Abstract
Electrophoresis continues to be a mainstay in molecular genetic laboratories for checking, sizing and separating both PCR products, nucleic acids derived from in vivo or in vitro sources and nucleic acid–protein complexes. Many genomic and genetic applications demand high throughput, such as the checking of amplification products from many loci, from many clones, from many cell lines or from many individuals at once. These applications include microarray resource development and expression analysis, genome mapping, library and DNA bank screening, mutagenesis experiments and single nucleotide polymorphism (SNP) genotyping. PCR hardware compatible with industry standard 96 and 384 well microplates is commonplace. We have previously described a simple system for submerged horizontal 96 and 192 well polyacrylamide or agarose microplate array diagonal gel electrophoresis (MADGE) which is microplate compatible and suitable for PCR checking, SNP typing (restriction fragment length polymorphism or amplification refractory mutation system), microsatellite sizing and identification of unknown mutations. By substantial redesign of format and operations, we have derived an efficient ‘dry’ gel system that enables direct 96 pin manual transfer from PCR or other reactions in microplates, into 768 or 384 well gels. Combined with direct electrode contact in clamshell electrophoresis boxes which plug directly to contacts in a powered stacking frame and using 5–10 min electrophoresis times, it would be possible (given a sufficient supply of PCRs for examination) for 1 million gel tracks to be run per day for a minimal hardware investment and at minimal reagent costs. Applications of this system for PCR checking and SNP genotyping are illustrated.
INTRODUCTION
Molecular genetic studies have entered an era when whole genomes, large clone and cell line collections and large population samples are under analysis. Individual PCRs of specific loci in specific samples remain the commonest first step for such analyses. This has led to demands in many laboratories for increased throughput and for economy of analysis both in time and reagents.
The well known industry standard 96 well microplate has evolved to form the basis of many high throughput approaches in the biosciences over the past 30 years (1). However, microplates are only permissive of liquid phase reactions or solid phase (binding) separations, whereas electrophoresis can derive information about parameters such as size, shape and charge of molecular moieties as well as acting as a highly resolving separation approach for complex mixtures. Slab gel electrophoresis gives convenient alignment of tracks. However, throughput using polyacrylamide slab gels has been particularly limited because acrylamide will not polymerise in the presence of air. Therefore, the standard slab format involves pouring the mix between two glass plates separated by spacers with insertion of a well-forming comb at one end of the gel, the rest of the gel being inaccessible for sample loading.
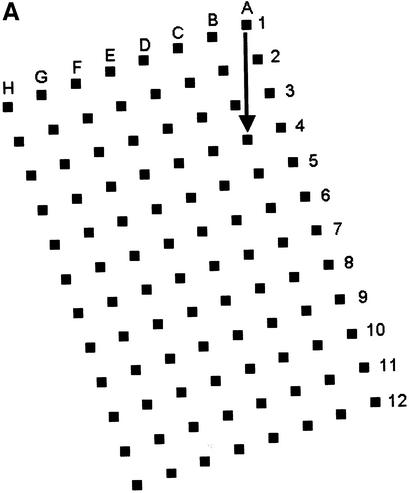
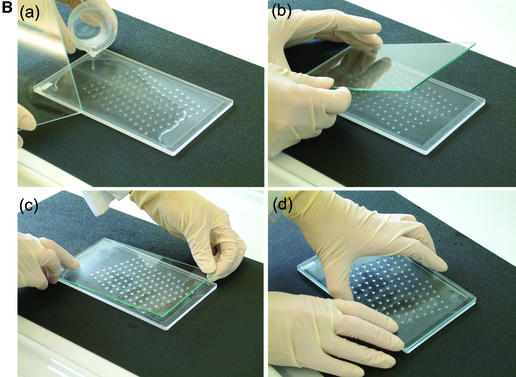
We described a generalised system, microplate array diagonal gel electrophoresis (MADGE), in 1994 which opens a number of advantages for both polyacrylamide and agarose gel electrophoresis in the molecular genetics laboratory (2,3). MADGE uses 96 track origins (wells) with 8 × 12 array locations at the 9 mm pitch identical with microplates. The long axis of the array is at an acute angle of 18.4°, relative to the direction of electrophoresis (Fig. 1A) so that with cubic wells of side 2 mm a track length of 26.5 mm is available, the sample passing diagonally between wells belonging to two other rows. The two piece kit consisting of two dimensional gel former and piece of backing glass or plastic (Fig. 1B) for the gel, both excludes air and gives a perfectly flat open face to the resultant 96 well gel, simplifying the production of agarose, PAGE, and other gel matrix or composite gels. For PAGE gels, standard glass (e.g. 2 mm thick float glass) is wiped on one surface with gamma-methacryloxypropyltrimethooxysilane (Sticky Silane). Acrylamide mix is poured into the gel former and the glass plate laid directly onto the teeth and sides of the former. When the gel has set, the glass plate is prised off: the gel is adherent to the glass on account of the chemical crosslinks to the Sticky Silane formed during polymerisation. This novel approach of PAGE gel forming can also be used for the simplified production of open faced horizontal PAGE gels (2) but with standard row(s) of wells.
Figure 1.
96 well MADGE. (A) Schematic plan view of 96 well MADGE. Direction of electrophoresis is arrowed relative to well A1. (B) Steps a–d for pouring a single 96 well PAGE MADGE gel. Gels can also be poured in batches in a purpose built box, or gel mix can be poured through the small gap at one end (see Materials and Methods) into a preassembled sandwich of gel former and glass plate.
