclease hypersensitive sites and a DNA topoisomerase site (30). Regulatory proteins that control rRNA expression are likely to bind this region while the repetitive sequences may be important for packaging of the element.
Direct sequencing of the palindromic element near the telomeres recognized the four nearly perfect tandem repeats with the 29 bp sequence reported by Emery and Weiner (18). The distal sequence also agrees with that reported by Emery and Weiner including three copies of an AG7 repeat (18). Although the strains used in this study and that of Emery and Weiner were independently cultured for many years, the terminal sequence of the palindrome has remained essentially constant. By sequencing independent clones carried in bacterial vectors, Emery and Weiner also were able to characterize more distal portions and found a variable number of repeats with the form AG1–8 that can account for the micro-heterogeneity that they observed at the ends of the linear extrachromosomal elements (18).
Previous reports have suggested that a copy of the palindrome is present in one of the chromosomes. Emery and Weiner probed digests of nuclear DNA with the terminal EcoRI fragment and found the expected high-copy fragments of the extrachromosomal copies as well as a single-copy fragment, which they ascribed to a chromosomal copy (18). In addition, Hofmann et al. reported a chromosomally encoded pseudogene corresponding to the 5S rRNA gene (13). By probing YACs carrying mapped regions of the Dictyostelium chromosomes, we found rRNA coding regions closely linked to the erkB gene on chromosome 4. Although YAC libraries are known to contain some chimeric inserts, the linkage of erkB to rDNA on six independently isolated YACs provides strong evidence for the presence of a chromosomally encoded rDNA copy on chromosome 4. Using DNA from these YACs as templates for PCR, we found that the central region of asymmetry was also present and flanked by at least 1.6 kb of palindromic sequence on both sides. Unlike the micronuclear integrated copy in Tetrahymena, which is a half-element that is opened and replicated to form the macronuclear extrachromosomal element (10,29), the chromosome 4-linked copy of the rDNA in Dictyostelium appears to contain more than a half-element. However, a complete description this chromosomal locus will only become apparent as the sequence of chromosome 4 is completed during the course of the ongoing genome project. An additional rDNA locus has been identified on chromosome 2, but its exact structure has not been determined (28).
There is genetic evidence that extrachromosomal copies of the rDNA elements replicate autonomously in Dictyostelium, and that their stable transmission to daughters cells does not require a chromosomal master copy (20). However, these studies do not preclude the requirement for a chromosomal copy under certain conditions or for maintaining the sequence integrity of the rRNA genes over time. If the extrachromosomal elements accumulate deleterious mutations, fresh copies generated from the chromosomal locus could be added to the population of elements. Over time, this mechanism could homogenize the population. While such a mechanism could account for the sequence identity between individual extrachromosomal rDNA elements, it is not sufficient to account for the strict sequence identity of the arms. Some rectification mechanism must function to keep the arms identical. One possibility is that one or the other arm of the chromosomal locus is periodically replaced by one of the hundred extrachromosomal copies by homologous recombination. Since the arms are indistinguishable, either the right arm or the left arm could pair with one of the locus arms. Subsequent copies generated by the rectified chromosomal locus would then come to predominate. The creation of defined modifications of the rDNA locus on chromosome 4 would be required to test these ideas.
The sequence of the palindrome can be affected through recombination. Cole and Williams found size heterogeneity in the extrachromosomal palindromes of certain mutant strains. In these strains, the extrachromosomal elements were present at discrete sizes up to 300 kb (34). The ladder of sizes resulted from a variable number of tandem duplications of a 34 kb region from the center of the palindrome. A dominant mutation that mapped to chromosome 4 was responsible for this phenotype. The simplest explanation for such a mutation would be an alteration in the structure of the rDNA locus on chromosome 4, and that would suggest that this locus acts as a master copy for the extrachromosomal elements. One possible scenario is that the original chromosomal mutation resulted in an insertion near the central region of the palindrome that generated asymmetric extrachromosomal elements. When an extrachromosomal element with an expanded arm recombined with the other arm of the chromosomal locus, a large symmetric duplication was formed. Further offset recombination could lead to expansion of the 34 kb region. When the enlarged locus generated extrachromosomal copies that replaced previous copies, the palindrome would be observed to increase in size. Such a mechanism would account for the dominance of the original mutation since modified versions would continue to appear even in the presence of a wild-type copy. After many generations, one allele or the other would be expected to predominate as has been found in diploid cell lines generated from strains showing restriction fragment length polymorphisms of the palindrome (20).
