Abstract
Here we devise a new method for high-throughput comparative sequence analysis. The developed protocol comprises a homogeneous in vitro transcription/RNase cleavage system with the accuracy and data acquisition speed of matrix-assisted laser desorption/ionization coupled with time-of-flight mass spectrometry (MALDI-TOF MS). In summary, the target region is PCR amplified using primers tagged with promoter sequences of T7 or SP6 RNA polymerase. Using RNase T1, the in vitro transcripts are base-specifically cleaved at every G-position. This reaction results in a characteristic pattern of fragment masses that is indicative of the original target sequence. To enable high-throughput analysis, samples are processed with automated liquid handling devices and nanoliter amounts are dispensed onto SpectroCHIP arrays for reliable and homogeneous MALDI preparation. This system enables rapid automated comparative sequence analysis for PCR products up to 1 kb in length. We demonstrate the feasibility of the devised method for analysis of single nucleotide polymorphisms (SNPs) and pathogen identification.
INTRODUCTION
The amount of available genomic sequence information from various organisms is growing dramatically. This knowledge enables the correlation of DNA sequence to protein function, phenotype or identity (1). In particular, the analysis of single nucleotide polymorphisms (SNPs) will have a significant impact on identification of human disease susceptibility genes and facilitate development of new drugs and patient care strategies. The demand for high-throughput methods enabling large scale analysis of genetic variability and implementation of dense SNP maps is therefore eminent (2).
The identification of pathogens, to name a further fast developing field, involved in bacterial and viral infections requires fast and accurate technologies for comparative sequence analysis (3). Not only their identification, but also the fast assessment of newly emerging sequence variations is an important aspect for the field of molecular diagnostics.
The increasing need for those comparative tools has furthered the development of several new approaches. Among those, mass spectrometry-based strategies have been extensively studied. Electrospray ionization and matrix- assisted laser desorption/ionization (MALDI) are the most common soft ionization techniques for the mass spectrometric analysis of nucleic acids (4–6). The use of MALDI coupled with time-of-flight mass spectrometry (MALDI-TOF MS) has become one of the leading technologies for SNP-scoring and determination of allelic frequencies (7). MALDI-TOF MS of nucleic acids is accurate, extremely fast and the inherent amenability to automation makes it suitable for high- throughput analysis. However, most of the current methods are based on sequence-specific extension and termination and limited to the analysis of short nucleic acids stretches (<60 nt) (8,9).
Further strategies for comparative sequence analysis includ ing other enzymatic and also chemical cleavage methods of DNA or RNA have been devised.
One example for an enzymatic DNA-based fragmentation approach is the uracil-DNA-glycosylase (UDG)-treatment of a PCR product, where dUTP completely replaces dTTP. After strand separation and UDG-treatment of the dU-containing PCR product, alkaline and heat treatment facilitates DNA cleavage at each T-position. Fragment detection is performed either by gel electrophoretic separation coupled with detection of fluorescent labeled products (10) or by MALDI-TOF MS (11,12). The strand separation necessary to obtain single-stranded DNA is currently performed by a bead-based approach utilizing a biotinylated primer reversibly immobilized to streptavidin-coated magnetic beads. However, automatic handling of a magnetic bead-based assay is more complicated than assay formats requiring only subsequent addition of reagents. In most cases, the use of a solid-phase separation is more expensive unless significant volume miniaturization can be obtained.
An approach based on chemical cleavage utilizes P3′-N5′-phosphoramidate-containing DNA (13). Either dCTP or dTTP are replaced by their analog P-N modified nucleoside triphosphates and are introduced into the target sequence during a primer extension reaction subsequent to PCR. Acidic reaction conditions produce base-specific cleavage fragments, which are analyzed by MALDI-MS. However, the required acidic conditions produce unwanted depurination by-products. A base loss of adenine and guanine is routinely observed and needs to be suppressed by incorporating 7-deaza analogs of dA and dG.
Although both of these DNA methods are robust and reasonably easy to handle, each approach is limited by the relatively low yield of single-stranded DNA products, which prevents minimizing the reaction volumes without a significant loss of sensitivity. This is of special importance for applications, where target regions of different length need to be isolated under uniform reaction conditions. The recovery yield of immobilized DNA template longer than 200 bp can decrease dramatically with increasing length and may require individual optimization for the immobilization step.
The use of post-PCR in vitro transcription systems provides an elegant solution for some of the issues encountered in the classical DNA amplification and primer extension combinations (14,15). Due to the additional amplification step, the amount of analyte available for mass spectrometric analysis is typically 50–100 times higher. Additionally, the process generates a single-stranded nucleic acid, eliminating the need for strand separation. Lastly, the RNA transcription process is beneficial especially for MALDI-TOF MS analysis due to the intrinsic property of RNA: the stability of RNA during the desorption/ionization process is higher than that of DNA, because of the balancing effect of the 2′-hydroxy group on polarization of the N-glycosidic bond of protonated bases (16).
In general, RNA cleavage methods can be divided into base-specific cleavage reaction and limited endo-/exonucleolytic cleavage reaction. Limited endonucleolytic cleavage of RNA with e.g. RNase T1 (G-specific) or RNase U2 (A-specific) was previously used in combination with time-limited alkaline hydrolysis to deduce sequence information from a synthetic 25mer RNA (17). However, the reading frame of the digested RNA was limited to 50 nt or less, because mass resolution as well as detection sensitivity decrease rather significantly with increasing analyte mass.
While this MALDI-MS-specific feature can efficiently be counteracted if only one or at most a few large analytes are present, it usually imposes insurmountable problems if smaller analytes in a mixture effectively suppress the high-mass ion signals. This is a limitation for many methods, such as incomplete endonuclease digestion. Mass resolution is compromised in this case by several factors. Efficient desalting requires elaborate sample conditioning and gets increasingly difficult with an increasing number of phosphates. Mass resolution can also be severely compromised if components of the preceding transcription and/or cleavage reaction such as enzymes or particularly unsuitable buffers remain in the solution and deteriorate the matrix crystallization during sample preparation. Hence, the buffers used in preceding enzymatic reactions need to be assessed in this respect. Only those can be used that have a minimal influence on the crystallization and do not hamper the MALDI process.