Whilst we noted in our original paper (2) that a variety of different configurations might be used for the 96 well gels and types of tank, much usage in other laboratories has continued to be as submerged gels. In part this has reflected the existent availability of horizontal tanks, mainly for conventional agarose electrophoresis. For submerged gel usage, sample loading is typically by 8 or 12 channel air displacement pipetting. Equivalent 96 channel pipettes are heavy and difficult to use manually and require an associated compressed air cylinder. 96 pin passive replicators cannot be used for submerged gels because they cannot retain their samples on passage through the electrophoresis buffer surface. We have previously published a description of 192 well gels (in which a second well is located half-way along each track of a 96 well gel, to form a pair, for example A1 and A1′, G7 and G7′, generalising Xn and Xn′) combined with MADGE-specific software, for semi-automatic single nucleotide polymorphism (SNP) genotyping (4). In this arrangement, a pair of allele-specific PCRs for a particular sample are examined in the concatenated ‘half-tracks’ Xn and Xn′ which can be recovered as a single ‘virtual’ track in software for purposes of convenient data calling. Alternately, all 192 wells could contain 192 independent rather than paired reactions. We have also described a thermostatted, higher resolution submersion format where 2 mm long × 2 mm deep × 1.5 mm wide wells and thus 1.5 mm wide tracks are used, with a 2 × 6 instead of 2 × 4 diagonal angle, in order to achieve longer tracks and higher resolution. [By ‘2 × 4 diagonal angle’, we mean the track length being a diagonal of a virtual rectangle formed by drawing imaginary lines between two adjacent wells (as width) and four adjacent wells (as length).] This approach was developed for resolving bands of >1.5% size and hence >1.5% mobility differences such as PCR products representing alleles of minisatellite, tetranucleotide and trinucleotide microsatellite loci (5,6). A system for identification of unknown mutations (meltMADGE) using thermal ramp electrophoresis (7) and partially denaturing conditions has been described in principle (8) but not in detail. In the latter formats, 96 well gels are covered by a second glass plate after loading, the assembly secured with clips or bands, and gels submerged in a rack holding up to 12 gels.
However, we have recognised that for simple PCR checking and SNP genotyping involving resolution of a small number of readily resolved bands, much higher well densities might be used, following the trend toward increased density PCR microplates such as 384 well format. Here, we describe a complete reconfiguration of MADGE format and associated procedures, which enables simple, manual use of gel well densities at least up to 768 well gels. Critically and also conveniently, the revised configuration has to use ‘dry’ electrophoresis. (By ‘dry,’ we mean ‘bufferless tank’.) There are several consequent advantages, which, cumulatively, have substantial implications for throughput increments and cost reduction for gel-based approaches to PCR checking and SNP genotyping.
MATERIALS AND METHODS
Format of wells for 384 well MADGE and 768 well MADGE
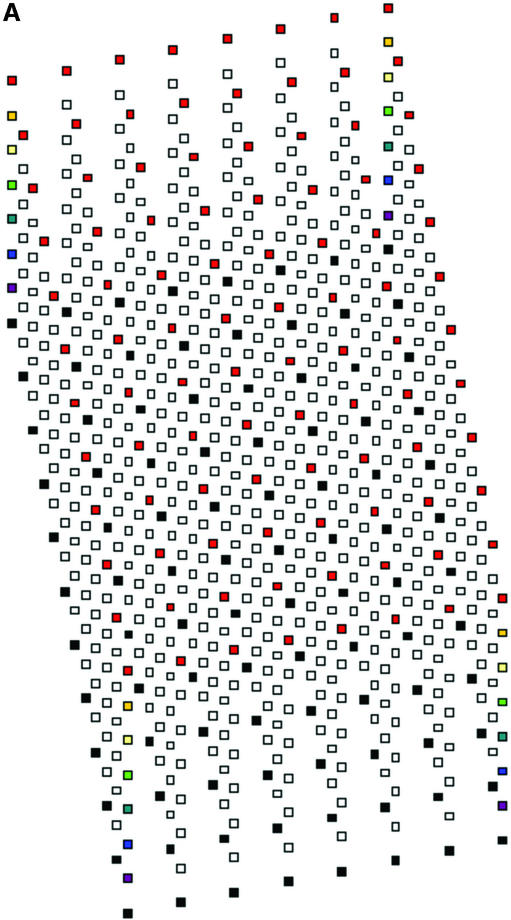
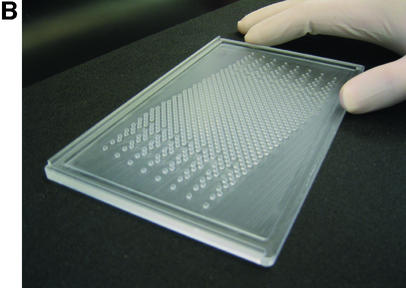
Various issues related both to manufacture of formers and to relationships between wells and tracks were considered or tried. Some of these are mentioned in the Discussion. A design was adopted which uses an 11.3° angle between the 12 well rows of each intercalated 96 well component array and the direction of electrophoresis (Fig. 2). Each well is cubic of side 1.5 mm, the eventual formed gel being 1.5 mm thick. Thus, as for 96 well polyacrylamide MADGE, the base of the well is the glass plate to which the gel is anchored. For 768 well gels, the eight sets of 96 well arrays are intercalated in a linear arrangement relative to the line of electrophoresis (Fig. 2). Nomenclature of wells follows the pattern A11, A12, A13, A14, A15, A16, A17, A18 representing location A1 of each of the eight arrays; more generally, location Xnm, X ranging from A to H representing the eight rows of a 96 well microplate, n ranging from 1 to 12 representing the 12 columns of a 96 well microplate; and m ranging from 1 to 8 representing the eight 96 well microplate array compatible sets of wells intercalated into one gel. A sample electrophoresed from well A11 would eventually meet well A12 if run for too long, that from A12 would meet A13, etc. A sample migrating from well A18 would meet well B61, because an 11.3° diagonal has been used corresponding with a 2 × 6 diagonal of the wells (2), which was also the angle chosen for longer track length 96 well MADGE for microsatellite sizing (6). Track width in this 768 well format is 1.5 mm corresponding with width of the wells, track length is 4.24 mm and gel thickness is 1.5 mm. Relative to 768 well gels, for 384 well gels the wells A12, A14, A16, and A18 would not be present (and more generally, arrays m = 2, 4, 6 and 8 are not present). Thus, in 384 well gels, the tracks are longer, track dimensions being 1.5 mm wide, 1.5 mm deep and 