There are numerous reports that haploid Dictyostelium nuclei contain seven chromosome-sized DNA bodies (26,35–39). However, genetic analysis over the past 35 years established six large linkage groups and physical mapping of the genome has identified only six large chromosomes (40–42). One small additional linkage group containing two genes has been described (43). Given the large number of genes that have been mapped over the years, it is unlikely that this linkage group represents a large chromosome unless it is very gene-poor. Thus, one chromosome-sized DNA of 4–8 Mb that can be seen in nuclei has not been detected by physical analyses. Curiously, the 8 Mb of rDNA within each cell which represents 20% of the nuclear DNA has remained unaccounted for in the microscopic examinations of mitotic nuclei described in the literature. rDNA monomers were assumed to be not detectable above the background staining in any DNA visualization technique using light microscopy, but the fact that the monomers are known to form higher order structures suggests that the rDNA should be visible (16).
We used rDNA-specific FISH to look at extrachromosomal rDNA localization and found that rDNA monomers associate to form rings within interphase nuclei, probably surrounding the nucleoli, as seen previously (16). However, when cells were treated with nocodazole, the rDNA ring structure tends to be disrupted and instead one sees linear aggregates. In many cases, the rDNA aggregate appeared as large as the other chromosomes. Thus, the 8 Mb of rDNA can condense into chromosome-sized DNA under certain conditions. These results suggest that the seventh large DNA body observed in the metaphase nucleus of Dictyostelium is probably a higher order aggregate of extrachromosomal rDNA. These rDNA aggregates may be artificially induced by nocodazole treatment or they may form naturally during mitosis, possibly serving as a segregation unit for the rDNA.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge the assistance of the staff of the Human Genome Sequencing Center at Baylor College of Medicine. We would like to thank Laura Cortez for technical assistance and Brian Desany for informatics assistance. We thank Robert Insall and Jason King (University of Birmingham) for showing us their unpublished images of seven chromosome-sized DNAs in nocodazole-treated cells. We thank the Dictyostelium Genome Sequencing Consortium for access to genomic DNA sequences prior to publication. The Consortium’s principal members include: Marie-Adele Rajandream and Bart Barrell (Sanger Institute, Cambridge); Angelika Noegel and Ludwig Eichinger (University of Cologne); Gernot Glockner and Matthias Platzer (Institute of Molecular Biotechnology, Jena); Jeffrey Williams (University of Dundee); Paul Dear and Robert Kay (MRC Laboratory of Molecular Biology, Cambridge); and Edward Cox (Princeton University). This work was supported by a grant from the Institute of Child Health and Development, National Institutes of Health, to A.K. (RO1 HD35925).
DDBJ/EMBL/GenBank accession nos+ AY171065–AY171067
REFERENCES
- 1.Gall J.G. (1974) Free ribosomal RNA genes in the macronucleus of Tetrahymena. Proc. Natl Acad. Sci. USA, 71, 3078–3081. [DOI] [PMC free article] [PubMed]
- 2.Karrer K.M. and Gall,J.G. (1976) The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J. Mol. Biol., 104, 421–453. [DOI] [PubMed]
- 3.Maizels N. (1976) Dictyostelium 17S, 25S, and 5S rDNAs lie within a 38,000 base pair repeated unit. Cell, 9, 431–438. [DOI] [PubMed]
- 4.Cockburn A.F., Newkirk,M.J. and Firtel,R.A. (1976) Organization of the ribosomal RNA genes of Dictyostelium discoideum: mapping of the non transcribed spacer regions. Cell, 9, 605–613. [DOI] [PubMed]
- 5.Vogt V.M. and Braun,R. (1976) Structure of ribosomal DNA in Physarum polycephalum. J. Mol. Biol., 106, 567–587. [DOI] [PubMed]
- 6.Cockburn A.F., Taylor,W.C. and Firtel,R.A. (1978) Dictyostelium rDNA consists of non-chromosomal palindromic dimers containing 5S and 36S coding regions. Chromosoma, 70, 19–29. [DOI] [PubMed]