In practice, purification and sample conditioning of the nucleic acid is required to obtain reproducible mass spectra quality with sufficient signal/noise (S/N) ratios. Purification schemes for RNA transcripts employ for example 5′ biotinylation followed by isolation via streptavidin-coated magnetic beads (17). Alternatively, non-tagged RNA has been purified via a reversible hybridization process, where synthetic oligonucleotides, complementary to the 3′end the RNA target, are covalently bound to magnetic beads and enable the specific capturing and purification of RNA strands (18). However, the resulting recovery yield of both immobilization methods is not quantitative. An efficient high-throughput method would largely benefit from a format allowing homogeneous transcription and cleavage without any intermediate immobilization-based purification.
In this study, we report a semi-automated protocol for a homogeneous RNA transcription and a G-specific endonucleolytic cleavage reaction with RNase T1 to analyze and verify the sequence identity of a target region of interest. The cleavage reaction produces a characteristic pattern of fragment masses, which is indicative for the individual target sequence of interest and sequence changes thereof.
MATERIALS AND METHODS
Oligonucleotides were purchased from Metabion (Germany). 5-Methylcytidine 5′-triphosphate lithium salt (Me-CTP) and 5-methyluridine 5′-triphosphate lithium salt (Me-UTP) were obtained from Trilink (USA).
PCR primer sequences
Each forward PCR primer carries a T7 promotor site attached to the 5′ side of the gene-specific primer sequence, each reverse PCR primer carries a SP6 promotor site. Apolipo protein B-100 (ApoB-100): forward PCR primer 5′-gtaata cgactcactatagggCTTACTTGAATTCCAAGAGC-3′; reverse PCR primer 5′-atttaggtgacactatagaaGGCTGACTTGCATGGACC-3′. UCP2: forward PCR primer 5′-gtaatacgactcactatagggtcttggccttgcagatccaag-3′; reverse PCR primer 5′-att taggtgacactatagaaccatcacaccgcggtactg-3′. Cholesteryl-ester-transfer protein: forward PCR primer 5′-gtaatacgactcactataggg tccagggaggactcaccatg-3′; reverse PCR primer 5′-atttaggtgacactatagaatgactgcaggaagctctgg-3′.
PCR amplification
A 5 µl PCR contained 5 ng of genomic DNA, 0.1 U of HotStarTaq (Qiagen, Germany), 1 pmol forward and reverse primer, respectively, 0.2 mM of each dNTP and 1× Hot Star Taq PCR buffer (Qiagen) as supplied by the enzyme manufacturer [contains 1.5 mM MgCl2, Tris–Cl, KCl, (NH4)2SO4 pH 8.7]. The temperature profile consisted of an initial enzyme activation performed at 94°C for 15 min, followed by 45 cycles of 94°C for 20 s, 56°C for 30 s, 72°C for 60 s and a final incubation at 72°C for 3 min.
RNA transcription and RNase T1 cleavage
A 2.4 µl aliquot of the obtained 5 µl PCR product was utilized for a 6 µl transcription reaction containing 10 U of T7 (or SP6) RNA polymerase (Epicentre), 0.5 mM of each NTP and 1× transcription buffer (Epicentre; contains 6 mM MgCl2, 10 mM DTT, 10 mM NaCl, 10 mM Spermidine, 40 mM Tris–Cl pH 7.9 at 20°C). In cases of transcription with Me-UTP or Me-CTP, one of the corresponding natural nucleotide triphosphate is completely replaced by its chemically modified analog. Transcription reactions were incubated at 37°C for 2 h. After completion, 20 U of RNase T1 were added to the reaction mixture and incubated for 30 min at 30°C.
Sample conditioning and mass spectrometry
Each sample was diluted with 21.0 µl ddH2O. Conditioning of the phosphate backbone was achieved using 6 mg SpectroCLEAN™ resin (SEQUENOM, USA). Sixteen nanoliters of the supernatant were robotically dispensed onto a silicon chip (SpectroCHIP™, SEQUENOM) as described (19). All mass spectra were recorded with a Biflex III mass spectrometer (Bruker Daltonik, Germany). Exclusively, positive ions were analyzed and approximately 50 single-shot spectra were accumulated. All samples were analyzed in linear TOF mode using delayed ion extraction and a total acceleration voltage of 20 kV.
Bacterial typing
Details of the strains and reference sequences used for bacterial typing are described by von Wintzingerode et al. (11). In brief, primers TPU1 (5′-AGAGTTTGATCMTGGCTCAG-3′, corresponding to Escherichia coli 16S rRNA positions 8–27) and RTU334a (5′-TGCCACCCGTAGGTGTATGG-3′, corresponding to E.coli 16S rRNA positions 334–353) were synthesized with a T7 and SP6 promotor tag, respectively (as described above). PCR, in vitro transcription, base-specific cleavage and sample preparation on chip arrays were performed as described above. For the spectra displayed in Figure 7, transcription was performed from the reverse direction using SP6 RNA polymerase. Discrimination of bacterial strains was performed based on their base-specific cleavage-derived mass signal patterns.
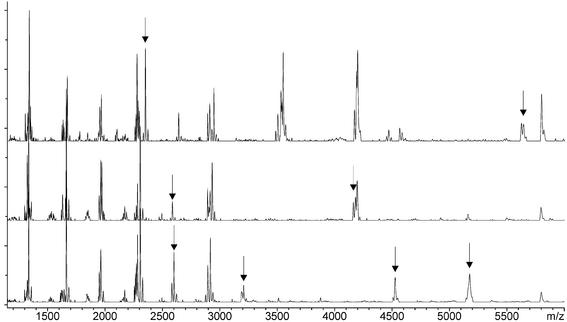
Figure 7.