9.98 mm long. As an additional option, extra wells outside each 96 well array can be included in the design of the former. Our preference has been an arrangement in which every 96 well array has one control for each column, those for columns 1–6 being located adjacent to row A and in keeping with the 9 mm pitch of the array, those for columns 7–12 being located adjacent to row H again corresponding with the column pitch (Fig. 2). However, these extra wells can cause some confusion in identification of the critical locations A1m and H12m and may preferably be omitted. The design machined in perspex (plexiglas) on a programmable computerised CNC cutter is shown in Figure 2. The finished perspex has outer dimensions 5 mm × 110 mm × 180 mm, with a 1.5 mm deep ‘swimming pool’ containing 768 or 384 pegs or ‘teeth’ which will form the gel wells; and sides of the ‘swimming pool’ 5 mm wide which are in the same plane as the flat 1.5 mm square tops of the teeth. The float glass used to make a PAGE gel is 2 mm × 110 mm × 170 mm. This fits the edges of the gel former perfectly for simple location (alternately a locating L square at one corner can be used), but leaves a small gap at one end so that the glass-gel can be prised off the former by inserting a blunt edge such as a spatula under the edge of the glass where it does not quite achieve complete closure over the end of the ‘swimming pool.’ Gels are prepared as described previously (2), using Sticky Silanised glass to support polyacrylamide gels or hydrophilic plastic to support agarose gels. 768 well and 384 well formers and related apparatus are now available from MadgeBio Ltd, Huntingdon, UK (www.madgebio.com).
Figure 2.
768 well MADGE. (A) Schematic plan view of a 768 well MADGE gel. Corner wells (clockwise from top left: H1, A1, A12 and H12) are shown in a different colour for each array. Direction of electrophoresis is (on the page) vertically down. ‘dry’ gels rest horizontally in the box/frame system (Fig. 4B). The first and eighth 96 well arrays (Xn1 and Xn8 according to our designation; see Materials and Methods) have been coloured red and black, respectively, at all array positions. Each well is cubic (1.5 mm sides), and the pitch between wells of each set of 96 is the microplate standard (9 mm). (B) Photograph of a 768 well MADGE gel former. 768 1.5 mm cubic teeth are visible within the ‘swimming pool’ into which the gel mix is poured prior to covering the gel former with a glass plate coated with Sticky Silane.
Gel loading
Both the simplest and also the most feasible means of manual transfer of PCR product from 384 well (or four or eight 96 well) PCR plates to 768 or 384 well MADGE gels, is to use a 96 pin passive replicator. Such replicators (e.g. MULTI-BLOT replicator, V&P Scientific, USA, VP408S2; or from MadgeBio, UK, www.madgebio.com) are constituted of pins with split ends, which will draw up sample by capillarity, but will release the sample upon touching a solid or gel surface. Split pins cannot pick up aqueous phase from under mineral oil, so oil-free PCR must be undertaken. This methodology is prevalent now and is achieved using rubber mats or film and lid compression to seal PCR plates: condensation during PCR is prevented by use of a heated lid covering the PCR block.
Gels can either be made with ethidium bromide incorporated, or can be prestained immediately prior to use, or poststained. The first option, or handling of gels in batches, will be advantageous to the high throughput laboratory. The gel is wiped with a glass rod so that a small residual volume of buffer remains in the well. The PCR plate is spun briefly before removing the sealing film. Five microlitres of sample loading buffer is added to each well using a repeating pipette. The gel is placed on a coloured grid (available from http://www.sgel.humgen.soton.ac.uk) to aid sample loading. The gel is aligned so that the first column (column 1) of wells lies over the red squares of the orientation grid.
Samples are transferred from the PCR plate to the MADGE gel using a 96 slot pin replicator. The slot pin replicator is located and pressed firmly into an appropriate 96 well array, either a 96 well plate of PCR or post-PCRs, or into a 96 well array integral to a 384 well plate. Samples of ∼2 µl are picked up: the volume is determined by the size of the slot in the pin. Two microlitres is ideal for the 1.5 mm cubic wells of 768 or 384 well MADGE gels. The replicator is then lined up over the gel-template and touched down, which discharges the samples into the wells (Fig. 3). Not quite all of the 2 µl is discharged. A second pick up and discharge can be undertaken to fill the wells almost completely, if a greater loading of sample is desired or if a second reaction or sample with different sized fragment(s) is to be duplexed into the same track. The replicator is blotted clean, rinsed in deionised water and blotted dry. The next 96 well array is then transferred, positioning the replicator over the next orientation colour (e.g. blue squares in our 384 well MADGE template) prior to sample discharge to gel wells by touch-off. This is repeated totalling eight transfers for 768 well gels or four transfers for 384 well gels.
Figure 3.