- 7.Worton R.G., Sutherland,J., Sylvester,J.E., Willard,H.F., Bodrug,S., Dube,I., Duff,C., Kean,V., Ray,P.N. and Schmickel,R.D. (1988) Human ribosomal RNA genes: orientation of the tandem array and conservation of the 5′ end. Science, 239, 64–68. [DOI] [PubMed]
- 8.Daniel D.C. and Johnson,E.M. (1989) Selective initiation of replication at origin sequences of the rDNA molecule of Physarum polycephalum using synchronous plasmodial extracts. Nucleic Acids Res., 17, 8343–8362. [DOI] [PMC free article] [PubMed]
- 9.Blomberg P., Randolph,C., Yao,C.H. and Yao,M.C. (1997) Regulatory sequences for the amplification and replication of the ribosomal DNA minichromosome in Tetrahymena thermophila. Mol. Cell Biol., 17, 7237–7247. [DOI] [PMC free article] [PubMed]
- 10.Yao M.C. and Gall,J.G. (1977) A single integrated gene for ribosomal RNA in a eucaryote, Tetrahymena pyriformis. Cell, 12, 121–132. [DOI] [PubMed]
- 11.Vogt V.M. and Braun,R. (1977) The replication of ribosomal DNA in Physarum polycephalum. Eur. J. Biochem., 80, 557–566. [DOI] [PubMed]
- 12.Ferris P.J., Vogt,V.M. and Truitt,C.L. (1983) Inheritance of extrachromosomal rDNA in Physarum polycephalum. Mol. Cell Biol., 3, 635–642. [DOI] [PMC free article] [PubMed]
- 13.Hofmann J., Winckler,T., Hanenkamp,A., Bukenberger,M., Schumann,G., Marschalek,R. and Dingermann,T. (1993) The Dictyostelium discoideum 5S rDNA is organized in the same transcriptional orientation as the other rDNAs. Biochem. Biophys. Res. Commun., 191, 558–564. [DOI] [PubMed]
- 14.Kimmel A.R. and Gorovsky,M.A. (1978) Organization of the 5S RNA genes in macro- and micronuclei of Tetrahymena pyriformis. Chromosoma, 67, 1–20. [DOI] [PubMed]
- 15.Simon I. and Olins,D.E. (1994) Higher-order association of extrachromosomal rDNA genes in Dictyostelium discoideum. Cell Biol. Int., 18, 1091–1094. [DOI] [PubMed]
- 16.Parish R.W., Schmidlin,S., Fuhrer,S. and Widmer,R. (1980) Electrophoretic isolation of nucleosomes from Dictyostelium nuclei and nucleoli. FEBS Lett., 110, 236–240. [DOI] [PubMed]
- 17.Ness P.J., Labhart,P., Banz,E., Koller,T. and Parish,R.W. (1983) Chromatin structure along the ribosomal DNA of Dictyostelium. Regional differences and changes accompanying cell differentiation. J. Mol. Biol., 166, 361–381. [DOI] [PubMed]
- 18.Emery H.S. and Weiner,A.M. (1981) An irregular satellite sequence is found at the termini of the linear extrachromosomal rDNA in Dictyostelium discoideum. Cell, 26, 411–419. [DOI] [PubMed]
- 19.Knecht D.A., Cohen,S.M., Loomis,W.F. and Lodish,H.F. (1986) Developmental regulation of Dictyostelium discoideum actin gene fusions carried on low-copy and high-copy transformation vectors. Mol. Cell. Biol., 6, 3973–3983. [DOI] [PMC free article] [PubMed]
- 20.Welker D.L., Hirth,K.P. and Williams,K.L. (1985) Inheritance of extrachromosomal ribosomal DNA during the asexual life cycle of Dictyostelium discoideum: examination by use of DNA polymorphisms. Mol. Cell. Biol., 5, 273–280. [DOI] [PMC free article] [PubMed]
- 21.Kuspa A., Maghakian,D., Bergesch,P. and Loomis,W.F. (1992) Physical mapping of genes to specific chromosomes in Dictyostelium discoideum. Genomics, 13, 49–61. [DOI] [PubMed]
- 22.Andersson B., Wentland,M.A., Ricafrente,J.Y., Liu,W. and Gibbs,R.A. (1996) A ‘double adaptor’ method for improved shotgun library construction. Anal. Biochem., 236, 107–113. [DOI] [PubMed]
- 23.Kuspa A. and Loomis,W.F. (1996) Ordered yeast artificial chromosome clones representing the Dictyostelium discoideum genome. Proc. Natl Acad. Sci. USA, 93, 5562–5566. [DOI] [PMC free article] [PubMed]
- 24.Bandyopadhyay R., McQuillan,C., Page,S.L., Choo,K.H. and Shaffer,L.G. (2001) Identification and characterization of satellite III subfamilies to the acrocentric chromosomes. Chrom. Res., 9, 223–233. [DOI] [PubMed]
- 25.Sussman M. (1987) Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol., 28, 9–29. [DOI] [PubMed]
- 26.Welker D.L. and Williams,K.L. (1980) Mitotic arrest and chromosome doubling using thiabendazole, cambendazole, nocodazole, and ben late in the slime mould Dictyostelium discoideum. J. Gen. Microbiol., 116, 397–407.