Overlay of three fragmentation mass spectra of different Bordetella strains (16S rDNA). RNA in vitro transcription was performed with SP6 RNA polymerase wherein CTP was replaced by Me-CTP. RNase T1 fragmentation reaction of 16S rDNA of yet-uncultured rDNA clone SHA 110 (upper spectrum), bacterium B-52 (middle spectrum) and Bordetella trematum (lower spectrum). Unique signals, marked by an arrow, were used for strain identification.
RESULTS AND DISCUSSION
Method evaluation
The schematic of our devised approach is documented in Figure 1. For evaluation and optimization of the protocol a 306 bp PCR-product derived from the ApoB-100 gene, tagged with T7 RNA and SP6 RNA promotor sequence, was used. The post-PCR reaction was divided into two tubes; transcription of the forward strand was performed with T7 RNA polymerase in one tube and RNA transcripts of the reverse strand were produced in the other tube using SP6 RNA polymerase (Fig. 1).
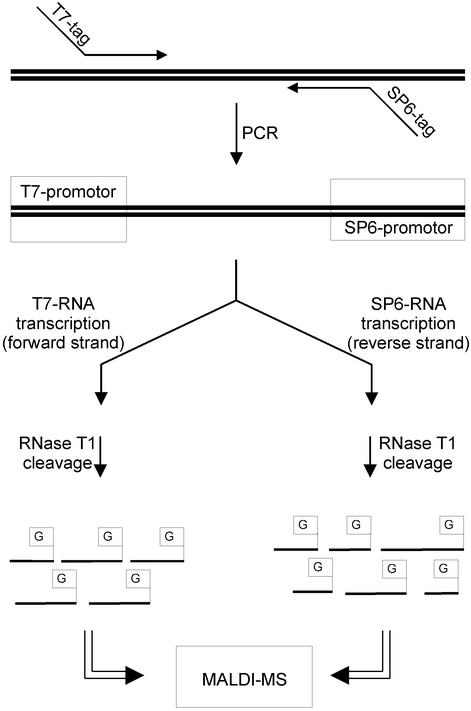
Figure 1.
Scheme depicting assay design for comparative sequence analysis by base-specific RNA cleavage reaction. Promotor sequences of T7 and SP6 RNA polymerase are tagged at the PCR primers adjacent to the target region. For post-PCR amplification, the sample is split and forward and reverse strands of the PCR product are transcribed into RNA by T7 and SP6 RNA polymerases, respectively. G-specific RNA cleavage reaction is performed by RNase T1 and obtained fragments are analyzed by MALDI-TOF MS.
In contrast to previous studies (17), RNase T1 cleavage was driven to completion. RNase amounts were optimized to yield complete cleavage for a large range of transcript product yields. Denaturing reagents, such as urea or formamide, which are known to disturb analyte/matrix crystallization, were avoided. An advantage of the present homogeneous approach over a limited/incomplete digestion is that it can be extended to template regions of 500 nt or more, without loss of information due to suppression of mass signals in higher mass range (>12 000 Da). The highest mass signal (and its corresponding cleavage product) in complete digests is sequence dependent and determined by the largest distance between two base-specific cleavage positions, but independent of the length of template (RNA transcript).
Since a homogenous single-well assay format does not allow any washing steps or removal of liquids, all reagents and reagent components have an influence on the downstream MALDI-TOF MS analysis. The evaluation and optimization of all these reagents and parameters was therefore required. Best performance was achieved with small, 5 µl PCR set-ups. One PCR reaction provides sufficient volume to carry out two separate transcription reactions to analyze both the forward and reverse strand. Sufficient PCR product yield and quality is achieved with 5 ng genomic DNA and 1 pmol of each gene-specific primer. An increase of DNA concentration resulted only in slight increases of yields. Increased primer concentration, however, led in some cases to a significant generation of primer dimers (data not shown), which results in unpredictable cleavage products interfering with the subsequent analysis. The optimized PCR conditions provide a universal protocol that we successfully applied to a wide range of target regions (data not shown). To reduce the costs of the RNA transcription and cleavage reaction per sample, the total volume of each reaction was minimized without loss in data quality of individual mass spectra, i.e. S/N ratio of all fragment signals and their mass accuracy. Reproducible in vitro transcript yields were obtained by using as little as 8 U of T7 RNA or SP6 RNA polymerase in a 6 µl transcription reaction, independent of the PCR-amplified target region and template length. To determine reproducibility, 26 individual samples were processed in replicates with automated liquid handling devices using 384-well MTP format. The optimal amount of RNase T1 required to generate exclusively RNA fragments with 3′-phosphate groups was 20 U. The incubation temperature of the cleavage reaction was optimized as well. In contrast to cleavage reactions at 37°C or even higher temperatures, which commonly generated a 1:3 mixture of 2′-3′-cyclic phosphates and 3′-phosphates, incubation at 30°C was found to force the cleavage reaction towards the 3′-phosphate group. This eliminated complexity generated by multiple mass signals for each given parent fragment in the mass spectrum. Furthermore, ribonucleoside triphosphate concentration, transcription buffer composition and the amount of RNA polymerase were optimized to guarantee reproducibility of the homogeneous RNA-based cleavage assay.
Miniaturized MALDI sample preparation with nanodispensing devices that transfer the sample onto a chip array (19), has been shown to surpass the standard 3-HPA macro-preparation. Non-homogeneous analyte distribution in the MALDI sample (hot spot formation), which is almost always observed in 3-HPA macro preparations and hampers automated MALDI measurement, is substantially less pronounced by the miniaturized and homogeneous sample crystallization on the chip array. Also, analyte disproportioning in the sample leading to either suppressed low or high mass windows of the full spectrum of analyte masses was not observed with this preparation method. As a result of the miniaturized MALDI sample, the acquisition time for the automatic mass spectrometry measurement could be reduced to 5 s for any single sample (accumulation of 50 single spectra at 10 Hz laser repetition rate).