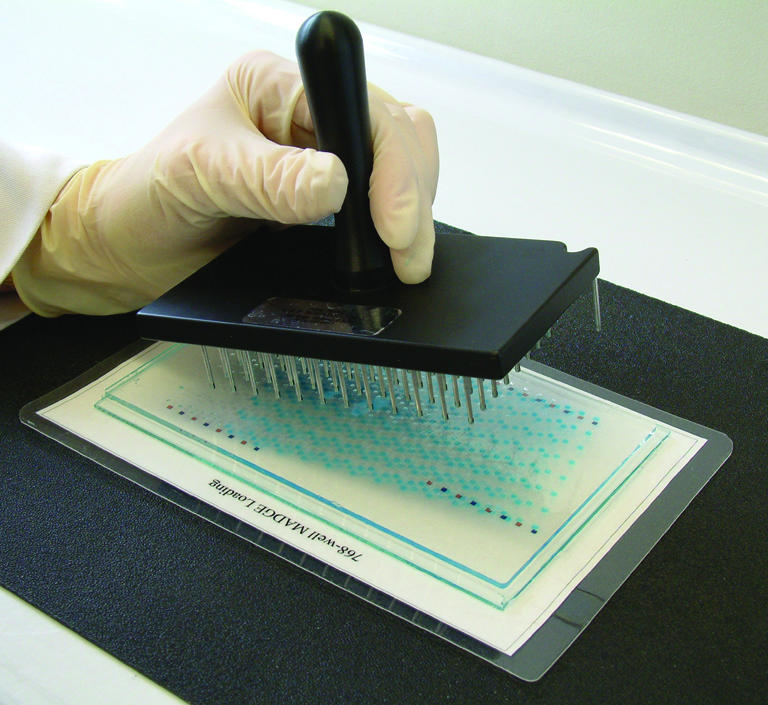
96 pin passive replicator loading of high well density (384 or 768 well) MADGE gels.
Electrophoresis system: dry tanks and stacking frames
Each 768 and 384 well polyacrylamide gel supported by and attached to a glass plate is then placed in a bufferless electrophoresis box and voltage applied directly across the gel. This system is schematised in Figure 4. The gel is located in the base of the box and a moving guillotine bar, bearing a cathodal platinum wire on its undersurface, is located onto the cathodal end of the gel, i.e. at the end of the gel near well A1. The lid of the box bears a bar which represents the anode, so that closure of the clamshell box brings the anode down onto the opposite end of the gel. Electrophoresis at 150 V for 5–10 min allows the resolution of most bands in PCR genotyping reactions. A frame system with electrical contacts for each box enables the convenient stacking of boxes and simultaneous connection in parallel to one power supply.
Figure 4.
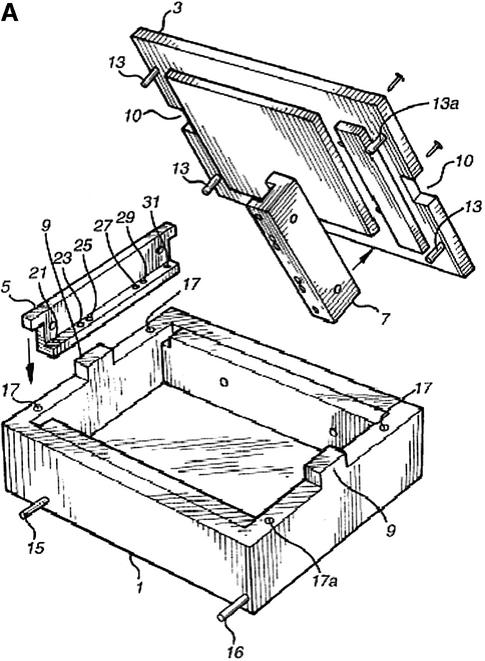
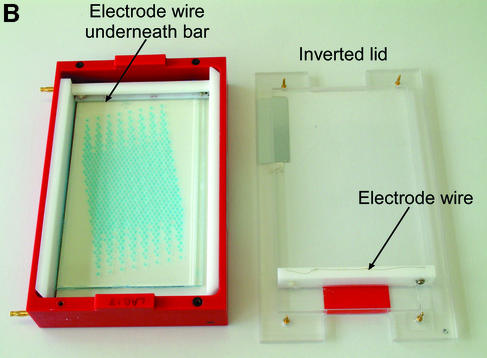
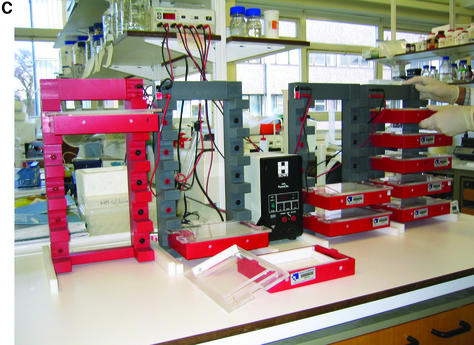
(A) Schematic of ‘dry’ electrophoresis box. Platinum electrodes in direct contact with the gel pass an electrical current through the matrix resulting in migration of nucleic acids towards the anode. Not to scale. Components are: 1, base; 3, lid; 13 and 17, locating pin and socket; 13a and 17a, conducting pin and socket leading to electrode in base; 9 and 10, guides locating top to base; 15 and 16, conducting pins for connecting box directly to power supply sockets in stacking frame (C); 7, support bar to which electrode in lid is threaded via holes 21, 23, 25, 27, 29, 31; 5, guillotine to the underside of which an electrode is threaded—this guillotine is lifted to admit gel and then rests directly on gel surface. (B) ‘Dry’ gel box electrophoresis 768 well MADGE gel. (C) ‘Dry’ box gel stack frames for multiple ‘dry’ box gel electrophoresis.