- 27.Henderson C. (1989) Aminoalkylsilane: an inexpensive, simple preparation for slide adhesion. J. Histotechnol., 12, 123–124.
- 28.Glöckner G., Eichinger,L., Szafranski,K., Pachebat,J., Dear,P., Lehmann,R., Baumgart,C., Abril,J.F., Parra,G., Guigó,R., Kumpf,K., Tunggal,B., Cox,E., Quail,M.A., the Dictyostelium Genome Sequencing Consortium, Platzer,M., Rosenthal,A. and Noegel,A.A. (2002) Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature, 418, 79–85. [DOI] [PubMed]
- 29.Yasuda L.F. and Yao,M.C. (1991) Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell, 67, 505–516. [DOI] [PubMed]
- 30.Ness P.J., Parish,R.W. and Koller,T. (1986) Mapping of endogenous nuclease-sensitive regions and of putative topoisomerase sites of action along chromatin of Dictyostelium ribosomal RNA genes. J. Mol. Biol., 188, 287–300. [DOI] [PubMed]
- 31.Roos U.-P., Bottini,F. and Jenni,V. (1992) Morphology of the nucleolus in undifferentiated amoebae of Dictyostelium discoideum. Eur. J. Protistol., 28, 94–101. [DOI] [PubMed]
- 32.Kiss G.B. and Pearlman,R.E. (1981) Extrachromosomal rDNA of Tetrahymena thermophila is not a perfect palindrome. Gene, 13, 281–287. [DOI] [PubMed]
- 33.Kan N.C. and Gall,J.G. (1981) Sequence homology near the center of the extrachromosomal rDNA palindrome in Tetrahymena. J. Mol. Biol., 153, 1151–1155. [DOI] [PubMed]
- 34.Cole R.A. and Williams,K.L. (1992) Tandem repeats in extrachromosomal ribosomal DNA of Dictyostelium discoideum, resulting from chromosomal mutations. Genetics, 130, 757–769. [DOI] [PMC free article] [PubMed]
- 35.Ross I.K. (1960) Studies on diploid strains of Dictyostelium discoideum. Am. J. Bot., 47, 54–59.
- 36.Sussman R.R. (1961) A method for staining the chromosomes of Dictyostelium discoideum myxamoebae in the vegetative stage. Exp. Cell Res., 24, 154–155.
- 37.Brody T. and Williams,K.L. (1974) Cytological analysis of the parasexual cycle in Dictyostelium discoideum. J. Gen. Microbiol., 82, 371–383.
- 38.Robson G.E. and Williams,K.L. (1977) The mitotic chromosomes of the cellular slime mould Dictyostelium discoideum: a karyotype based on Giemsa banding. J. Gen. Microbiol., 99, 191–200.
- 39.Zada-Hames I.M. and Ashworth,J.M. (1978) The cell cycle during the vegetative stage of Dictyostelium discoideum and its response to temperature change. J. Cell Sci., 32, 1–20. [DOI] [PubMed]
- 40.Newell P.C., Williams,K.L., Kuspa,A. and Loomis,W.F. (1993) Genetic map of Dictyostelium. In O’Brien,S.J. (ed.), Genetic Maps: Locus Maps of Complex Genomes, 6th Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 3.01–3.10.
- 41.Cox E.C., Vocke,C.D., Walter,S., Gregg,K.Y. and Bain,E.S. (1990) Electrophoretic karyotype for Dictyostelium discoideum. Proc. Natl Acad. Sci. USA, 87, 8247–8251. [DOI] [PMC free article] [PubMed]
- 42.Loomis W.F., Welker,D., Hughes,J., Maghakian,D. and Kuspa,A. (1995) Integrated maps of the chromosomes in Dictyostelium discoideum. Genetics, 141, 147–157. [DOI] [PMC free article] [PubMed]
- 43.Darcy P.K., Wilczynska,Z. and Fisher,P.R. (1993) Phototaxis genes on linkage group V in Dictyostelium discoideum. FEMS Microbiol. Lett., 111, 123–127. [DOI] [PubMed]
- 44.Pustell J. and Kafatos,F.C. (1982) A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res., 10, 4765–4782. [DOI] [PMC free article] [PubMed]