Important for the success of the homogeneous base-specific cleavage approach was the investigation of sample conditioning methods, which suit the needs of reproducible, high-throughput MALDI sample preparation. Despite the optimization of reaction components described above, homogeneous sample crystallization on the chip-arrayed matrix required a final dilution of the sample. This dilution was accomplished by simply adding ddH2O to the reaction well after completion of this cleavage. Without dilution, buffer ingredients and detergent inhibit the crystallization process of the MALDI sample. As a result, no fragment signals were detected in the MALDI-TOF spectra in these cases (data not shown). Sample dilution and addition of SpectroCLEAN, an ion-exchange resin loaded with ammonium ions, to the final solution proved sufficient to condition the phosphate backbone of nucleic acid fragments for chip-array based MALDI-TOF MS analysis. Similar approaches for conditioning of the phosphate backbone prior to MALDI-TOF MS analysis have been described earlier (20).
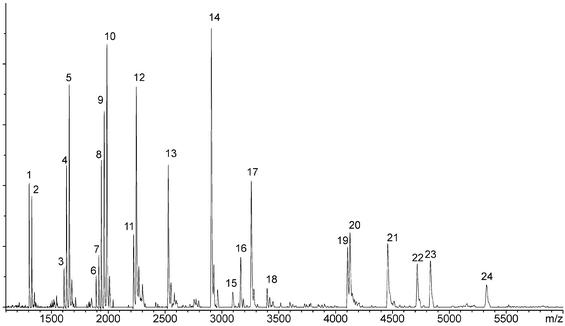
Figure 2 shows a representative fragmentation spectrum of the ApoB-100 (21) target region. All observed cleavage products are consistent with 5′-OH and 3′-phosphate groups. Fragments corresponding to the 2′-3′-cyclic phosphate group, known to be stable intermediates under limited cleavage conditions, were not detected (17). Signals 15 and 18 (Table 1) have been shown to be the very 3′-fragment of the full-length RNA transcript and, accordingly, posses a 3′-hydroxy group. We observed non-templated nucleotide addition by the RNA polymerase during transcription with several amplicon sequences. Mass signal 18 listed in Table 1 represents such a product, where either CTP or UTP have been added template-independently to the 3′-end of the transcript. In contrast to Taq DNA polymerases, we could not see a preference for adenine incorporation.
Figure 2.
MALDI-TOF fragmentation spectrum of ApoB-100 target region. RNA in-vitro transcription was performed with T7 RNA polymerase utilizing unmodified NTPs. Each mass signal is labeled with its reference number and was assigned to the corresponding RNA fragments calculated in silico (see Table 1). The masses of the individual fragments indicate a complete RNase cleavage reaction because all RNA fragments except for the 3′ terminal ones possess only a 5′-hydroxy and 3′-phosphate group.
Table 1. Comparsion of calculated RNase T1 fragments and experimental data of the ApoB-100 target region.
| No. | Sequence | Location | Calculated mass in Da | Experimental mass in Da |
|---|---|---|---|---|
| 1 | 5OH-UCAG-3p, 5OH-UACG-3p, 5OH-UCAG-3p, 5OH-CUAG-3p | @262, @145, @236, @164 | 1304.80 | 1305.03 |
| 2 | 5OH-CAAG-3p | @258 | 1327.84 | 1328.18 |
| 3 | 5OH-UUCAG-3p | @246 | 1610.96 | 1611.18 |
| 4 | 5OH-CACAG-3p | @140 | 1633.02 | 1633.27 |
| 5 | 5OH-CAAAG-3p | @202 | 1657.05 | 1657.35 |
| 6 | 5OH-CUCUUG-3p | @240 | 1893.11 | 1893.01 |
| 7 | 5OH-UCCAUG-3p | @252 | 1916.15 | 1916.11 |
| 8 | 5OH-CACACG-3p | @22 | 1938.21 | 1938.50 |
| 9 | 5OH-CAAAUG-3p | @230 | 1963.22 | 1963.32 |
| 10 | 5OH-UAAAAG-3p | @84 | 1987.24 | 1987.42 |
| 11 | 5OH-UCUUCAG-3p | @29 | 2222.32 | 2222.01 |
| 12 | 5OH-AACCUUG-3p | @74 | 2245.36 | 2245.83 |
| 5OH-AUAUCUG-3p | @66 | 2246.34 | ||
| 13 | 5OH-CCACCCUG-3p | @207 | 2525.53 | 2526.30 |
| 5OH-CUUACUUG-3p | @3 | 2528.49 | ||
| 14 | 5OH-AAUUCCAAG-3p | @11 | 2903.78 | 2905.01 |
| 5OH-AAAAUUUUG-3p | @90 | 2905.75 | ||
| 15 | 5OH-UCACCUAAAU-3OH | @278 | 3089.94 | 3091.61 |
| 16 | 5OH-CCUUCUAUAG-3p | @266 | 3162.88 | 3162.79 |
| 17 | 5OH-AACAUACAAG-3p | @192 | 3256.03 | 3256.57 |
| 18 | 5OH-UCACCUAAAU-C-3OH | @278 | 3395.13 | 3394.93 |
| 5OH-UCACCUAAAU-U-3OH | @278 | 3396.11 | ||
| 19 | 5OH-AACUCUCUCCAUG-3p | @216 | 4102.46 | 4103.08 |
| 20 | 5OH-CCACACUCCAACG-3p | @108 | 4123.53 | 4123.82 |
| 21 | 5OH-CACUUCCAAAAUUG-3p | @49 | 4455.69 | 4456.00 |
| 22 | 5OH-CAUAUAUUCCCUCUG-3p | @121 | 4714.80 | 4714.49 |
| 23 | 5OH-AAAAACCACUUACAG-3p | @149 | 4830.99 | 4832.07 |
| 24 | 5OH-CCUCUUUUUCACCAACG-3p | @172 | 5325.17 | 5325.42 |
RNA in vitro transcription was performed with T7 RNA polymerase utilizing unmodified NTP. For simplicity, less informative fragments (≤3 nt) are not shown.