Visualisation and analysis of MADGE gels
Given the complexity of the image, 768 and 384 well MADGE gel images benefit from use of software for analysis. However, overlay of a guide template onto a photograph or printout does make it feasible to read gels directly, for example in the laboratory immediately after pilot experiments have been undertaken. The image can be analysed in the MADGE module of Phoretix (Newcastle, UK) 1D Advanced image analysis software. Present versions of software are designed to handle standard 96 well MADGE gels but the higher density formats can be examined by identifying and analysing each of the four intercalated 96 well arrays separately. The general features of software for MADGE analysis have been described previously but in brief involve user specified or templated identification of 96 well/track arrays; rearraying to a directly aligned format; background subtraction; band identification and characterisation; rule-based or user based classification of band arrays in tracks; and export of primary data to spreadsheets for further analysis.
Further protocol details
Further protocol details and URL links related to specific gel mixes, other reagent and equipment components and to specific applications, can be accessed via Southampton Genetic Epidemiology Laboratories web site (www.sgel.humgen.soton.ac.uk).
RESULTS
Use of the 768 well and 384 well gel format
It is straightforward to prepare, load and electrophorese 768 well MADGE gels by the methods described. Within our laboratory, at least 15 staff and students of widely differing skills and ages have used either the 384 well or 768 well system. Use of 96 pin passive replicators in conjunction with simple colour template underlays for gels, simplifies the process of recognition of specific subarrays and locations within the apparently dense, confusing set of diagonally arrayed, intercalated set of 768 wells. The main characteristics of the system should be appreciable from Figures 1–4.
Gel images
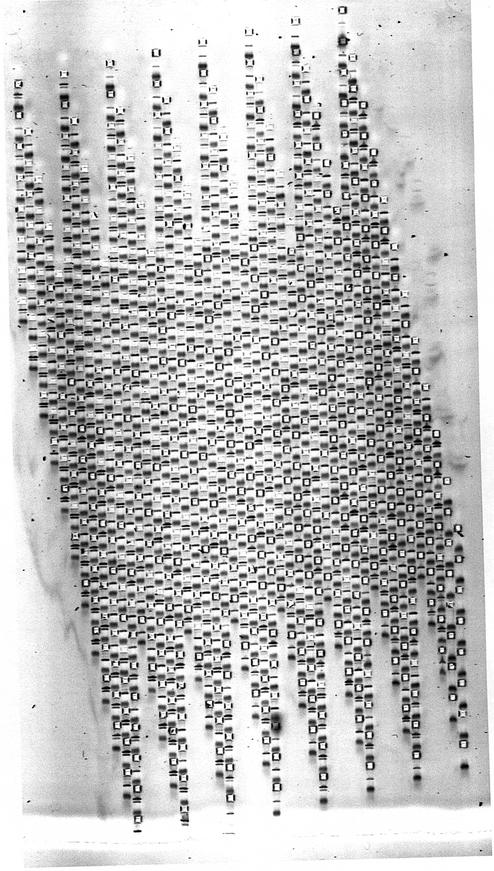
In Figure 5, we show the first proof-of-principle experiment using a 768 well gel. In Figure 6 we show a SNP analysis by allele-specific PCR examined on a 384 well gel. In Figure 7, we show a SNP analysis by allele-specific PCR examined on a 768 well gel. In Figure 8, we show the examination of a duplex PCR, with different sized amplicons representing X and Y chromosomes in arrays from a DNA bank containing subjects of both genders, randomly mixed. Further details are given in the figure legends. The main points to note for each of these analyses are that sample loading is adequate and consistent between array positions; that resolution on 768 well PAGE MADGE is sharp and readable; that good gel quality is achieved without problems from air bubbles within the dense array of wells; and that there are no significant migration distortions induced in tracks on account of the adjacency of wells containing other samples which have been loaded directly from PCR in PCR buffers. A few tracks in the images are blank, representing either template negative controls from the PCR step, or occasional PCR failure. Sample transfer using pin replicators is 100% efficient in that every microplate sample is represented on the gel: this is readily judged and monitored by observing that following a transfer step, marker dye is evident in every gel well.
Figure 5.
768 well MADGE. In each track of this gel, presence of one band denotes amplification of control amplicon only, whereas two bands denote positive amplification also for a specific allele in an ARMS PCR test. Track width, real size, is 1.5 mm. Gel set-up and loaded by 96-pin passive replicator. This image represents the first proof of principle experiment with a 768 tooth gel former and gel—minor artefacts related to sample entry into the gel are evident, which can be avoided as described in the Discussion.
Figure 6.
384 well gel used for dual-reaction ARMS assay. On the main gel image the red rectangle indicates reactions a and b for sample A1, array 1, while the blue rectangle indicates reactions a and b for sample A1, array 2. The enlargement shows the three possible genotype calls as pairs of lanes with either two bands in each for heterozygote or two bands in one and one in the other for each category of homozygote.
Figure 7.
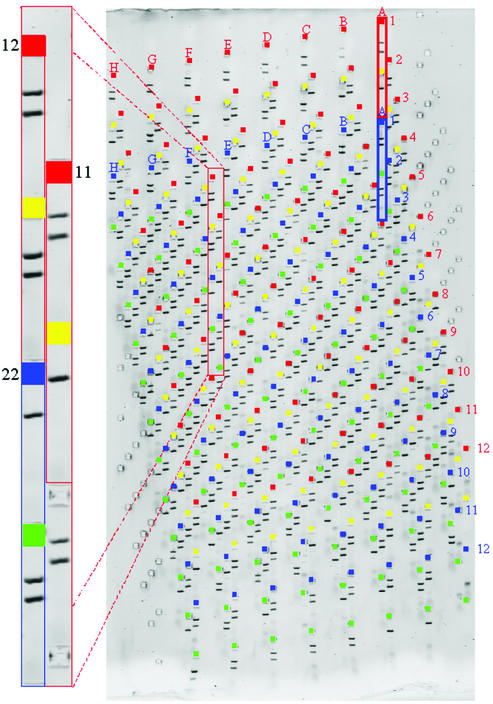
768 well SNP assay. The wells for each component 96 well array are highlighted in a different colour. Reactions examined in each track represent a three-oligonucleotide ARMS assay as described in the text. Two bands in a lane indicate allele present, absence of the smaller band indicates allele absent. The larger band represents a ‘constant’ amplicon surrounding the SNP locus. The smaller band is only present if the ‘constant’ amplicon contains the appropriate base from which an allele-specific primer for that base can prime. Preferred sizes of amplicons are in the range 70–300 bp for examination on 5 or 7.5% PAGE MADGE gels.
Figure 8.
A test for presence/absence of X and Y chromosomes in a mixed gender DNA bank. Duplex PCR yields two bands, one X-specific and one Y-specific in males, but only yields the X-specific band in females.
DISCUSSION
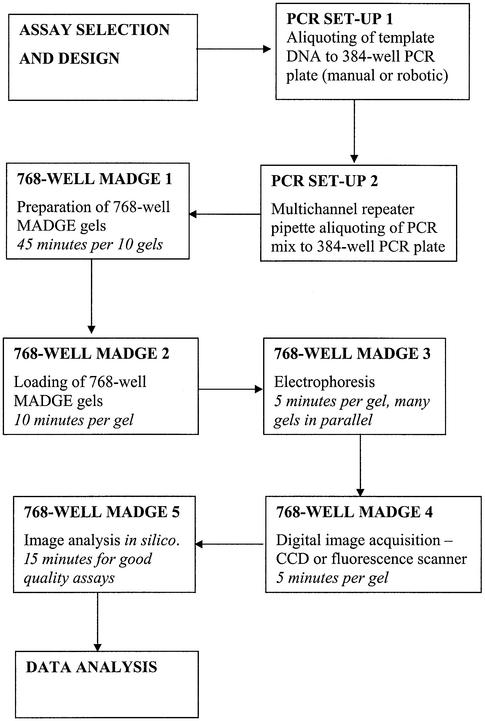
The 768 and 384 well protocol described here depends on use of oil-free PCR, manual transfers from PCR plates to gel using passive 96 split pin devices, and use of ‘dry’ rather than submerged gels to enable use of passive pin transfers. The total reconfiguration of operations to passive transfers and ‘dry’ gels was both necessary, as discussed below, and also advantageous. The use of ‘dry’ gels and the box/stacking frame system leads to a clean, safe and compact laboratory arrangement for high throughput electrophoresis. The electrical current taken per gel depends on the total cross-sectional area of gel and submersion buffer. Since there is no submersion buffer, current taken is low, typically ∼20 mA per gel. Thus, one small electrophoresis power pack can readily supply power to all the boxes in one frame. 768 well gel run times are typically 5 min. Using four frames, with five boxes per frame, and one 96 pin passive replicator, gels could be run at a rate of 12 runs per hour × 4 frames × 5 boxes × 768 well gels per hour. In an 8 h day, 1 474 560 tracks could be run. It takes several minutes to load each gel because eight 96 pin transfers are necessary to load a 768 well gel from two 384 well PCR plates. Thus, one technician can handle about 12 gels per hour, or 96 gels per day (73 728 tracks). However, PCRs on standard 384 well blocks such as the MJ Tetrad instruments in use in our laboratory, typically take 1–2 h, thus demand for gel tracks comes at a rate corresponding with the need to set up, run and image two 768 or four 384 well gels per hour per Tetrad PCR machine. Figure 9 is a flowchart of the various steps and estimated times. It should be evident that for a modest number of 768 or 384 well gel formers, boxes, frames and passive replicators, the total PCR capacity and demands from a number of users in even a large laboratory can be met with 768 well MADGE electrophoresis equipment with a footprint no greater than 1 m2. The cost of the necessary equipment is far less than that of the associated PCR hardware, so the methodology should be accessible within the budget of any standard molecular genetics laboratory.
Figure 9.
Flowchart of steps and guideline of time from assay design, through PCR and high throughput electrophoresis to data calling and analysis. An estimation of time for each step is given. It should be noted that all steps can be executed without reliance on large, fixed base automation and can therefore be deployed in essentially any laboratory.
384 well microplates have become an established new standard in higher throughput genomics laboratories for clone operations (arrayed libraries, gridding, spotting, storage, etc.) and for PCR. 384 well microplates have wells in a 16 × 24, 4.5 mm pitch array. A diagonal turn of this array would result in MADGE wells too narrow (<1 mm) for manual access, but we have devised 768 and 384 well MADGE formats that combine eight or four 96 well arrays in a linear (rather than tetradic) intercalation. This format enables use of 1.5 mm cubic wells which can be readily accessed by human hand and eye. All possible formats were considered. A diagonal of 2 × 4 relative to the wells (Fig. 1) represented the original format but tracks were considered unnecessarily wide and are insufficiently long to divide into eight subtracks each with its own well of origin. Other diagonals to consider were 2 × 5, 2 × 6, 2 × 7, 2 × 8 and 2 × 9. 2 × 9 would enable tracks of infinite length if the diagonal were relative to the columns of 8 rather than the rows of 12 relative to an 8 × 12 microplate. However, maximal track width is 1.48 mm for 2 × 7 position, 1.27 mm for 2 × 8 and 1.12 mm for 2 × 9 diagonal (2). Allowing for tolerance between tracks and adjacent wells and considering the pin sizes available and suitable for passive transfers manually, we preferred a minimum well size of 1.5 mm in each dimension. 2 × 6 was preferable over 2 × 5 diagonal on account of the total track length available. Furthermore, for a 384 well linear arrangement of four 96 well arrays, a 2 × 5 diagonal was unsuitable because the fourth intercalated well would be juxtaposed (fused to) the fourth well in each 2 × 5 set. In contrast, the 2 × 6 diagonal gave favourable separation of all wells/teeth both for 768 and 384 well formats (Fig. 2). The array is rather dense and confusing for loading by 8 or 12 channel pipettes, which would be inefficient anyway, but 96 pin passive or air displacement transfer is not a problem for human hand and eye. However, air displacement 96 channel pipettes such as the SPLATT from Intelligent Automation Systems, USA, are heavy, costly and require an attached air cylinder and thus are not very suitable for the general laboratory. Higher specification equipment such as the Robbins Hydra which is a fixed base instrument, represents an even greater departure from the versatile and distributable equipment repertoire found in the general molecular genetics laboratory. Cleaning of fixed pin air displacement systems is a further issue, whereas disposable tip systems become costly in consumables on account of the total tip consumption in the high throughput setting. In contrast, passive transfer split pin devices are simple, light, easy to clean and economical.