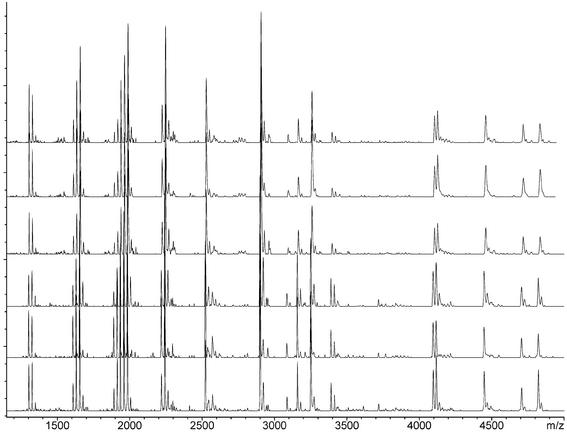
As demonstrated by the comparison between the theoretical and experimental fragment masses (Table 1), all major signals in the spectrum could be unambiguously assigned to expected fragments and all of the expected signals were identified in the spectrum. Solutions for discrimination of fragments with very similar mass (beyond the resolution of the linear axial TOF instrument used here) will be discussed below. In general, the method provides highly reproducible mass signal patterns that facilitate automated peak pattern interpretation and allow the inclusion of S/N ratio changes of mass signals as supporting evidence for sequence changes. Figure 3 shows an example of six independent reactions of the ApoB-100 assay processed on different days from several individuals of the same sequence and genotype.
Figure 3.
Reproducibility of comparative sequence analysis via base-specific RNase T1 cleavage reaction. The G-specific cleavage patterns of six independent reactions of the ApoB-100 target region are shown. All samples were processed with automated liquid handling devices on different days with several individuals of the same sequence and genotype.
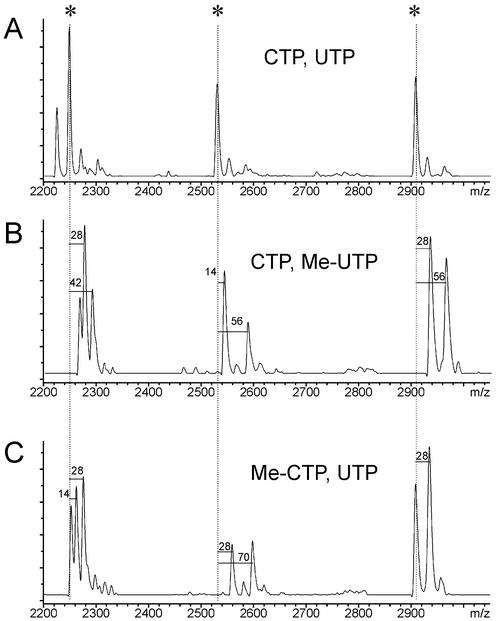
A serious limitation for this RNA-based fragmentation approach is caused by the small mass difference (1 Da) between uracil (U) and cytosine (C). In a few cases, two RNA fragments with identical length differing by only one or a few U or C could not be separated with the current resolution of the linear MALDI-TOF instrument and only the combined signal of both fragments was detected. The differentiation between these is difficult unless a substantially higher mass resolution could be achieved. Approaches have been described to overcome the instrument-related limitation in the discrimination between uracil and cytosine (22). We developed an alternative, where the pyrimidine residue of 1 nt is completely replaced by a chemically modified base during the transcription reaction. We successfully exchanged either UTP or CTP by its 5-methyl base-modified ribonucleotide triphosphate analog without loss in transcription yield. The nucleotide mass of either U or C is increased by 14 Da. Therefore, the mass difference between the two formerly overlapping fragments, differing by one U or C was increased to 15 and 13 Da, respectively. Figure 4 demonstrates the increased information content of a base-specific cleavage reaction, when base-modification is employed.
Figure 4.
During the RNA in vitro transcription either U or C was replaced by its 5-methyl ribonucleotide triphosphate analog to circumvent the small mass difference between U and C that inhibits the differentiation between individual RNA fragments. (A) A subview into the base-specific cleavage spectrum of ApoB-100 target region where the transcription was performed with normal CTP and UTP. Three relevant signals at m/z = 2245.8, 2526.3 and 2905.0 occur. A G-specific cleavage spectrum of the same ApoB target is shown in the middle spectrum (B) where UTP is replaced by 5-Me UTP during the transcription reaction. The mass difference to the relevant original fragment is labeled and indicates the number of U bases within this fragment. (C) The result on the fragment masses if CTP is replaced by its Me-CTP analog. For both base modifications, each previous unresolved signal was separated into two individual peaks.
Figure 4A shows a selected mass window of an ApoB-100 sample generated under standard transcription conditions, with three relevant signals at m/z = 2245.8, 2526.3 and 2905.0. The theoretical mass calculation predicts that each of those signals consists of two individual fragments with different sequences. However, the difference of the calculated fragment mass is <4 Da and a separation of these signals could not be achieved. Figure 4B and C show the mass signal pattern obtained from the same ApoB-100 sequence region, now processed with either Me-CTP instead of the normal CTP or with 5-Me UTP instead of UTP during the RNA transcription reaction. Now, the previously unresolved signal at m/z = 2245.8 is separated into two signals at 2260.4 and 2273.4 (for Me-CTP). Similarly, by the utilization of 5-Me UTP instead of UTP, the previously overlapping signals at 2245.8 are separated into two signals. All three signals were successfully separated into their individual fragments by base modification of the nucleotides. The observed mass shifts correspond to the number of modified nucleotides present in the analyzed cleavage product.
Another advantage of the mass modification method derives from the fact that without any previous sequence information the A, C, U-composition of any RNase T1 fragment can be calculated. Three different RNase T1 cleavage reactions are needed utilizing (i) CTP, UTP, (ii) Me-CTP, UTP and (iii) CTP, Me-UTP. For any RNA fragment, the mass difference between a given fragment of reaction (i) and (ii) and the difference between reaction (i) and (iii) are utilized to calculate the number of U’s and C’s in the fragment. Since it is known that each fragment (except the last fragment) contains only one G if cleavage is performed with RNase T1, the number of A’s can be derived.
Applications
Detection of sequence variations. To explore and verify the capabilities of the RNase T1 fragmentation method, we investigated a wider range of target regions in various individuals. Sequence regions of the following genes known to carry polymorphisms have been used as model systems for comparative sequence analysis: CETP (23), uncoupling protein 2 (UCP2) (24) and ApoB-100 (21). All of the polymorphisms were analyzed with a sample collection of 26 randomly chosen in-house control DNAs. In most cases the predicted fragmentation pattern was obtained. In a few samples one additional/missing signal occurred, which is the result of a SNP. Based on the changes of fragmentation pattern signals, the location and nature of the sequence change was determined. All results were confirmed independently using the MassEXTEND™ procedure (previously published under PROBE) (25). The potential of the described method for detection of sequence changes is exemplified in Figure 5. Here, the sequence variation of CETP polymorphism I405V was investigated.
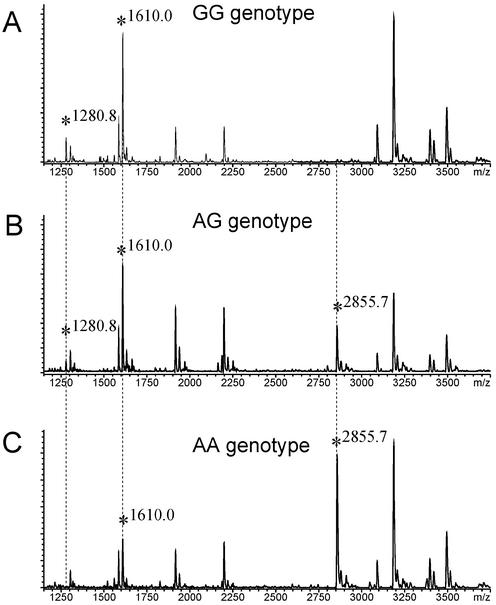
Figure 5.
G-specific cleavage reactions applied to the I405V polymorphism of the CETP gene, analyzed in three different individuals. RNA transcription with T7 RNA polymerase was performed with unmodified NTPs. Unique signals corresponding to the individual genotype are marked with a vertical dotted line.
The upper spectrum shows the characteristic pattern of fragment masses for a homozygous G-sample (Fig. 5A). A new, unique signal at m/Z = 2855.7 appears due to loss of a cleavage site if a heterozygous individual is analyzed (Fig. 5B). The intensity of the unique signal increases if the sample is homozygous A (Fig. 5C). An additional, unique identifier can be assigned in the lower mass range at m/z = 1280.8 for the homozygous G-sample. The signal decreases for a heterozygous sample and vanishes for the homozygous A-sample. Please observe, that the signal intensity at 1280.8 Da in the lower mass range, indicative for the homozygous G-sample, is generally reduced due to instrument setting-related signal suppression in the low mass range. A further observation is a S/N ratio change for the mass signal at 1610 Da. However, as can be deduced from the AA genotype, this signal is not unique for the sequence change. The corresponding cleavage product is generated twice in the CETP amplicon. Such S/N changes can only be used as supporting information for the presence of a sequence change if the overall S/N distribution of mass signals is highly reproducible between multiple spectra, a requirement that is strongly linked to sample preparation and the use of miniaturized arrays.
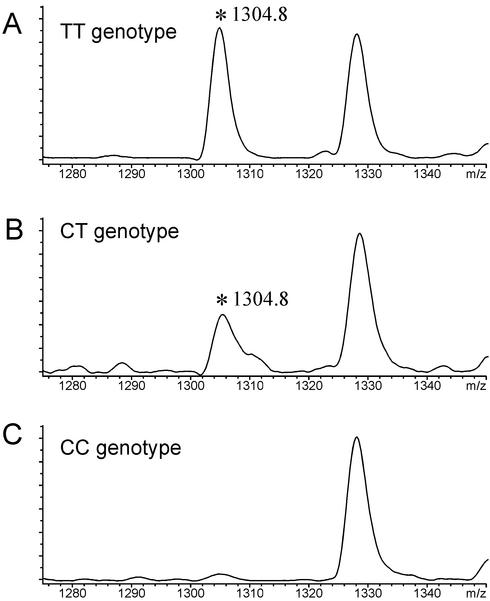
The C/T-polymorphism of UCP2-marker (A55V) is illustrated in Figure 6. Analysis of the reverse strand is performed with SP6 RNA polymerase and RNase T1 cleavage. The unique identifier for the selected SNP is the signal at m/z = 1304.8.
Figure 6.
G-specific cleavage reactions applied to a polymorphism of the UCP2 gene, analyzed in three different individuals. RNA transcription was performed with SP6 RNA polymerase using unmodified NTP. Shown is a sub-view into the full spectrum. The abundance of a unique signal at m/z = 1304.8 corresponds to the individual genotype.
In summary, the use of complete base-specific cleavage enables the detection of small mass changes as they may occur at polymorphic sites over a large dynamic mass range between 1200 and 8000 Da. A PCR assay design with two different promoter sequences tagged to the forward and reverse PCR primer, respectively, allows the usage of only one PCR-product for two complementary cleavage reactions.