The present approach to machining of the gel formers is near its limit because the space between ‘teeth’ is small, but other arrangements of teeth, or other approaches to gel former production, may enable yet higher densities. We considered various different diagonal angles for the intercalations of 96 well arrays. A restricted number of possibilities exist, because 96 well array format must be preserved for each intercalated array, and neither the wells nor the tracks of any array should interfere with the wells or tracks of any other array. Furthermore, wells of 1 mm dimensions or less would be unsuitable for manual operations. Also, wells with <1 mm between their edges or corners would tend to cause difficulties in machining of formers (and possibly also in plastic moulding of formers if that were ever attempted). Such narrow gaps would also risk tearing of the narrow strips of gel matrix between closely spaced teeth, during separation of gel from gel former. Higher precision cutting of gel formers and passive replicators made to higher specifications, etc., might permit future step up to 1536 well formats. However, this might force the transition from economy of start up and versatility of the human operator, to more expensive hardware configurations. In these circumstances, complete departure from industry standard microplate format may be appropriate, both for PCR and post-PCR procedures including electrophoresis.
The small (1.5 mm cubic) wells of 768 and 384 well gels have one further advantage. Artefacts such as dumbbell gel bands can be caused by failure of amplicon in the presence of high salt in the centre of a well to enter the gel. We have previously noted that waiting for 10 min between loading sample into the well and starting the electrophoresis, prevents artefacts attributable to ionic differences between sample and gel (9). Smaller wells permit this ‘dialysis’ to occur faster than with larger wells such as the 2 mm cubic wells adopted for the earlier 96 well MADGE formats.
PCR and post-PCR reactions underpin very many analyses in molecular genetics. The simplest test is presence or absence of PCR product. For example, radiation hybrid mapping depends on testing by PCR for the presence of specific loci in a panel of radiation hybrid cell lines. Each cell line contains a set of chromosomal subfragments, fragmented by radiation, representing a partial complement of the genome under study. Loci close together will tend to give PCR products in the same cell lines. This is an efficient means to determine the physical relationships (linkage, order and distance apart) of a set of hundreds or thousands of loci belonging to the genome being studied. For the rat genome, a complete radiation hybrid genome map was constructed using PCR in conjunction with an adaptation of 96 well MADGE (10). However, PCR checking has numerous other uses, for example in checking sets of PCR products for use in constructing microarrays; and in medium throughput expression analysis by RT–PCR (11); in checking amplicons prior to more time consuming procedures such as sequencing or high resolution sizing; and in screening cell lines or clones for a specific mutation or marker. Our laboratory is focussed on molecular genetic epidemiology. PCRs in this context commonly derive from DNA banks representing thousands of subjects analysed in parallel. The amplicon may be for SNP analysis, for gene dosage analysis, for microsatellite typing or for scanning for possible unknown mutations. The ‘amplicon’ may also be preparatory, for example long PCR (12) or whole genome amplification (13) which will serve as a template for a range of secondary reactions. Some SNP assays are now efficiently and economically served by highly multiplexed assays (both at PCR and post-PCR steps) of which perhaps mass spectrometry (http://www.sequenom.com/stage1/) and minisequencing (http://www.orchid.com/) so far offer the most promise. However, in the study of specific genomic regions and new markers there remain many contexts in which set-up and checking of singleton assays is needed. This occurs both in specialised purpose-built high throughput facilities and in traditional laboratories. In our laboratories, singleton SNP assays by both liquid phase [e.g. improved TaqMan methodology (14)] and gel-based methods are in use. Singleton SNP assays can rapidly and readily be achieved by traditional tools such as allele-specific PCR or restriction fragment length polymorphism (RFLP) analysis, used in conjunction with electrophoresis. Allele-specific PCR, also called amplification refractory mutation system (ARMS) PCR, is the most general. We use two gel-based ARMS formats (4,15). Optimisation must be performed for new SNP assays but, once established, assay procedure does not incur the additional step inherent in restriction site analysis. Some restriction enzymes are economical and assays using natural or PCR-induced RFLPs may be easier to set up, but some enzymes will multiply the assay cost many-fold relative to PCR and gel costs. The reagent and plastics cost of a 384 well plate of 10 µl PCRs is ∼£10 whereas the cost of a 768 well or 384 well MADGE gel is £0.50. Use of 768 well gels substantially reduces the number of gels to be prepared and it reduces electrophoresis tank needs 8-fold relative to 96 well gels. The use of passive replicators rather than 8 channel air-displacement pipetting almost eliminates disposable tip consumption and reduces the number of transfer steps and potential errors 12-fold. The ‘dry’ stacking system without tank buffers also offers substantial safety, cleanliness and space saving benefits in the laboratory.