Bacteria typing. The identification of pathogens involved in bacterial as well as viral infections requires fast and accurate technologies for sequence analysis. Such tools are important for a timely selection of appropriate treatments as well as monitoring and identification of new, for example antibiotic-resistant, sub-species. Furthermore, the discrimination of environmental from pathogenic strains on a molecular basis gains increasing importance with the growing number of phenotypically similar strains identified. To explore the potential of the described method for bacteria typing, eight different Bordetella strains were analyzed by amplifying their variable 16S RNA gene region with universal non-selective primers (11). The small mass difference of 1 Da between rC- and rU-nucleotides was circumvented by replacement of one rC with its 5-methyl analog, without detectable loss of transcription yield. G-specific cleavage with RNase T1 produces a characteristic pattern of fragment masses that is indicative for the individual 16S rRNA gene target sequences. All eight individual Bordetella strains were identified unambiguously and the results are concordant with those obtained by fluorescent dideoxy sequencing, the current standard method. Examples are shown in Figure 7, which represents an overlay of three RNase T1 cleavage reactions of different Bordetella strains. Arrows mark the discriminative mass signals.
Conclusions
We have introduced an improved method for comparative DNA sequence analysis via base-specific cleavage of in vitro transcribed RNA. Accurate determination of the produced G-specific fragments is achieved by MALDI-TOF mass spectrometry. This allows the rapid and simultaneous identification of all RNA fragments in a single measurement. In contrast to DNA-based fragmentation approaches, our method has the advantages of additional amplification during transcription and the increased stability of ribo-oligonucleotides in the MALDI process.
We have demonstrated that the described RNase cleavage approach produces highly accurate results in a 384-well MTP format processed with automated liquid handling devices. In contrast to previously reported DNA methods for comparative sequence analysis relying on bead purification methods (11,12) we found that the final cleavage product can be analyzed by mass spectrometry without any solid-phase purification by capture via biotin/streptavidin or oligonucleotide hybridization. The high sample-to-sample reproducibility is based on the improved performance using a RNA fragmentation assay and on the miniaturization of the MALDI sample preparation, which enables automated acquisition and analysis of mass spectra. The introduced improvements allowed us to perform a homogenous high-throughput reaction that is more flexible and robust than previous base-specific reactions utilizing mass spectrometry. The developed settings are universal enough to facilitate application of the method to a variety of different target regions. A concern for the presented approach might be the stability of the RNA transcripts and transcription-based processes in general. In our experiments, transcription and cleavage with RNase were not separated by >24 h, as microtiter plates were usually processed through post-PCR within a day. In case overnight storage was required, samples were frozen to –20°C and no interfering degradation signals were observed, although no special precautions were taken. The RNase T1 treated transcription reactions could even be stored for weeks. Repeated nano-transfer of samples on different days/weeks with subsequent MALDI-TOF MS analysis did not reveal degradation. However, it should be noted that handling of multiple RNases of different cleavage specificity might impose a challenge when the process is performed without special precaution (tip washes/changes on pipetting robots) and when these other RNases are very stable (e.g. RNase A).
In contrast to chemical base degradation methods or chemical cleavage reactions, the RNase approach proceeds under mild cleavage conditions. No acid-induced adenine or guanine loss was detected and, therefore, no purine-modified nucleotides, used for acidic cleavable fragmentation such as P3′-N5′ phosphoramidate-containing DNA, were required. The RNase T1 approach is, however, limited to the identification of polymorphisms that are detectable by G-specific cleavage of either the forward or the reverse strand. To render this approach more universal, there is still a demand for further base-specific endo-ribonucleases, which show the same specificity and reliability as RNase T1.
The utilization of 5-methyl modified pyrimidine bases increases the power of the fragmentation method to recognize C/U sequence changes, which normally would not be detectable with the current resolution of the linear TOF mass spectrometer used herein.
We have demonstrated the great potential of base-specific cleavage for various applications, including sequence validation, targeted SNP discovery and bacteria typing. Examples for applications amenable to this method include identification of mutant viral strains, analysis of methylation patterns and length determination of microsatellites. Analysis of genomic methylation patterns could be enabled by tracking changes in base-specific cleavage patterns when bisulphate treated genomic DNA is used for amplification. MALDI-TOF MS-based repeat length determination in microsatellites has always been hindered by the length of the DNA product. The use of base-specific cleavage might allow for an intelligent trimming of the final amplicon and for extraction of the core information, the repeat structure itself, if cleavage is performed at a nucleotide not present in the repeat sequence. The cleavage procedure suggested here results in shorter products, which should be more amenable to mass spectrometric analysis.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank their colleagues Drs Marc Zabeau and Patrick Stanssens from Methexis Genomics and Charles R. Cantor from Sequenom for their valuable contributions to the work presented. We are grateful to Friedrich von Wintzingerode and Ulf B. Goebel for providing us with suitable samples for bacteria typing. This work was supported by BMBF grant 0311747.