Gel-based methods enable checking of amplicons in ways not possible in liquid phase. Amplicon size and purity can be evaluated, useful also in troubleshooting some liquid phase methodologies. The assay shown in Figure 8 for presence of an X and a Y chromosome marker used a duplex PCR with two different amplicon sizes. It was used as a DNA bank quality control to test for sample swaps, verifying gender in relation to phenotypic records. Sizing information substantially confirms the veracity of an amplicon, and is thus useful in any tests for specific loci such as the radiation hybrid mapping application described above.
There is intense activity to systematise the genomic, genetic, transcriptomic and proteomic descriptions of organisms. A supplement to Nature Genetics, ‘The Chipping Forecast’, considered in detail the shift from hypothesis-led to descriptive hypothesis-forming research in the biosciences (16). The 768 well, ‘dry’ gel, stacking-frame electrophoresis system that we have described here incurs no substantial capital costs. Reagent costs are minimal and, in general, gel-based assays represent well understood, widely available, adaptable methods. The system is thus readily accessible to almost any standard laboratory. With a capacity up to 1 million tracks per day for minimal investment, the achievable throughputs for low resolution electrophoresis by ‘dry’ gel 768 well PAGE MADGE match the current shift of philosophy in biosciences research toward higher throughput, systematic description.
Acknowledgments
ACKNOWLEDGEMENTS
Sylvia Diaper, Shuwen Huang and Manch Patel are thanked for assistance. These developments were made in the course of research funded by the Medical Research Council, UK, British Heart Foundation and Wessex Medical Trust (Hope). The Biotechnology and Biological Sciences Research Council, UK is thanked for support. I.N.M.D. was a Lister Institute Research Professor.
REFERENCES
- 1.Ye S. and Day,I.N. (2003) Microarrays and Microplates: Applications in the Biosciences. BIOS, Oxford, UK.
- 2.Day I.N. and Humphries,S.E. (1994) Electrophoresis for genotyping: microtiter array diagonal gel electrophoresis on horizontal polyacrylamide gels, hydrolink, or agarose. Anal. Biochem., 222, 389–395. [DOI] [PubMed]
- 3.Day I.N. and Humphries,S.E. (1994) Electrophoresis for genotyping: devices for high throughput using horizontal acrylamide geles (H-PAGE) and microtitre array diagonal gel electrophoresis (MADGE). Nature Product Rev. [June 36–37].
- 4.O’Dell S.D., Gaunt,T.R. and Day,I.N. (2000) SNP genotyping by combination of 192-well MADGE, ARMS and computerized gel image analysis. Biotechniques, 29, 500–506. [DOI] [PubMed]
- 5.O’Dell S.D., Chen,X. and Day,I.N. (2000) Higher resolution microplate array diagonal gel electrophoresis: application to a multiallelic minisatellite. Hum. Mutat., 15, 565–576. [DOI] [PubMed]
- 6.Chen X.H., O’Dell,S.D. and Day,I.N. (2002) Microplate array diagonal gel electrophoresis for cohort studies of microsatellite loci. Biotechniques, 32, 1080–1082, 1084,, 1086. [DOI] [PubMed]
- 7.Day I.N., O’Dell,S.D., Cash,I.D., Humphries,S.E. and Weavind,G.P. (1995) Electrophoresis for genotyping: temporal thermal gradient gel electrophoresis for profiling of oligonucleotide dissociation. Nucleic Acids Res., 23, 2404–2412. [DOI] [PMC free article] [PubMed]
- 8.Day I.N., Spanakis,E., Palamand,D., Weavind,G.P. and O’Dell,S.D. (1998) Microplate-array diagonal-gel electrophoresis (MADGE) and melt-MADGE: tools for molecular-genetic epidemiology. Trends Biotechnol., 16, 287–290. [DOI] [PubMed]
- 9.Voropanov A.M. and Day,I.N. (2000) Elimination of dumbbell bands and enhancement of resolution in MADGE using delayed start electrophoresis. Biotechniques, 28, 32–34. [DOI] [PubMed]
- 10.Watanabe T.K., Bihoreau,M.T., McCarthy,L.C., Kiguwa,S.L., Hishigaki,H., Tsuji,A., Browne,J., Yamasaki,Y., Mizoguchi-Miyakita,A., Oga,K., Ono,T., Okuno,S., Kanemoto,N., Takahashi,E., Tomita,K., Hayashi,H., Adachi,M., Webber,C., Davis,M., Kiel,S., Knights,C., Smith,A., Critcher,R., Miller,J. and James,M.R. (1999) A radiation hybrid map of the rat genome containing 5,255 markers. Nature Genet., 22, 27–36. [DOI] [PubMed]
- 11.Zhang J., Day,I.N. and Byrne,C.D. (2002) A novel medium throughput quantitative competitive PCR technology to simultaneously measure mRNA levels from multiple genes. Nucleic Acids Res., 30, e20. [DOI] [PMC free article] [PubMed]
- 12.Gu D.F., Hinks,L.J., Morton,N.E. and Day,I.N. (2000) The use of long PCR to confirm three common alleles at the CYP2A6 locus and the relationship between genotype and smoking habit. Ann. Hum. Genet., 64, 383–390. [DOI] [PubMed]
- 13.Cheung V.G. and Nelson,S.F. (1996) Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc. Natl Acad. Sci. USA, 93, 14676–14679. [DOI] [PMC free article] [PubMed]
- 14.Lunge V.R., Miller,B.J., Livak,K.J. and Batt,C.A. (2002) Factors affecting the performance of 5′ nuclease PCR assays for Listeria monocytogenes detection. J. Microbiol. Methods, 51, 361–368. [DOI] [PubMed]
- 15.Ye S., Dhillon,S., Ke,X., Collins,A.R. and Day,I.N. (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res., 29, e88. [DOI] [PMC free article] [PubMed]
- 16.Lander E.S. (1999) Array of hope. Nature Genet., 21 (Suppl. 1), 3–4. [DOI] [PubMed]