REFERENCES
- 1.Blackwell J.M. (2001) Genetics and genomics in infectious disease susceptibility. Trends Mol. Med., 7, 521–526. [DOI] [PubMed]
- 2.Chamberlain J.C. and Joubert,P.H. (2001) Opportunities and strategies for introducing pharmacogenetics into early drug development. Drug Discov. Today, 6, 569–574. [DOI] [PubMed]
- 3.Hungnes O., Jonassen,T.O., Jonassen,C.M. and Grinde,B. (2000) Molecular epidemiology of viral infections. How sequence information helps us understand the evolution and dissemination of viruses. APMIS 2000, 108, 81–97. [DOI] [PubMed]
- 4.Muddiman D.C., Anderson,G.A., Hofstadler,S.A. and Smith,R.D. (1997) Length and base composition of PCR-amplified nucleic acids using mass measurements from electrospray ionization mass spectrometry. Anal. Chem., 69, 1543–1549. [DOI] [PubMed]
- 5.Null A.P., Hannis,J.C. and Muddiman,D.C. (2001) Genotyping of simple and compound short tandem repeat loci using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem., 73, 4514–4521. [DOI] [PubMed]
- 6.Buetow K.H., Edmonson,M., MacDonald,R., Clifford,R., Yip,P., Kelley,J., Little,D.P., Strausberg,R., Koester,H., Cantor,C.R. and Braun,A. (2001) High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry mass spectrometry based strategies and studies among those approaches. Proc. Natl Acad. Sci. USA, 98, 581–584. [DOI] [PMC free article] [PubMed]
- 7.Amexis G., Oeth,P., Abel,K., Ivshina,A., Pelloquin,F., Cantor,C.R., Braun,A. and Chumakov,K. (2001) Quantitative mutant analysis of viral quasispecies by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl Acad. Sci. USA, 98, 12097–12102. [DOI] [PMC free article] [PubMed]
- 8.Nordhoff E., Luebbert,C., Thiele,G., Heiser,V. and Lehrach,H. (2000) Rapid determination of short DNA sequences by the use of MALDI-MS. Nucleic Acids Res., 28, e86. [DOI] [PMC free article] [PubMed]
- 9.Kwon Y., Tang,K., Cantor,C.R., Koester,H. and Kang,C. (2001) DNA sequencing and genotyping by transcriptional synthesis of chain-terminated RNA, sequencing and genotyping by transcriptional synthesis of chain-terminated RNA. Nucleic Acids Res., 29, e11. [DOI] [PMC free article] [PubMed]
- 10.Vaughan P. and McCarthy,T.V. (1998) A novel process for mutation detection using uracil DNA-glycosylase. Nucleic Acids Res., 26, 810–815. [DOI] [PMC free article] [PubMed]
- 11.von Wintzingerode F., Boecker,S., Schlotelburg,C., Chiu,N.H., Storm,N., Jurinke,C., Cantor,C.R., Goebel,U.B. and van den Boom,D. (2002) Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: a tool for rapid bacterial identification. Proc. Natl Acad. Sci. USA, 99, 7039–7044. [DOI] [PMC free article] [PubMed]
- 12.Elso C., Toohey,B., Reid,G.E., Poetter,K., Simpson,R.J. and Foote,S.J. (2002) Mutation detection using mass spectrometric separation of tiny oligonucleotide fragments. Genome Res., 12, 1428–1433. [DOI] [PMC free article] [PubMed]
- 13.Shchepinov M.S., Denissenko,M.F., Smylie,K.J., Woerl,R.J., Leppin,A.L., Cantor,C.R. and Rodi,C.P. (2001) Matrix-induced fragmentation of P3′-N5′ phosphoramidate-containing DNA: high-throughput MALDI-TOF analysis of genomic sequence polymorphisms. Nucleic Acids Res., 25, 3864–3872. [DOI] [PMC free article] [PubMed]
- 14.Kirpekar F., Nordhoff,E., Kristiansen,K., Roepstorff,P., Lezius,A., Hahner,S., Karas,M. and Hillenkamp,F. (1993) Matrix assisted laser desorption/ionization mass spectrometry of enzymatically synthesized RNA up to 150 kDa. Nucleic Acids Res., 22, 3866–3870. [DOI] [PMC free article] [PubMed]
- 15.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed]
- 16.Tang W., Zhu,L. and Smith,L.M. (1997) Controlling DNA fragmentation in MALDI-MS by chemical modification. Anal. Chem., 69, 302–312. [DOI] [PubMed]
- 17.Hahner S., Lüdemann,H.-C., Kirpekar,F., Nordhoff,E., Roepstorff,P., Galla,H.J. and Hillenkamp,F. (1997) Matrix-assisted laser desorption/ionization mass spectrometry of endonuclease digests of RNA. Nucleic Acids Res., 25, 1957–1964. [DOI] [PMC free article] [PubMed]
- 18.Spottke B., Gross,J., Finne,E., Reiss,J., Gross-Hardt,S. and Hillenkamp,F. (2001) Reverse sanger sequencing of α-thiophosphate-containing RNA in vitro transcripts by MALDI-MS after solid-phase purification. Proceedings of the 49th ASMS Conference on Mass Spectrometry, Chicago, IL.
- 19.Little D.P., Braun,A., O’Donnell,M.J. and Koester,H. (1997) Mass spectrometry from miniaturized arrays for full comparative DNA analysis. Nature Med., 12, 1413–1416. [DOI] [PubMed]
- 20.Harksen A., Ueland,P.M., Refsum,H. and Meyer,K. (1999) Four common mutations of the cystathionine beta-synthase gene detected by multiplex PCR and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin. Chem., 45, 1157–1161. [PubMed]
- 21.Soria L.F., Ludwig,E.H., Clarke,H.R.G., Vega,G.L., Grundy,S.M. and McCarthy,B.J. (1989) Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc. Natl Acad. Sci. USA, 86, 587–591. [DOI] [PMC free article] [PubMed]
- 22.Faulstich K., Woerner,K., Brill,H. and Engels,J.W. (1997) A sequencing method for RNA oligonucleotides based on mass spectrometry. Anal. Chem., 69, 4349–4353. [DOI] [PubMed]
- 23.Agellon L.B., Quinet,E.M., Gillette,T.G., Drayna,D.T., Brown,M.L. and Tall,A.R. (1990) Organization of the human cholesteryl ester transfer protein gene. Biochemistry, 29, 1372–1376. [DOI] [PubMed]
- 24.Walder K., Norman,R.A., Hanson,R.L., Schrauwen,P., Neverova,M., Jenkinson,C.P., Easlick,J., Warden,C.H., Pecqueur,C., Raimbault,S., Ricquier,D., Harper,M., Silver,K., Shuldiner,A.R., Solanes,G., Lowell,B.B., Chung,W.K., Leibel,R.L., Pratley,R. and Ravussin,E. (1998) Association between uncoupling protein polymorphisms (UCP2-UCP3) and energy metabolism/obesity in Pima Indians. Hum. Mol. Genet., 7, 1431–1435. [DOI] [PubMed]
- 25.Jurinke C., van den Boom,D., Cantor,C.R. and Koester,H. (2001) The use of MassARRAY technology for high throughput genotyping. Adv. Biochem. Eng. Biotechnol., 77, 57–74. [DOI] [PubMed]